Abstract
Background
Microdeletions within chromosome 15q13.3 are associated both with a recently recognised syndrome of mental retardation, seizures, and dysmorphic features, and with schizophrenia.
Methods and results
Based on routine diagnostic testing of ~8200 samples using array comparative genomic hybridisation, we identified 20 individuals (14 children and six parents in 12 families) with microdeletions of 15q13.3. Phenotypes in the children included developmental delay, mental retardation, or borderline IQ in most and autistic spectrum disorder (6/14), speech delay, aggressiveness, attention deficit hyperactivity disorder, and other behavioural problems. Both parents were available in seven families, and the deletion was de novo in one, inherited from an apparently normal parent in four, and inherited from a parent with learning disability and bipolar disorder in two families. Of the 14 children, six in five families were adopted, and DNA was available for only one of these 10 biological parents; the deletion was very likely inherited for one of these families with two affected children. Among the unavailable parents, two mothers were described as having mental retardation, another mother as having “mental illness”, and one father as having schizophrenia. We hypothesise that some of the unavailable parents have the deletion.
Conclusions
The occurrence of increased adoption, frequent autism, bipolar disorder, and lack of penetrance are noteworthy findings in individuals with deletion 15q13.3. A high rate of adoption may be related to the presence of the deletion in biological parents.
Unconfirmed histories of antisocial behaviours in unavailable biological parents raise the concern that future research may show that deletion 15q13.3 is associated with such behaviours.
The availability of the human genome sequence and the use of array based hybridisation to identify genomic copy number variations (CNVs) are catalysing the rapid discovery of novel microdeletion and microduplication syndromes.1–3 Presently, there is routine clinical utilisation of genome-wide analysis of copy number using either single nucleotide polymorphism (SNP) based arrays or array comparative genomic hybridisation (aCGH) without SNP analysis. The extensive distribution, high frequency, and gene content of benign and pathogenic CNVs across the genome have provided a new perspective regarding genomic polymorphism and “genomic disorders”.4–6 These CNVs often result from rearrangements that are mediated by non-allelic homologous recombination (NAHR) between highly homologous segmental duplications (low copy repeats).7 Many of the genomic regions that are flanked by segmental duplications have a high mutability for gain and loss of copy number, and the frequency of de novo CNVs is much higher than that for de novo single base mutations.8 Genomic disorders include well characterised microdeletion/duplication syndromes such as DiGeorge/velocardiofacial syndrome (DGS/VCFS; MIM 188400/MIM 192430), Williams–Beuren syndrome (WBS; MIM 194050), and Smith–Magenis syndrome (SMS; MIM 182290), and the reciprocal duplication syndromes for these three entities.
It has long been known that microscopically detectable chromosomal abnormalities cause mental retardation and less commonly autism9 with virtually every chromosome potentially involved. However, array based methods have revealed a much higher frequency of submicroscopic deletions and duplications causing mental retardation (see review by Stankiewicz and Beaudet 10) and autism, and a much larger fraction of autism is now believed to be caused by genetic mutations, both CNVs and point mutations, than was previously appreciated.11–13 Very recently, homozygosity mapping has been used to identify autosomal recessive mutations/deletions causing autism.14 Although deletions of 22q11.2 DGS/VCFS have been known to cause schizophrenia for many years,15 there is broader emerging evidence that CNVs may also play a larger role in the aetiology of psychiatric disorders such as schizophrenia.16, 17
The recurrent 15q13.3 microdeletion syndrome resulting in loss of a ~1.5 Mb segment (3.95 Mb in one case) was first described using whole genome aCGH and quantitative polymerase chain reaction (PCR) screening of individuals with mental retardation, seizures, and/or congenital anomalies.18 The presence of segmental duplication blocks of sufficient size and orientation predict that this recurrent deletion is most likely mediated by NAHR between these segmental duplications.18, 19 The genomic region from 15q11 to 15q13 including BP1 to BP5 is extremely complex and polymorphic, but a recent publication goes a substantial way towards delineating the region.20 Two recent reports found deletions of 15q13 as one of the most common identifiable genetic causes of schizophrenia.21, 22 Recently, deletion 15q13.3 was reported to be associated with autism,23 and its association with epilepsy was confirmed and extended.24
SUBJECTS AND METHODS
Subjects
The Medical Genetics Laboratories (MGL) at Baylor College of Medicine (BCM) have performed aCGH on >14 000 cases referred from February 2004 to May 2008 with the 8200 most recent studies having coverage for 15q13.3; the most common reasons for testing are developmental delay, mental retardation, dysmorphic features, congenital anomalies, autism, and general suspicion of a chromosomal anomaly.25 These samples were analysed on consecutive versions of targeted arrays, initially bacterial artificial chromosome (BAC) based and later oligonucleotide based as described previously.25, 26 Coverage for the 15q13 BP3-BP4 region was present from very early in the series, but a BAC for the more important BP4-BP5 region was not introduced until the first 5800 samples had been analysed. The first deletion was detected in December 2006, and a total of 15 independent families (a 16th was found in a sample from the South Carolina Autism Project) deleted for 15q13 were detected for frequency of ~0.18% (15/(14 000–5800)) in samples submitted to the diagnostic laboratory. Sufficient clinical information and informed consent were available for 14 children and six parents with the deletion in 12 families reported here. All patients were of European descent (white) except that one father of an adopted child was African American.
Molecular studies
Clinical samples were analysed on a series of progressively more complex arrays from February 2004 to July 2008. Initial arrays included 366 BACs and progressed to arrays with 1475 BACs and then to 44K oligonucleotide based (Agilent Technologies, Santa Clara, California, USA) partly in a BAC emulation format as described previously25, 26; more recently a 105K custom Agilent array is being used. Initial identification of the cases by aCGH was based on loss of a genomic segment interrogated by BAC clones or by oligonucleotides within the BACs RP11-348B17 (chr15:29,067,933–29,289,407) only in 11/12 families and both by this BAC and by BAC clone RP11-143J24 (chr15:27,906,198–28,091,821) in 1/12 family. The deletions were confirmed in all of the cases by fluorescence in situ hybridisation (FISH).
A custom designed chromosome-15 specific microarray was used for the further definition of the deletions involving 15q11.2q14 in these patients. This was a 44K Agilent oligonucleotide array designed using the Agilent E array tool to achieve maximal density across chromosome 15q11.2q14 (35 600 probes distributed across the interval from chr15:19,798,964–32,949,987 (build of May 2004). Manufacturer’s protocols for hybridisation and data analysis were followed (Agilent Technologies) with minor modifications.27 Briefly, 1 μg of genomic DNA from test samples and gender matched reference controls was digested with AluI (5 units) and RsaI (5 units) (Promega, Madison, Wisconsin, USA) for 2 h at 37°C. Random primed DNA labelling with Cy3-dCTP and Cy5-dCTP was performed following the manufacturer’s recommended protocol (Perkin Elmer, Shelton, Connecticut, USA). Following microarray scanning, images were analysed using the Feature Extraction Software (version 9.5.3.1, Agilent Technologies) and imported into the Agilent CGH-Analytics V3.4 software for analysis. Statistically significant CNVs were determined using the aberration detection module (ADM)-2 algorithm with a threshold of 4.5–6. Breakpoint boundaries were determined by the position of the first and last oligonucleotide probes, respectively, included in the algorithmically determined region of aberration.
RESULTS
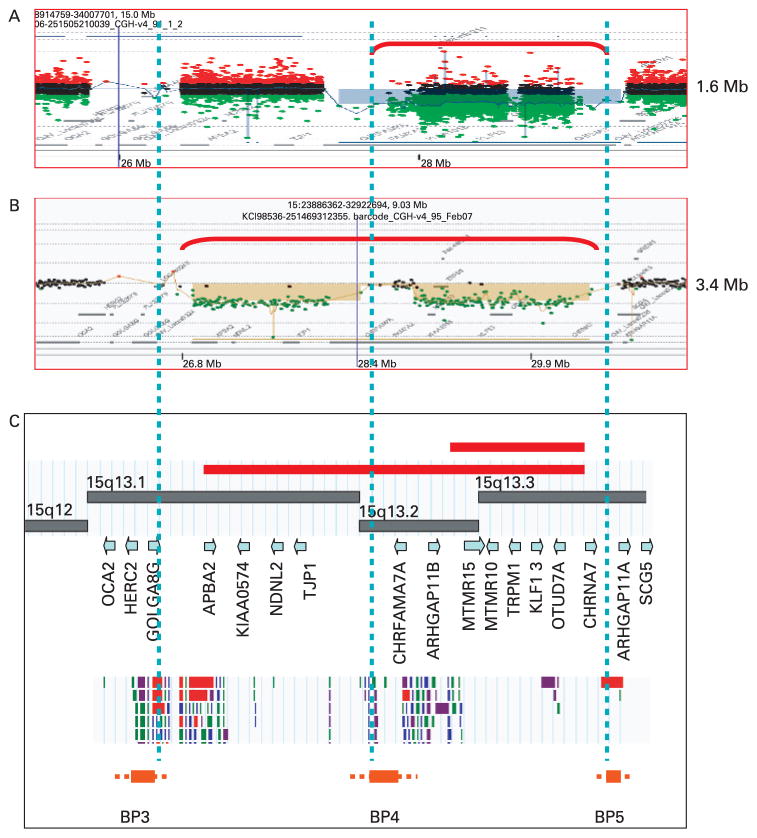
Examples of the detection of the deletions are shown in fig 1A,B using the custom designed chromosome 15-specific oligonucleotide array27 or using a commercial whole genome 244 k array (both from Agilent Technologies), respectively. The most common BP4-BP5 deletion of 1.6 Mb corresponds approximately to 15q13.3 and was seen in 11/12 families (fig 1A), while the larger BP3-BP5 deletion of 3.4 Mb corresponds to 15q13.1q13.3 (fig 1B) and was seen in one family. The genomic architecture of this region is shown in fig 1C. In all of the families, the BP4-BP5 critical region is deleted. We also detected four cases of duplication of the BP4-BP5 segment (data not shown) as might be anticipated based on the NAHR mechanism; it is not clear at present whether duplications in this region are benign or may cause phenotypic abnormalities as discussed below. Based on their size and breakpoints, these duplications likely represent the reciprocal recombination product to the ~1.6 Mb deletions.
Figure 1.
Array comparative genomic hybridisation (aCGH) analysis of 15q13 deletions. Chromosome 15 specific aCGH plots for the recurrent BP4-BP5 1.6 Mb deletion (A) and Agilent 244 k array plot for BP3-BP5 3.4 Mb deletion (B). (C) Physical and gene map of the region with the breakpoints involved in the recurrent and rare deletions and the position of breakpoints relative to the segmental duplication blocks (Database of Genomic Variants, http://projects.tcag.ca/variation). Regions of deletion are shown by red brackets and bars. Genes are indicated by gene symbols and breakpoint regions (BP3, BP4, and BP5) are indicated with low copy repeat blocks above each BP region.
The clinical descriptions of the families reported here are without photographs for reasons of confidentiality as explained below. Similar to the cases described previously, developmental delay, mental retardation, or borderline IQ was detected in 12/14 of the children (fig 2, table 1; IQ 82 in proband from family 2 and IQ unknown but possibly normal in proband from family 4), and all 14 had at least mild mixed expressive and receptive language delay. The ages and sex of the children are omitted to preserve confidentiality, but nine of the children were male and four were female counting the monozygous twins as one case. Importantly, at least 6/14 of the children detected with 15q13.3 microdeletion had symptoms within the range of autism spectrum disorder (ASD), one of them being diagnosed with Asperger syndrome. Interestingly, only one of the two siblings in family 8 is thought to have autism, although more detailed evaluations are planned. Autism was not frequent in the first report of 15q13.3 deletions with only one case with “mild autism” detected.18 Notably, even among the eight children in this report who did not have ASD, most were not tested with autism specific diagnostic instruments; language impairment was prominent and more severe than the developmental delay in the gross and fine motor skills, suggesting that additional patients in this report might meet criteria for autism if tested thoroughly. Abnormal behaviours, including aggressiveness, repeated head banging, and/or attention deficit hyperactivity disorder (ADHD), were detected in 9/14 children (table 1). Although aggressive behaviour was not frequent in the previous report of 15q13 deletions, one boy with the deletion and “mild autism” was hospitalised five times in psychiatric facilities due to aggression and rage.
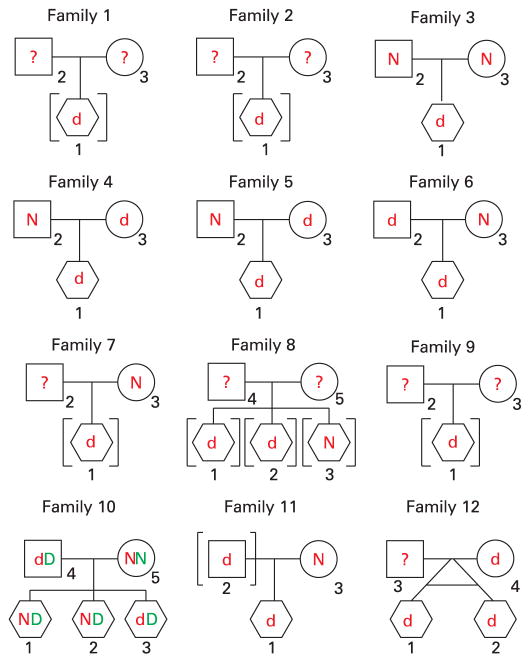
Figure 2.
Pedigrees for 10 families. All families have the 1.6 Mb deletion except family 1. Only biological parents are shown. Children in brackets are adopted or in legal custody of individuals other than the biological parents. The red lower case d and the red upper case N indicate the deletion or normal genotype, respectively, for 15q13.3. The green upper case D and the green upper case N indicate the deletion of normal genotype for 16q24.1 in family 10. All children are shown as hexagon symbols to mask sex for purposes of confidentiality.
Table 1.
Individuals with deletion 15q13.3
| Family | Child/parent* | DD or MR/autism | Language delay/seizures | Weight/height/FOC | Behaviour | Other | Inheritance |
|---|---|---|---|---|---|---|---|
| F1-1 | Child | DQ 58/autism disorder | Moderate/no | 50%/30%/≫97% | Mild stereotypic | Benign hydrocephalus, mild Chiari I | Adopted to unrelated family; little information on biological parents |
| F2-1 | Child | IQ 82/Asperger | Mild/no | 95%/>97%/90% | ADHD | Adopted to unrelated family; mother said to have mental retardation | |
| F3-1 | Child | Moderate DD/borderline | Severe/no | All 50–75% | None reported | Normal MRI | De novo |
| F4-1 | Child | Normal?/no | Mild/yes | 65%/7%/? | None reported | Normal MRI | Mother has deletion |
| F4-3 | Mother | Normal/no | No/no | Normal | Normal | Mother of F4-1; normal phenotype | |
| F5-1 | Child | Moderate DD/no | Moderate/no | 44%/11%/75% | None reported | Digital findings, arachnoid cyst | Mother has the deletion |
| F5-3 | Mother | Normal/no | No/no | Normal | Normal | Mother of F5-1; normal phenotype | |
| F6-1 | Child | Mild DD/no | Moderate/no | 92%/>89%/50% | Aggressive, self injurious | Father has deletion | |
| F6-2 | Father | Learning disabilities/no | Likely/no | Normal | None reported | Father of F6-1; learning disabilities, bipolar disorder | |
| F7-1 | Child | Moderate DD/no | Moderate/no | 25%/50%/50% | Head banging | Digital findings, MRI normal | Mother learning disorder but no deletion. Removed from parental care 15 months of age |
| F8-1 | Child | Mild MR/yes | Moderate/no | Normal range | ADHD | Long digits, facial dysmorphisms, sleep problems | Three sibs adopted to same unrelated family; 2 affected and 1 unaffected sib; mother said to have mental retardation; father schizophrenia |
| F8-2 | Child | Mild MR/no | Moderate/no | Normal range | Impulsive, ADHD, mood disorder | Long digits, facial dysmorphisms, sleep problems | Sibling of F8-1 |
| F9-1 | Child | IQ 27/autism disorder ADI-R | Severe/no | 50%/75% (10 y)/50% birth | Extremely hyperkinetic and very aggressive behaviours | Mild facial dysmorphism | In custody of paternal grandmother; mother deceased and had “mental illness”. Father normal |
| F10-1 | Child | Global delay/too young | Severe language delay/no | 2%/10%/25% | Rocking | MRI normal | Father has deletion. Mother learning disability but no deletion. Two sibs with learning disability but not deletion. Chromosome 16q24.1 duplication present in father and three siblings including F10-1 |
| F10-4 | Father | Normal/no | No/no | Normal | Normal | Father of F10-1; normal phenotype | |
| F11-1 | Child | Moderate DD/no | Severe/no | 95%/90%/>97% | None | Father has deletion | |
| F11-2 | Father | Learning disabilities/no | Not known/no | Normal | None | Father of F11-1; learning disabilities, bipolar disorder | |
| F12-1 | Child | IQ 70/autism | Moderate/no? | <5%/5%/2% | Impulsive, ADHD, mood disorder | Nystagmus | MZ twin of F12-2; mother has deletion |
| F12-2 | Child | IQ 70/autism | Moderate/no? | <5%/5%/2% | Impulsive, ADHD, mood disorder | Nystagmus | MZ twin of 12-1; mother has deletion |
| F12-4 | Mother | Normal/no | No/no | Normal | Normal | Mother of F12-1 and F12-2; normal phenotype |
ADHD, attention deficit hyperactivity disorder; ADI-R, Autism Diagnostic Interview-Revised; DD, developmental delay; FOC, frontal occipital circumference; MR, mental retardation; MRI, magnetic resonance imaging.
Age and sex of children is not provided for purposes of confidentiality.
The facial appearances were generally consistent with those already published. The appearance is variable and ranged from near normal to moderately dysmorphic. Common facial features in our patients as well as in the previously described patients include hypertelorism, short philtrum, and everted and thick upper lip. At least two of our patients were thought initially to have coarse facies that prompted further testing with normal results for disorders associated with facial coarseness, including mucopolysaccharidoses and Coffin–Lowry syndrome. Mild digital aberrations, including brachydactyly and clinodactyly, were observed in five of our patients, similar to the previously described cases.18
Epilepsy was observed in only one of the 14 children and in none of the five parents with the deletion. Another child had convulsive-like episodes not associated with abnormal electrical activity. The prevalence of seizures therefore is lower in our cohort than in the initial report of this syndrome. This difference was observed regardless of the fact that all of our families had deletion of CHRNA7, a gene that was postulated to mediate the epilepsy in 15q13.3 deletions. Nystagmus was present in monozygous twins. Abnormal eye movements are reported in schizophrenia.28
Both parents were available for deletion testing in only seven families, and analysis of the mother alone was possible in an eighth. The deletion was confirmed to be a de novo event in a single family (F3). The deletion was inherited in six of the seven families (F4, F5, F6, F10, F11, and F12) where both parents were available for analysis, and presumptively inherited in a family (F8) with affected siblings but parental samples not available, making the deletion inherited in seven of eight families where determinable, counting F8 as inherited. In two families (F6 and F11), the deletion was inherited from the fathers who both had learning disabilities by self report and were diagnosed with bipolar disorder. In four families (F4, F5, F10, and F12), the deletion was inherited from an apparently normal parent, and these parents were perceived by themselves, family members, and physicians as being normal based on education, employment, and rearing of a family. They did not have a history of cognitive impairment, psychiatric diagnosis, or convulsions, although formal assessments of cognition and behaviour were not performed. Dysmorphic features in these four parents were either entirely absent or extremely subtle. Deletion carriers with a normal phenotype were not observed for the BP4-BP5 deletion in the first report.18
In family 10, the index patient had two siblings with learning disabilities. The father and all three children have a duplication 16q24.1 (Chr.16:82.5–84.2 Mb) which may well contribute to the phenotype in the two siblings and in the patient. The mother in this family has learning disabilities and a normal result on a clinical array analysis. No other deletion 15q13.3 individuals had additional potentially pathological CNVs detected by array analysis.
DISCUSSION
The wide range and heterogeneity of phenotypic expression for the common 1.6 Mb deletion is remarkable and includes normal phenotype, mental retardation, borderline IQ, autism, seizures, bipolar disorder, and schizophrenia based on the families reported here and on publications of others. Combining the present cases with those reported previously by Sharp et al,18 only two out of the 18 families with 15q13.3 deletion syndrome did not have the common 1.6 Mb deletion, but instead had a larger ~3.4 Mb deletion extending from BP3 and BP5 (fig 1). The phenotype for the 1.6 Mb deletion can include individuals who function normally in society, although subtle cognitive deficits or behavioural susceptibilities have not been ruled out in these individuals. Also the mental retardation in the affected children was usually not severe, and may often allow living independently, particularly if many of the unavailable biological parents in our study prove to have the deletion. The 3.4 Mb deletion from BP3-BP5 was also far less frequent in one earlier report,18 and the reason for the lower frequency may relate to the relative homologies of the low copy repeats at BP3, BP4, and BP5. Deletions of chromosome 1q21 also appear to cause an unusually wide spectrum of paediatric and adult phenotypes including mental retardation, autism and schizophrenia.21, 22–29, 30
Notable aspects of this deletion are the high frequency of adoption or foster care, an autism spectrum diagnosis in six of 14 children, the presence of bipolar disorder in two fathers with the deletion, and apparent lack of penetrance in four parents with the deletion. The association with autism is in agreement with an earlier report.23 Although seizures were reported in only one of our 20 cases, a recent study of patients with epilepsy as the presenting diagnosis found deletion 15q13.3 in 12 of 1223 cases and 0 of 3699 controls.24 We found that 43% (six of 14; one unaffected sibling also adopted in fig 2) of the children were in adoptive care; some children were placed for adoption voluntarily in infancy through agencies and some were removed from the biological parents by child protection services. None of the parents were deceased at the time of adoption. The social information available, combined with evidence that this deletion is frequently inherited rather than de novo, suggest that the adoption in many cases may have been related to cognitive, psychiatric, and/or social issues in the biological parents. For comparison, in the general population of the USA about 2.5% of the children are adopted (http://www.census.gov/prod/2003pubs/censr-6.pdf), although a precise rate of adoption or custody by individuals other than the biological parents is not known for children with disabilities, and the rate may be higher than for normal children, because of both acquired or inherited disabilities in the biological parents. We hypothesise that some of the unavailable biological parents may have the deletion, but we have no certainty of this except that one or the other parent presumably has the deletion in the case of the affected siblings (F8). Our suspicion is based partly on the hypothesis that the phenotype caused by the deletion in a parent contributed to the outcome of adoption, but also on the observation that deletions of BP4-BP5 are frequently inherited rather than de novo, 7/8 of informative probands in our report and 2/4 in the previous report.18 Although mental retardation was reported for two unavailable biological mothers, “mental illness” for another mother, and schizophrenia for one biological father, we have no direct documentation of these diagnoses. There were unconfirmed reports from adoption agencies, family members, and adoptive parents of antisocial behaviour for numerous biological parents. Future research is warranted to determine if deletion 15q13.3 is associated with antisocial behaviours. Based on this possibility and because reporting of third party information is problematic from an informed consent perspective,31, 32 we have chosen to omit all family specific information about social behaviour and all photographs of children and parents in order to minimise any potential for stigmatisation, especially of minors.
The presence of a diagnosis of bipolar disorder in two parents with the deletion suggests that this condition may also be caused by deletion 15q13.3. The apparent lack of penetrance in four parents is consistent with the presence of this deletion at a low frequency in normal populations. In three reports, the 15q13.3 deletion was seen in 0 of 2962 controls,18 in 0 of 3181 controls,21 and in 8 of 39 800 controls.22 As smaller submicroscopic deletions and duplications are identified, it is likely that phenotypes will be milder, that penetrance will be less than 100%, and that a larger fraction of cases will be inherited. Lack of penetrance and variation in expression could be explained in part by polymorphisms in the level of expression of the key genes in the deleted region on the normal chromosome, but environmental factors may also be important, and the latter would have important implications for management of children found to have the deletion at a young age. It may be the case that duplications will confer milder phenotypes with lower penetrance as seems to be the case for duplications of the DGS/VCFS, WBS, and SMS regions. We observed four cases of duplication 15q13.3, and duplications were found in other reports as well,18–23, 24 but usually at a lower frequency. Although a recent study of patients with epilepsy found duplication 15q13.3 equal in frequency to deletion being present in 12 of 1223 cases, duplication was also present in 23 of 3699 controls, in sharp contrast to 0 deletions in 3699 controls.24 The association of duplication with epilepsy was not statistically significant. If duplications do cause phenotypic abnormalities, the penetrance is certainly lower than for deletions.
There is now very substantial evidence that the BP4-BP5 deletion of 1.6 Mb causes a mental retardation phenotype with high but not complete penetrance and can also cause schizophrenia. The genomic structure for this region is poorly defined in even recent assemblies of the human genome; the region is highly complex and repetitive and almost certainly is variable among normal chromosomes.20 This interval contains six well characterised genes; CHRNA7, OTUD7A, KLF13, TRPM1, MTMR10, and MTMR15, while CHRFAM7A and FAM7A(2) may be imbedded in BP4 and BP5 (fig 1C). The ARHGAP11B and ARHGAP11A genes appear to be imbedded in BP4 and BP5, respectively. The CHRNA7 gene encodes the α7 subunit of the neuronal nicotinic receptor, which is a homopentameric synaptic ion channel protein (MIM 118511). Although there are reports of genetic linkage to this region for juvenile myoclonic epilepsy33 and for rolandic epilepsy,34 these results do not implicate the CHRNA7 gene specifically any more that the immediate neighbouring genes. There are numerous publications suggesting a possible role for the CHRNA7 gene in schizophrenia as reviewed elsewhere.35, 36 There is a CHRFAM7A fusion gene embedded in BP4 with an additional copy possibly imbedded in BP5 on some chromosomes, but the function of this gene, if any, is unclear. The fusion gene includes exons 5–10 of CHRNA7 fused to five exons of gene family member 7A (FAM7A).37 FAM7A(2) appears to be imbedded in both BP4 and BP5.20 There is polymorphism both for copy number and for an inversion of CHRFAM7A in the human population.38 The presence and polymorphism of the fusion gene greatly complicates genotyping and sequence analysis for exons 5–10 of CHRNA7. If haploinsufficiency for CHRNA7 is the primary cause of the phenotype, treatment trials with nicotinic pharmacological agents would be of interest. The other genes in the BP4-BP5 region should not be neglected as candidates to cause a neurologic phenotype by virtue of an excessively narrow focus on CHRNA7.
In conclusion, we have found a diagnosis of autism to be common in the 15q13.3 deletion syndrome, and we have observed lack of penetrance, and two parents with the deletion and a diagnosis of bipolar disorder, in addition to the previously reported occurrence with mental retardation, seizures, dysmorphisms, and schizophrenia, Of the non-penetrant individuals, three were female parents and one was a male parent, while the majority (9/13 counting monozygous twins as one case) of the children with abnormal phenotypes were male. This raises the possibility that penetrance may be influenced by the sex of the individual. The deletion is more frequently inherited rather than de novo; affected children are often in the care of individuals other than the biological parents; and unconfirmed reports of antisocial behaviour in biological parents are of concern and deserve further investigation. Both behavioural intervention and pharmacotherapy should be investigated as treatments in a cohort of individuals known to have this deletion.
Acknowledgments
We thank the families participating in these studies for their cooperation. We thank the many faculty and staff who contribute to the activities of the Medical Genetic Laboratories at BCM where most of these patients were ascertained.
Funding: This work was supported by US National Institutes of Health grants HD-037283, M01-RR00188 (General Clinical Research Center), HD-024064 (Mental Retardation and Developmental Disabilities Research Center) and RR-019478 (Rare Disease Clinical Research Consortia) and by generous support from the William Stamps Farish Fund.
Footnotes
Competing interests: Many of the authors are faculty members in the Department of Molecular and Human Genetics at Baylor College of Medicine (BCM) which offers extensive genetic laboratory testing including use of arrays for genomic copy number analysis, and the department derives revenue from this activity.
Patient consent: All family specific information about social behaviour and all photographs of children and parents have been omitted from this article in order to minimise any potential for stigmatisation
References
- 1.Ballif BC, Hornor SA, Jenkins E, Madan-Khetarpal S, Surti U, Jackson KE, Asamoah A, Brock PL, Gowans GC, Conway RL, Graham JM, Jr, Medne L, Zackai EH, Shaikh TH, Geoghegan J, Selzer RR, Eis PS, Bejjani BA, Shaffer LG. Discovery of a previously unrecognized microdeletion syndrome of 16p11.2-p12.2. Nat Genet. 2007;39:1071–3. doi: 10.1038/ng2107. [DOI] [PubMed] [Google Scholar]
- 2.Ben-Shachar S, Ou Z, Shaw CA, Belmont JW, Patel MS, Hummel M, Amato S, Tartaglia N, Berg J, Sutton VR, Lalani SR, Chinault AC, Cheung SW, Lupski JR, Patel A. 22q11.2 distal deletion: a recurrent genomic disorder distinct from DiGeorge syndrome and velocardiofacial syndrome. Am J Hum Genet. 2008;82:214–21. doi: 10.1016/j.ajhg.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, Saemundsen E, Stefansson H, Ferreira MA, Green T, Platt OS, Ruderfer DM, Walsh CA, Altshuler D, Chakravarti A, Tanzi RE, Stefansson K, Santangelo SL, Gusella JF, Sklar P, Wu BL, Daly MJ. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–75. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 4.Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Månér S, Massa H, Walker M, Chi M, Navin N, Lucito R, Healy J, Hicks J, Ye K, Reiner A, Gilliam TC, Trask B, Patterson N, Zetterberg A, Wigler M. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–8. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 5.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–51. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 6.Lupski JR. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:145–9. doi: 10.1016/s0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- 7.Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- 8.Lupski JR. Genomic rearrangements and sporadic disease. Nat Genet. 2007;39:S43–7. doi: 10.1038/ng2084. [DOI] [PubMed] [Google Scholar]
- 9.Vorstman JAS, Staal WG, van DE, van EH, Hochstenbach PFR, Franke L. Identification of novel autism candidate regions through analysis of reported cytogenetic abnormalities associated with autism. Mol Psychiatry. 2006;11:18–28. doi: 10.1038/sj.mp.4001781. [DOI] [PubMed] [Google Scholar]
- 10.Stankiewicz P, Beaudet AL. Use of array CGH in the evaluation of dysmorphology, malformations, developmental delay, and idiopathic mental retardation. Curr Opin Genet Dev. 2007;17:182–92. doi: 10.1016/j.gde.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 11.Jacquemont M-L, Sanlaville D, Redon R, Raoul O, Cormier-Daire V, Lyonnet S, Amiel J, Le MM, Heron D, De Blois M-C, Prieur M, Vekemans M, Carter NP, Munnich A, Colleaux L, Phillipe A. Array-based comparative genomic hybridization identifies high frequency of cryptic chromosomal rearrangements in patients with syndromic autism spectrum disorders. J Med Genet. 2006;43:843–9. doi: 10.1136/jmg.2006.043166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yamrom B, Yoon S, Krasnitz A, Kendall J, Leotta A, Pai D, Zhang R, Lee YH, Hicks J, Spence SJ, Lee AT, Puura K, Lehtimaki T, Ledbetter D, Gregersen PK, Bregman J, Sutcliffe JS, Jobanputra V, Chung W, Warburton D, King MC, Skuse D, Geschwind DH, Gilliam TC, Ye K, Wigler M. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–9. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, Shago M, Moessner R, Pinto D, Ren Y, Thiruvahindrapduram B, Fiebig A, Schreiber S, Friedman J, Ketelaars CE, Vos YJ, Ficicioglu C, Kirkpatrick S, Nicolson R, Sloman L, Summers A, Gibbons CA, Teebi A, Chitayat D, Weksberg R, Thompson A, Vardy C, Crosbie V, Luscombe S, Baatjes R, Zwaigenbaum L, Roberts W, Fernandez B, Szatmari P, Scherer SW. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–88. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morrow EM, Yoo S-Y, Flavell SW, Kim T-K, Lin Y, Hill RS, Mukaddes NM, Balkhy S, Gascom G, Hashmi A, Al-Saad S, Ware J, Joseph RM, Greenblatt R, Gleason D, Ertelt JA, Apse KA, Bodell A, Partlow JN, Barry B, Yao H, Markianos K, Ferland RJ, Greenberg ME, Walsh CA. Identifying autism loci and genes by tracing recent shared ancestry. Science. 2008;321:218–23. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murphy KC. Schizophrenia and velo-cardio-facial syndrome. Lancet. 2002;359:426–30. doi: 10.1016/S0140-6736(02)07604-3. [DOI] [PubMed] [Google Scholar]
- 16.Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, Stray SM, Rippey CF, Roccanova P, Makarov V, Lakshmi B, Findling RL, Sikich L, Stromberg T, Merriman B, Gogtay N, Butler P, Eckstrand K, Noory L, Gochman P, Long R, Chen Z, Davis S, Baker C, Eichler EE, Meltzer PS, Nelson SF, Singleton AB, Lee MK, Rapoport JL, King MC, Sebat J. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–43. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 17.Xu B, Roos JL, Levy S, van Rensburg EJ, Gogos JA, Karayiorgou M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet. 2008;40:880–5. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- 18.Sharp AJ, Mefford HC, Li K, Baker C, Skinner C, Stevenson RE, Schroer RJ, Novara F, De GM, Ciccone R, Broomer A, Casuga I, Wang Y, Xiao C, Barbacioru C, Gimelli G, Bernardina BD, Torniero C, Giorda R, Regan R, Murday V, Mansour S, Fichera M, Castiglia L, Failla P, Ventura M, Jiang Z, Cooper GM, Knight SJ, Romano C, Zuffardi O, Chen C, Schwartz CE, Eichler EE. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat Genet. 2008;40:322–8. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharp AJ, Locke DP, McGrath SD, Cheng Z, Bailey JA, Vallente RU, Pertz LM, Clark RA, Schwartz S, Segraves R, Oseroff VV, Albertson DG, Pinkel D, Eichler EE. Segmental duplications and copy-number variation in the human genome. Am J Hum Genet. 2005;77:78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Makoff AJ, Flomen RH. Detailed analysis of 15q11-q14 sequence corrects errors and gaps in the public access sequence to fully reveal large segmental duplications at breakpoints for Prader-Willi, Angelman, and inv dup(15) syndromes. Genome Biol. 2007;8:R114. doi: 10.1186/gb-2007-8-6-r114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stone JL, O’Donovan MC, Gurling H, Kirov GK, Blackwood DH, Corvin A, Craddock NJ, Gill M, Hultman CM, Lichtenstein P, McQuillin A, Pato CN, Ruderfer DM, Owen MJ, St CD, Sullivan PF, Sklar P, Purcell Leader SM, Stone JL, Ruderfer DM, Korn J, Kirov GK, Macgregor S, McQuillin A, Morris DW, O’Dushlaine CT, Daly MJ, Visscher PM, Holmans PA, O’Donovan MC, Sullivan PF, Sklar P, Purcell Leader SM, Gurling H, Corvin A, Blackwood DH, Craddock NJ, Gill M, Hultman CM, Kirov GK, Lichtenstein P, McQuillin A, O’Donovan MC, Owen MJ, Pato CN, Purcell SM, Scolnick EM, St CD, Stone JL, Sullivan PF, Sklar LP, O’Donovan MC, Kirov GK, Craddock NJ, Holmans PA, Williams NM, Georgieva L, Nikolov I, Norton N, Williams H, Toncheva D, Milanova V, Owen MJ, Hultman CM, Lichtenstein P, Thelander EF, Sullivan P, Morris DW, O’Dushlaine CT, Kenny E, Waddington JL, Gill M, Corvin A, McQuillin A, Choudhury K, Datta S, Pimm J, Thirumalai S, Puri V, Krasucki R, Lawrence J, Quested D, Bass N, Curtis D, Gurling H, Crombie C, Fraser G, Leh KS, Walker N, St CD, Blackwood DH, Muir WJ, McGhee KA, Pickard B, Malloy P, Maclean AW, Van BM, Visscher PM, Macgregor S, Pato MT, Medeiros H, Middleton F, Carvalho C, Morley C, Fanous A, Conti D, Knowles JA, Paz FC, Macedo A, Helena AM, Pato CN, Stone JL, Ruderfer DM, Korn J, McCarroll SA, Daly M, Purcell SM, Sklar P, Purcell SM, Stone JL, Chambert K, Ruderfer DM, Korn J, McCarroll SA, Gates C, Gabriel SB, Mahon S, Ardlie K, Daly MJ, Scolnick EM, Sklar P. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–41. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, Fossdal R, Sigurdsson E, Sigmundsson T, Buizer-Voskamp JE, Hansen T, Jakobsen KD, Muglia P, Francks C, Matthews PM, Gylfason A, Halldorsson BV, Gudbjartsson D, Thorgeirsson TE, Sigurdsson A, Jonasdottir A, Jonasdottir A, Bjornsson A, Mattiasdottir S, Blondal T, Haraldsson M, Magnusdottir BB, Giegling I, Moller HJ, Hartmann A, Shianna KV, Ge D, Need AC, Crombie C, Fraser G, Walker N, Lonnqvist J, Suvisaari J, Tuulio-Henriksson A, Paunio T, Toulopoulou T, Bramon E, Di FM, Murray R, Ruggeri M, Vassos E, Tosato S, Walshe M, Li T, Vasilescu C, Muhleisen TW, Wang AG, Ullum H, Djurovic S, Melle I, Olesen J, Kiemeney LA, Franke B, Kahn RS, Linszen D, van OJ, Wiersma D, Bruggeman R, Cahn W, Germeys I, de HL, Krabbendam L, Sabatti C, Freimer NB, Gulcher JR, Thorsteinsdottir U, Kong A, Andreassen OA, Ophoff RA, Georgi A, Rietschel M, Werge T, Petursson H, Goldstein DB, Nothen MM, Peltonen L, Collier DA, St CD, Stefansson K. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–6. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller DT, Shen Y, Weiss LA, Korn J, Anselm I, Bridgemohan C, Cox GF, Dickinson H, Gentile J, Harris DJ, Hegde V, Hundley R, Khwaja O, Kothare S, Luedke C, Nasir R, Poduri A, Prasad K, Raffalli P, Reinhard A, Smith SE, Sobeih M, Soul J, Stoler J, Takeoka M, Tan WH, Thakuria J, Wolff P, Yusupov R, Gusella JF, Daly MJ, Wu BL. Microdeletion/duplication at 15q13.2q13.3 among individuals with features of autism and other neuropsychiatirc disorders. J Med Genet. 2009;46:242–8. doi: 10.1136/jmg.2008.059907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Helbig I, Mefford HC, Sharp AJ, Guipponi M, Fichera M, Franke A, Muhle H, de KC, Baker C, von SS, Kron KL, Steinich I, Kleefuss-Lie AA, Leu C, Gaus V, Schmitz B, Klein KM, Reif PS, Rosenow F, Weber Y, Lerche H, Zimprich F, Urak L, Fuchs K, Feucht M, Genton P, Thomas P, Visscher F, de Haan GJ, Moller RS, Hjalgrim H, Luciano D, Wittig M, Nothnagel M, Elger CE, Nurnberg P, Romano C, Malafosse A, Koeleman BP, Lindhout D, Stephani U, Schreiber S, Eichler EE, Sander T. 15q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nat Genet. 2009;41:160–2. doi: 10.1038/ng.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu X, Shaw CA, Patel A, Li J, Cooper ML, Wells WR, Sullivan CM, Sahoo T, Yatsenko SA, Bacino CA, Stankiewicz P, Ou Z, Chinault AC, Beaudet AL, Lupski JR, Cheung SW, Ward PA. Clinical implementation of chromosomal microarray analysis: summary of 2513 postnatal cases. PLoS ONE. 2007;2:e327. doi: 10.1371/journal.pone.0000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ou Z, Kang SH, Shaw CA, Carmack CE, White LD, Patel A, Beaudet AL, Cheung SW, Chinault AC. Bacterial artificial chromosome-emulation oligonucleotide arrays for targeted clinical array-comparative genomic hybridization analyses. Genet Med. 2008;10:278–89. doi: 10.1097/GIM.0b013e31816b4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sahoo T, del Gaudio D, German JR, Shinawi M, Peters SU, Person RE, Garnica A, Cheung SW, Beaudet AL. Prader-Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat Genet. 2008;40:719–21. doi: 10.1038/ng.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yee RD, Baloh RW, Marder SR, Levy DL, Sakala SM, Honrubia V. Eye movements in schizophrenia. Invest Ophthalmol Vis Sci. 1987;28:366–74. [PubMed] [Google Scholar]
- 29.Mefford HC, Sharp AJ, Baker C, Itsara A, Jiang Z, Buysse K, Huang S, Maloney VK, Crolla JA, Baralle D, Collins A, Mercer C, Norga K, de RT, Devriendt K, Bongers EM, de LN, Reardon W, Gimelli S, Bena F, Hennekam RC, Male A, Gaunt L, Clayton-Smith J, Simonic I, Park SM, Mehta SG, Nik-Zainal S, Woods CG, Firth HV, Parkin G, Fichera M, Reitano S, Giudice ML, Li KE, Casuga I, Broomer A, Conrad B, Schwerzmann M, Raber L, Gallati S, Striano P, Coppola A, Tolmie JL, Tobias ES, Lilley C, Armengol L, Spysschaert Y, Verloo P, De CA, Goossens L, Mortier G, Speleman F, van BE, Nelen MR, Hochstenbach R, Poot M, Gallagher L, Gill M, McClellan J, King MC, Regan R, Skinner C, Stevenson RE, Antonarakis SE, Chen C, Estivill X, Menten B, Gimelli G, Gribble S, Schwartz S, Sutcliffe JS, Walsh T, Knight SJ, Sebat J, Romano C, Schwartz CE, Veltman JA, de Vries BB, Vermeesch JR, Barber JC, Willatt L, Tassabehji M, Eichler EE. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl J Med. 2008;359:1685–99. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brunetti-Pierri N, Berg JS, Scaglia F, Belmont J, Bacino CA, Sahoo T, Lalani SR, Graham B, Lee B, Shinawi M, Shen J, Kang SH, Pursley A, Lotze T, Kennedy G, Lansky-Shafer S, Weaver C, Roeder ER, Grebe TA, Arnold GL, Hutchison T, Reimschisel T, Amato S, Geragthy MT, Innis JW, Obersztyn E, Nowakowska B, Rosengren SS, Bader PI, Grange DK, Naqvi S, Garnica AD, Bernes SM, Fong CT, Summers A, Walters WD, Lupski JR, Stankiewicz P, Cheung SW, Patel A. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat Genet. 2008;40:1466–71. doi: 10.1038/ng.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lounsbury DW, Reynolds TC, Rapkin BD, Robson ME, Ostroff J. Protecting the privacy of third-party information: recommendations for social and behavioral health researchers. Soc Sci Med. 2007;64:213–22. doi: 10.1016/j.socscimed.2006.08.035. [DOI] [PubMed] [Google Scholar]
- 32.Resnik DB, Sharp RR. Protecting third parties in human subjects research. IRB. 2006;28:1–7. [PMC free article] [PubMed] [Google Scholar]
- 33.Elmslie FV, Rees M, Williamson MP, Kerr M, Kjeldsen MJ, Pang KA, Sundqvist A, Friis ML, Chadwick D, Richens A, Covanis A, Santos M, Arzimanoglou A, Panayiotopoulos CP, Curtis D, Whitehouse WP, Gardiner RM. Genetic mapping of a major susceptibility locus for juvenile myoclonic epilepsy on chromosome 15q. Hum Mol Genet. 1997;6:1329–34. doi: 10.1093/hmg/6.8.1329. [DOI] [PubMed] [Google Scholar]
- 34.Neubauer BA, Fiedler B, Himmelein B, Kampfer F, Lassker U, Schwabe G, Spanier I, Tams D, Bretscher C, Moldenhauer K, Kurlemann G, Weise S, Tedroff K, Eeg-Olofsson O, Wadelius C, Stephani U. Centrotemporal spikes in families with rolandic epilepsy: linkage to chromosome 15q14. Neurology. 1998;51:1608–12. doi: 10.1212/wnl.51.6.1608. [DOI] [PubMed] [Google Scholar]
- 35.Flomen RH, Collier DA, Osborne S, Munro J, Breen G, St CD, Makoff AJ. Association study of CHRFAM7A copy number and 2 bp deletion polymorphisms with schizophrenia and bipolar affective disorder. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:571–5. doi: 10.1002/ajmg.b.30306. [DOI] [PubMed] [Google Scholar]
- 36.Martin LF, Freedman R. Schizophrenia and the alpha7 nicotinic acetylcholine receptor. Int Rev Neurobiol. 2007;78:225–46. doi: 10.1016/S0074-7742(06)78008-4. [DOI] [PubMed] [Google Scholar]
- 37.Gault J, Robinson M, Berger R, Drebing C, Logel J, Hopkins J, Moore T, Jacobs S, Meriwether J, Choi MJ, Kim EJ, Walton K, Buiting K, Davis A, Breese C, Freedman R, Leonard S. Genomic organization and partial duplication of the human alpha7 neuronal nicotinic acetylcholine receptor gene (CHRNA7) Genomics. 1998;52:173–85. doi: 10.1006/geno.1998.5363. [DOI] [PubMed] [Google Scholar]
- 38.Flomen RH, Davies AF, Di FM, Cascia CL, kie-Ogilvie C, Murray R, Makoff AJ. The copy number variant involving part of the alpha7 nicotinic receptor gene contains a polymorphic inversion. Eur J Hum Genet. 2008;16:1364–71. doi: 10.1038/ejhg.2008.112. [DOI] [PubMed] [Google Scholar]