Abstract
Rare germline variants are difficult to identify using traditional sequencing due to relatively high cost and low throughput. Using second-generation sequencing, we report a targeted, cost-effective method to quantify rare SNPs from pooled genomic DNA. We pooled DNA from 1,111 individuals and targeted four genes. Our novel base-calling algorithm, SNPSeeker, derived from Large Deviation theory, can detect SNPs present at frequencies below the raw error rate of the sequencing platform
The cumulative impact of rare variants on common disease is currently unknown, but recent studies have implicated rare genetic variants in many complex traits and diseases. Consequently, it has been suggested that the combined effects of rare deleterious mutations could explain a substantial fraction of the genetic susceptibility to many common diseases1, 2. Identifying rare variants necessitates genotyping large populations of individuals, either sequentially (e.g. the 1000 Genomes Project3) or, to minimize cost and time, as a pooled sample. However, it has proven difficult to quantify the prevalence of deleterious alleles in pooled samples. Sanger and array-based resequencing are expensive for the amount of sequencing coverage obtained, thus incompatible with large pools. Second-generation sequencing has lowered sequencing costs by over 100-fold, but high error rates have hindered the analysis of large pooled samples, since it is difficult to distinguish rare variants from sequencing errors.
Due to cost and time savings, pooled-sample sequencing should be useful for studying rare, human-specific genetic variation in large populations; characterizing deleterious alleles at multiple loci that may impact disease susceptibility and treatment; quantifying the abundance of rare somatic mutations; and identifying germline variants associated with disease state.
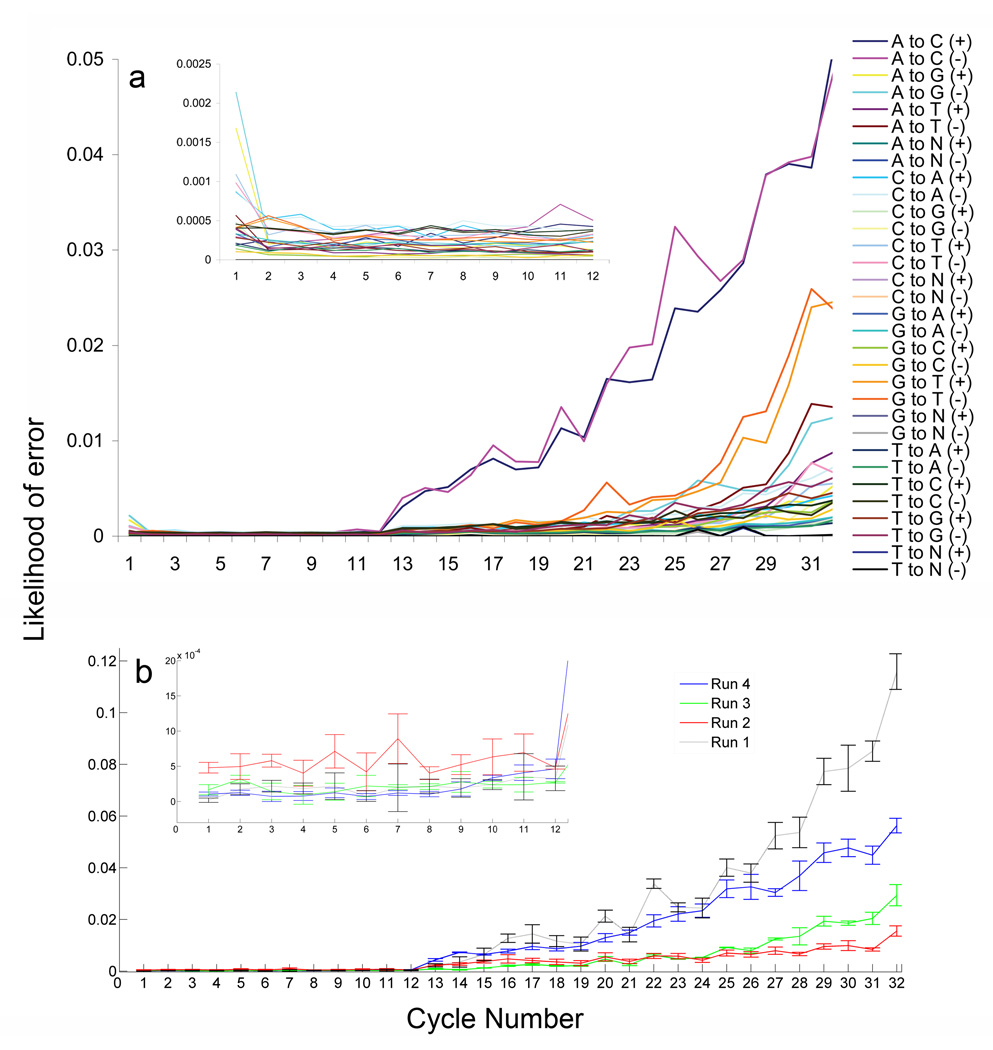
We have implemented a novel combination of molecular biology techniques and computational analysis to achieve targeted resequencing and rare variant detection in 13 kb per individual from 1,111 individuals using the Illumina Genome Analyzer I. We pooled a normalized amount of DNA isolated from dried blood on 1,111 de-identified Guthrie cards collected for newborn screening. The Missouri Department of Health and Senior Services Institutional Review Board and the Washington University Human Research Protection Office reviewed and approved use of de-identified DNA samples (Supplementary Methods). Using specific primers (Supplementary Table 1), we PCR-amplified 15 loci covering 14.5 kilobases of the surfactant protein B (SFTPB), TP53, APC and β-actin (ACTB) genes. Amplicons were ligated into long concatemers, randomly fragmented, and prepared for Illumina sequencing according to the manufacturer’s protocol. This generated 4.4 × 107 sequences, 83.4% (3.7 × 107 reads = 1.3 gigabases) of which aligned to the reference, allowing for up to 2 mismatches. To quantify sequencing errors, we included a 1,276 bp region of the pUC19 plasmid as an internal control. We used the first 800 bases to train our algorithm and the remaining 476 bases as a test set. We found that the first twelve bases of each Illumina read contained significantly fewer errors than later bases (Fig. 1a), so we used only these to identify sequence variation. However, sequencing errors were still present at a higher frequency than that of a single allele in the pool, making an accurate error model essential to distinguishing bona fide variants from sequencing errors. Since existing second-generation base-calling programs cannot detect and quantify rare variants in large pooled samples, we developed SNPSeeker, an algorithm based on Large Deviation Theory (www.genetics.wustl.edu/rmlab). SNPSeeker uses a 2nd order dependency error model for SNP identification and takes into account the position in the sequencing read (i.e. cycle number) and the identity of the two upstream bases. Consequently, only mismatches at bases 3–12 of each sequencing read were used to identify SNPs (this reduces the effective coverage per allele to 13.8-fold in these experiments). Unexpectedly, we found that incorporating quality scores did not improve results beyond these parameters (Supplementary Fig. 1 and Results). For each machine run, we trained a new error model using the internal pUC19 control, because we found that error rates varied significantly between machine runs (Fig. 1b).
Figure 1.
Error modeling. (a) The cumulative likelihood of every possible misincorporation event for sequencing cycles 1–32 is depicted for both the sense (+) and antisense (−) strands. The Illumina data filtering process truncated the data from two dates at 32 bases instead of 36, which is why only 32 cycles are represented here. INSET. Higher resolution of the error probability across cycles 1–12. (b) The intra- and inter-day variability for the A→C misincorporation event from four different sequencing dates. The error bars represent the standard deviation between different flowcell channels from the same date. INSET. Higher resolution of cycles 1–12.
The SNPSeeker algorithm significantly improved the specificity of SNP calling (Supplementary Fig. 2 and Results). Using SNPSeeker, no SNPs were called in the training sequence (bases 1–800), in the negative control (bases 801–1,276) or in 656 of the 658 bp without a known SNP in the SFTPB amplicon. This yields a specificity of 99.8% and demonstrates that our base-calling algorithm is specific and able to accurately model sequencing and PCR errors.
We validated called SNPs by comparing them to prior individual Sanger sequencing at the SFTPB locus in this cohort4 and performing additional Taqman assays (Supplementary Tables 2 and 3) To estimate our method’s sensitivity, we sequenced a 665 bp region of the SFTPB gene with 7 known SNPs. Our method identified all 7 SNPs at very similar frequencies to those found by Sanger. Three of these SNPs were present at < 1.5% in this population demonstrating that this method has the sensitivity to detect rare SNPs in this pooled sample (Supplementary Table 3a). We also identified two SNP positions within SFTPB that were not identified by Sanger. We performed individual Taqman assays on each of these positions and neither mutation was identified.
In addition to the 9 called SNPs in SFTPB, 55 additional SNPs were called in ACTB, TP53, and APC (Supplementary Table 4). Of these, 37 (67%) were previously described in dbSNP (build 128). By chance, one would expect less than 1 SNP, on average, to be shared between these two sets. Using Fisher’s Exact Test, we found that the observed degree of overlap is highly significant (P < 1.3 × 10−56). Therefore, it is highly likely that the 37 SNPs identified by SNPSeeker and found in dbSNP are bona fide variants. Many of these SNPs were rare: 26 of the 37 dbSNPs that were identified had estimated allele frequencies of less than 1.5% in our population (Supplementary Table 4). We also performed a comparative genomic analysis of the deleterious nature of the non-synonymous SNPs identified (Supplementary Results and Table 5).
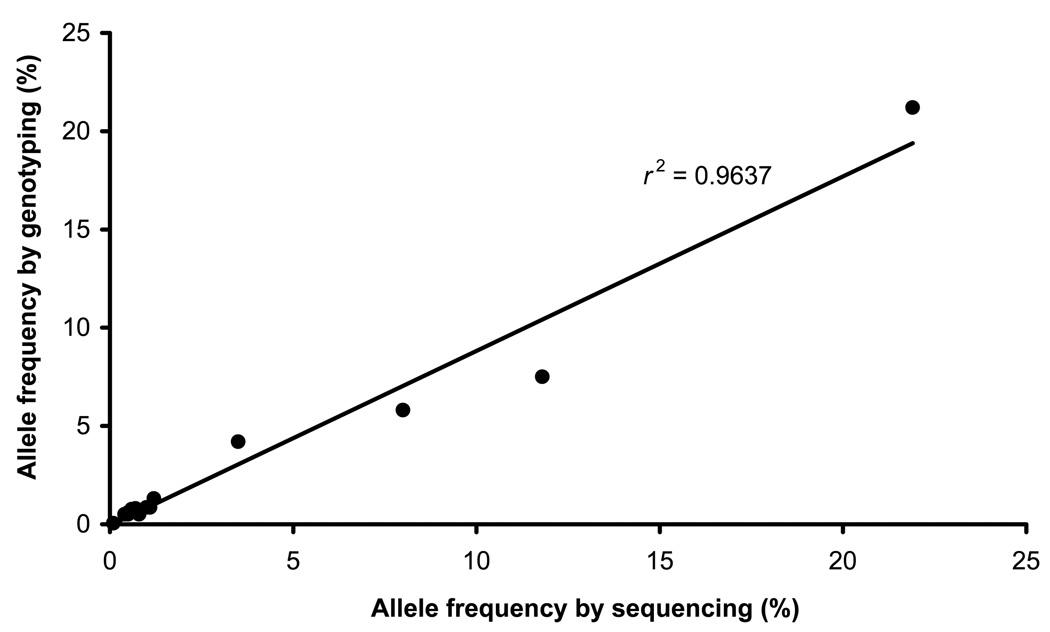
To estimate the positive predictive value of our method, we chose seven called SNPs in these genes for independent validation by individual Taqman genotyping. All seven SNPs were predicted to be rare, with estimated minor allele frequencies (MAF) ranging from 0.5–1.2%. Three of the selected SNPs were previously reported in dbSNP, though not in our population, and the remaining four have not been previously described. Taqman genotyping validated all seven called rare variants (Supplementary Table 3b). When combined with SFTPB results, we validated 14 of 16 predicted SNPs, giving a positive predictive value of 87%. To determine if the pooled sample sequencing method could accurately quantify allele frequencies, we plotted the predicted versus true MAFs for each of the 14 validated SNPs (Fig. 2). The observed and predicted frequencies were highly correlated (r2 = 0.96) across a wide range of frequencies: from a single allele (0.05%) up to several hundred alleles (21.2%). When we plotted predicted SNP average heterozygosities against reported average heterozygosity values for all SNPs in common with dbSNP, the correlation remained strong at r2 = 0.82 (Supplementary Fig. 3). These findings indicate that pooled sample sequencing is able to accurately determine the population frequency of common and rare alleles.
Figure 2.
Allele frequency by sequencing vs. genotyping. The allele frequency as determine by sequencing is plotted against the actual frequencies as determined by individual Taqman assay for the 14 validated SNPs in our dataset (correlation coefficient r2 = 0.96).
A deeper understanding of genetic variation in the human population will allow us to dissect the causative factors that contribute to a wide array of human disease, understand the genetic characteristics that make us uniquely human, and quantify the impact of selection across our genome throughout history. We have successfully resequenced 13,237 bases per 1,111 individuals at approximately 2% of the cost of the original analysis by Sanger sequencing.4 Importantly, this cost savings did not come at the price of sensitivity or accuracy.
The positive predictive value of 87% obtained in this study is consistent with previously published values of 85%5 and 92%6 in assays identifying SNPs from multiple organisms via second-generation sequencing, but we analyzed over an order of magnitude more individuals (1,111 vs. 2(ref5) or 66(ref6)) than these studies. The ability to pool larger numbers of individuals enables the discovery of rare SNPs, which is important, since most deleterious SNPs are unlikely to be present at frequencies greater than 1%. Furthermore, our method more accurately estimates MAFs (r2 = 0.96 vs. 0.676), which is important to accurately identify disease-associated alleles when comparing disease and normal cohorts.
At the SFTBP locus, we successfully detected a single mutant allele in a background of 2,221 wild-type alleles; however, there were not enough private mutations in our validation set to determine the sensitivity of our method for the detection of private SNPs (MAF < 0.05%) in this population. For applications where it is important to detect singleton SNPs with a high sensitivity, we recommend choosing a pool size such that private mutations are present at frequencies similar to those of the rare SNPs validated here (MAF = 0.5–1.2%).
There are various applications for this method. Sequencing large, random populations at various genetically significant loci would enable the study of human-specific variation and selection. Quantification of rare somatic mutations in tumors and precancerous lesions would facilitate improved understanding of tumorigenesis. Finally, sequencing case-control or matched sample cohorts will enable identification of rare mutations associated with complex diseases.1, 2 Candidate genes can be selected based on prior knowledge1, 2 or they can be informed by genome wide association studies. Combining pooled-sample sequencing with genomic selection strategies,7–9 makes it possible to move beyond the candidate gene approach and perform a more systematic survey of protein-coding DNA. Such knowledge would be a valuable tool for disease screening, assigning risk stratification, providing longitudinal preventative care, and tailoring risk-appropriate therapy.
Supplementary Material
Acknowledgments
This work was supported in part by the National Institutes of Health under the Ruth L. Kirschstein National Research Service Award T32 HD 007499 from the NICHD (T.E.D.), the NHLBI (RO1HL065174, RO1HL082747, F.S.C.), the Children’s Discovery Institute Fellowship Award MC-F-2006-1 (T.E.D.), the Children’s Discovery Institute Grant MC-II-2006-1 (R.D.M.), and the Saigh Foundation (F.S.C., R.D.M., T.E.D.).
Reference List
- 1.Cohen JC, et al. Science. 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 2.Ji W, et al. Nat. Genet. 2008;40:592–599. doi: 10.1038/ng.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayden EC. Nature. 2008;451:378–379. doi: 10.1038/451378b. [DOI] [PubMed] [Google Scholar]
- 4.Hamvas A, et al. Pediatr. Res. 2001;50:666–668. doi: 10.1203/00006450-200111000-00021. [DOI] [PubMed] [Google Scholar]
- 5.Barbazuk WB, Emrich SJ, Chen HD, Li L, Schnable PS. Plant J. 2007;51:910–918. doi: 10.1111/j.1365-313X.2007.03193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Tassell CP, et al. Nat. Methods. 2008;5:247–252. doi: 10.1038/nmeth.1185. [DOI] [PubMed] [Google Scholar]
- 7.Albert TJ, et al. Nat. Methods. 2007;4:903–905. doi: 10.1038/nmeth1111. [DOI] [PubMed] [Google Scholar]
- 8.Hodges E, et al. Nat. Genet. 2007;39:1522–1527. doi: 10.1038/ng.2007.42. [DOI] [PubMed] [Google Scholar]
- 9.Porreca GJ, et al. Nat. Methods. 2007;4:931–936. doi: 10.1038/nmeth1110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.