Abstract
Childhood leukemia, which accounts for > 30% of newly diagnosed childhood malignancies, is one of the leading causes of death for children with cancer. Genome-wide studies using microarray chips to identify copy number changes in human cancer are becoming more common. In this pilot study, 45 pediatric leukemia samples were analyzed for gene copy aberrations using novel molecular inversion probe (MIP) technology. Acute leukemia subtypes included precursor B-cell acute lymphoblastic leukemia (ALL) (n = 23), precursor T-cell ALL (n = 6), and acute myeloid leukemia (n = 14). The MIP analysis identified 69 regions of recurring copy number changes, of which 41 have not been identified with other DNA microarray platforms. Copy number gains and losses were validated in 98% of clinical karyotypes and 100% of fluorescence in situ hybridization studies available. We report unique patterns of copy number loss in samples with 9p21.3 (CDKN2A) deletion in the precursor B-cell ALL patients, compared with the precursor T-cell ALL patients. MIPs represent an attractive technology for identifying novel copy number aberrations, validating previously reported copy number changes, and translating molecular findings into clinically relevant targets for further investigation.
1. Introduction
Childhood leukemia is the most common cancer among persons under 15 years of age, accounting for > 30% of all childhood malignancies [1]. In developed countries, 8 of 10 children diagnosed with acute lymphoblastic leukemia (ALL) will survive 5 years or longer due to treatment assignments based on presenting age, white blood cell count, extramedullary disease, blast cytogenetics, and initial treatment response [2]. These risk stratification categories for ALL are still evolving, however, and treatment failures occur in 10–15% of lower risk patients [3]. Like-wise, acute myeloid leukemia (AML) patients are often risk-stratified based on molecular cytogenetics and treatment response; nonetheless, the most recent Children’s Oncology Group trial reported a 5-year survival of 58% for pediatric AML [4]. Even with aggressive retrieval therapy, most children for whom leukemia treatment fails will die [5], and relapsed leukemia remains one of the leading causes of mortality for children with cancer. Robust biomarkers for risk of relapse are needed, so that children with leukemia can be risk-stratified as accurately as possible at the time of diagnosis [3,6].
Aneuploidy and translocation events are gross indicators of genomic instability events associated with ALL. Array comparative genomic hybridization methods have significantly higher resolution in identifying copy number aberrations (CNAs), compared with conventional cytogenetic analysis, and as a result their application in ALL have led to the discovery of CNAs within specific intervals of the genome [7–9]. Single nucleotide polymorphism (SNP) mi-croarrays have even higher resolution of CNAs in pediatric leukemia. Mullighan et al. [10] reported multiple deletions, amplifications, point mutations, and structural rearrangements in genes regulating B lymphocyte development and differentiation in 40% of B-precursor ALL, and two other studies [11,12] also reported successful use of SNP arrays to characterize recurring CNAs in childhood ALL. All three of these SNP array studies identified the well-described loss of 9p21.3, encompassing CDKN2A and CDKN2B, genes that are known to be frequently deleted in ALL. The CDKN2A gene encodes two distinct proteins, p16INK4a and p14ARF which are involved in cell cycle regulation. This region is more frequently deleted in precursor T-cell than precursor B-cell ALL [13], with variable prognostic significance [14–16].
To survey CNAs in childhood leukemia at the resolution of individual gene sequences, we applied a novel genomic technology (molecular inversion probes, or MIPs) that has been adapted for gene copy analysis in cancer [17,18]. In this technology, the probe is a single oligonucleotide that recognizes and hybridizes to a specific genomic target sequence with two recognition sites [19]. After the probe hybridizes to the target DNA, a single base-pair gap exists in the middle of the two recognition sequences. This gap can be either a SNP or a nonpolymorphic nucleotide. The reaction is split into four tubes, with each tube containing polymerase, ligase, and a single nucleotide. In the presence of the appropriate nucleotide and with specific annealing to the target sequence, a circularization event occurs, making the probe amplifiable in subsequent steps. The amplified probes are ultimately detected and quantitated on microarrays that have barcode sequences complementary to those in the individual MIP probes. Because a barcode intermediate is used instead of direct genomic DNA hybridization to an array, it is possible to query any unique sequence without the hybridization constraints of array comparative genomic hybridization or oligonucleotide arrays. Thus far, MIPs have been validated and used for CNA detection in breast [18], ovarian [20,21], and colorectal cancer [22].
Here, we report on use of MIPs to detect novel areas of gene CNAs and allelic imbalance in childhood leukemia. For this analysis, we used a MIP cancer panel with a resolution of specific gene sequences for > 1,000 cancer genes. We identified a number of novel deletions and amplifications of specific genes, including unique patterns of loss of heterozygosity in precursor B-cell ALL, compared with precursor T-cell ALL. This pilot study demonstrates the feasibility of using MIP technology to analyze childhood leukemia specimens.
2. Materials and methods
2.1. Patients and samples
All patient material was obtained with informed consent from the Lucile Packard Children’s Hospital at Stanford University. The study was previously approved by the institutional review board at Stanford University School of Medicine.
Genomic DNA was extracted from 45 pediatric leukemia samples obtained at diagnosis in the form of bone marrow aspirates, pheresis products, or peripheral blood with blasts > 85%. Normal genomic DNA was extracted from 20 of the same patients, from peripheral leukocytes obtained after documented remission that included negative bone marrow studies. Samples were collected from patients diagnosed and treated between April 1999 and June 2007. See Table 1 for characteristics of patients analyzed in this study.
Table 1.
Clinicodemographic characteristics of 45 childhood leukemia cases
| Pre-B ALL | Pre-T ALL | AML | Other (BL, CML) | Total | |
|---|---|---|---|---|---|
| Sample size, no. | 23 | 6 | 14a | 2 | 45 |
| Mean age, yr (range) | 4.8 (1.3–16.9) | 7.8 (2.6–14.2) | 8.9 (0.3–18.4) | 10.8 (8.2–13.3) | 6.7 (0.3–18.4) |
| Male, no. (%) | 14 (61) | 5 (83) | 9 (64) | 2 (100) | 30 (67) |
| Female, no. (%) | 9 (39) | 1 (17) | 5 (36) | 0 | 15 (33) |
| Sample source, no. | 22 BMA; 1 Pher | 6 BMA | 10 BMA; 2 Pher; 2 PB | 1 BMA; 1 Pher | 39 BMA; 4 Pher; 2 PB |
| Mean blasts, % (range) | 88 (26–100) | 75 (39–88) | 82 (42–100) | 90 (80–100) | 84 (26–100) |
| Dx sample, no. (%) | 21 (91) | 6 (100) | 12 (86) | 2 (100) | 41 (91) |
| Relapse sample, no. (%) | 2 (9) | 0 | 2 (14) | 0 | 4 (9) |
| Mean WBC, no. × 109/L (range) | 36.2 (0.4–252) | 191.4 (13.3–491.4) | 147.9 (0.5–513.6) | 361 (22–700) | 106.1 (0.4–700) |
| CNS 2 or 3, no. (%) | 10 (44) | 2 (33) | NA | NA | 12 (41)b |
| Relapse, no. (%) | 4 (19) | 2 (33) | 4 (40)c | 0 | 10 (26) |
| Survival, no. (%) | 21 (91) | 5 (83) | 7 (54)d | 2 (100) | 35 (81) |
| Hispanic, no. (%) | 5 (22) | 0 | 6 (43) | 1 (50) | 12 (27) |
Abbreviations: ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; BL, Burkitt lymphoma; BMA, bone marrow aspirate; CML, chronic myeloid leukemia; CNS, central nervous system disease; Dx, diagnosis; PB, peripheral blood; Pher, pheresis; WBC, white blood cell count.
Includes paired diagnosis and relapse samples from a single patient (counted separately).
For only ALL (pre-B + pre-T).
Does not include 2 relapsed samples, 1 refractory sample, and 1 early death.
Includes paired diagnosis and relapse samples from a single patient (counted as one).
2.2. Genomic DNA preparation
Genomic DNA was isolated from leukemia samples and peripheral leukocytes using a genomic DNA preparation kit (Gentra Systems, Minneapolis, MN). The DNA quantitation was done using a double-stranded assay (PicoGreen, P7589; Invitrogen, Carlsbad, CA).
2.3. Molecular inversion probe design
A cancer panel of 24,037 SNPs was chosen for the MIP panel synthesis (Affymetrix, CA). Each probe required genomic sequences of ~20 nucleotides on either flank of the SNP position, which were incorporated into the molecular inversion probe design. In addition to probes across the genome, extra probes were chosen from intragenic sequences of > 1,000 genes that have been reported to be involved in cancer development (Supplemental Table 1). Each gene was represented on average by three to six probes. SNP locations for each probe refer to human genome build NCBI 35.1 (hg17; May 2004).
2.4. Molecular inversion probe assay
The MIP assay was performed as described previously [17,18]. The initial step involved an overnight annealing of 4.7 µL of DNA samples (75 ng total) in a pool containing 24,037 probes (200 amol/µL per probe) and 0.045 µL of enzyme A mixed in a 384-well plate on ice. The reaction was incubated at 20°C for 4 minutes, at 95°C for 5 minutes, and then at 58°C overnight. The MIP probes were circularized with the addition of 4 µL of the appropriate nucleotide at 58°C for 10 minutes. The uncircularized probes and genomic DNA were eliminated by addition of 4 µL of exonucleases and incubation at 37°C for 15 minutes, followed by heat inactivation. The circularized probes were linearized by restriction enzyme digest at 37°C for 15 minutes, followed by universal primer amplification for 18 cycles at 95°C for 20 seconds, 64°C for 40 seconds, and 72°C for 10 seconds. For the labeling reaction, the product was further amplified with the label primers for 10 cycles.
For barcode array hybridization, MIP polymerase chain reaction (PCR) products were mixed with hybridization cocktail, denatured, and hybridized to 30 k universal tag arrays (Affymetrix, Santa Clara, CA) at 39°C for 16 hours with two arrays for each allele. The overnight hybridized arrays were washed on a standard Affymetrix fluidic station and stained with streptavidin–phycoerythrin at 5 ng/mL (Invitrogen). Copy number estimation was obtained from the barcode hybridization signals as described previously [17].
2.5. Statistical analysis
To facilitate the identification of recurrent CNAs in pediatric leukemia, we relied on cross-sample segmentation, a generalization of the circular binary segmentation (CBS) method of Olshen et al. [23] for performing a simultaneous segmentation of multiple samples. The CBS method, which is widely used for segmentation of single samples, works through the recursive application of a chi-square statistic for change-point detection. The multisample segmentation method sums the chi-squared statistics from each sample at each location of the chromosomal breakpoints and scans the genome using the summed chisquare statistics for the optimal location of breakpoints across all of the samples (Zhang et al, Technical Report no. 2008-1, available at http://stat.stanford.edu/reports/papers2008.html). The cross-segmentation method avoids the ad hoc post-processing steps required to summarize the segmentation results of individual samples into a sparse multisample summary.
Cross-sample segmentation was separately applied to the leukemia and to the matched normal germline samples. A prefilter was then applied by deleting any CNA intervals reported in the normal samples that had overlap with intervals reported in the leukemia sample set. The remaining intervals (present in leukemia samples only) were further restricted after segmentation using the following criteria: > 1 SNP and copy number cutoff of > 0.8 deviation from baseline (loss = smoothed value < 1.2 and gain = smoothed value > 2.8). The absolute deviation value of > 0.8 was chosen to detect true CNAs with greater stringency. There was no minimum basepair restriction length on classifying gain or loss, but instead each region had a minimum two-probe restriction (i.e., ≥2 consecutive MIPs needed to make the call). Finally, only those regions were called that contained individual probes with a quality control call rate of > 90% or relative standard deviation of < 20% [18].
2.6. Clinical associations
Significant associations between CNAs and clinical parameters were identified using the computer program PaGE5.1, which relies on a modified two-tailed t-statistic for class comparisons with random permutations of class labels to estimate a false discovery rate [24]. P-values were calculated on the identified significant genes using Student’s t-test with two-tailed distribution. Clinical parameters were based on current Children’s Oncology Group risk features and included age at presentation, sex, white blood cell count at diagnosis, central nervous system disease, leukemia subtype, risk category, initial treatment response, and relapse status.
2.7. G-banding karyotype and FISH analysis
Clinical cytogenetic findings were extracted from the medical records for each patient and were compared with the MIP findings. Karyotyping was performed on leukemia sample metaphase chromosome preparations with slide preparations stained and analyzed by a modified G-banding method using Wright’s stain. Interphase FISH analysis was performed on cell preparations using a dual-color probe combination specific for centromeres of chromosomes 4 and 10 (CEP4 and CEP10; Abbott Molecular, Des Plaines, IL) and with the TEL/AML1 dual-color fusion probe for ETV6/RUNX1 (Abbott Molecular), designed for identification of the ALL-associated cryptic translocation t(12;21).
2.8. Quantitative PCR
Quantitative PCR was performed in triplicate on 1 ng of genomic DNA from our clinical samples using Platinum SYBR Green qPCR Supermix-UDG with ROX (Invitrogen) or Taqman universal PCR master mix (Applied Biosystems, Foster City, CA). RNase P, was used to normalize copy number. Human genomic DNA (0.05 ng to 12.8 ng, Roche Diagnostics, Indianapolis, IN) was used to generate standard curves for each primer set from which the quantity of each gene was calculated. Quantitative analysis was performed on a sequence detection system (ABI PRISM 7900-HT; Applied Biosystems). Copy number was determined by the formula Quantitygene/QuantityRNaseP. Primers were designed for CDKN2A and for exons 3, 6, and 8 of PAX5 (Supplemental Table 2), genes involved in two of the most common deletions in childhood ALL.
3. Results
In 45 childhood leukemia samples, 69 genomic regions of recurring amplifications or deletions were identified in at least 2 samples each (Table 2; Supplemental Table 3–4). Also identified were 13 regions of gain or loss that were unique to a single sample. Among the 24,000 loci analyzed, the ALL samples (pre-B and pre-T) averaged a total of 15 copy number changes (7.7 amplifications and 7.3 deletions per sample) and the AML samples averaged a total of 12.5 copy number changes (6.5 amplifications and 6 deletions per sample). See Table 3 for mean number of recurring copy number changes per individual leukemia subcategory. Ninety-eight percent of our ALL and AML samples had at least one recurring deletion and 77% of our samples had at least one recurring gain detected by MIPs.
Table 2.
The 20 most frequent recurring copy number aberrations in childhood leukemia, as detected by molecular inversion probes
| Lineage-specific cases, % | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Cases, no. | Typea | Chr | Location | Ref.b | Length bp | B-ALL (n = 23) | T-ALL (n = 6) | AML (n = 14) | Genes in regionc |
| 23 | Loss | 16 | 16q22.3 | – | 10352 | 44 | 83 | 50 | MLKL |
| Gain | 16 | 16q22.3 | – | – | 4 | – | – | MLKL | |
| 22 | Loss | 10 | 10q25.2 | – | 2490 | 57 | 67 | 36 | ADRA2A |
| 19 | Loss | 7 | 7q21.13 | – | 1363 | 35 | 67 | 43 | FZD1 |
| Gain | 7 | 7q21.13 | – | – | 4 | – | – | FZD1 | |
| 19 | Loss | 17 | 17p13.1 | [10] | 1978 | 17 | 17 | 36 | none known |
| Gain | 17 | 17p13.1 | – | – | 26 | 17 | 14 | none known | |
| 17 | Loss | 14 | 14q24.2 | [10] | 270 | 30 | 50 | 36 | none known |
| Gain | 14 | 14q24.2 | – | – | 9 | – | – | none known | |
| 16 | Loss | 4 | 4p16.3 | – | 79037 | 9 | – | 14 | TACC3 |
| Gain | 4 | 4p16.3 | – | – | 26 | 33 | 29 | TACC3 | |
| 15 | Loss | 21 | 21q22.11 | [11] | 7219 | 4 | – | 7 | OLIG2 |
| Gain | 21 | 21q22.11 | – | – | 30 | 17 | 36 | OLIG2 | |
| 15 | Loss | 5 | 5q35.3 | – | 18679 | 17 | 67 | 50 | CLK4 |
| 15 | Loss | 1 | 1p36.12 | – | 203480 | 44 | 50 | 14 | EIF4G3, HP1BP3, SH2D5, KIF17, DDOST |
| 14 | Loss | 6 | 6p22.1 | – | 717 | 9 | – | 7 | none known |
| Gain | 6 | 6p22.1 | – | – | 26 | 17 | 29 | none known | |
| 14 | Loss | 9 | 9p21.3 | [10,11] | 24424 | 35 | 100 | – | CDKN2A |
| 13 | Loss | 11 | 11q13.2 | – | 3454 | 22 | 33 | 36 | none known |
| Gain | 11 | 11q13.2 | – | – | 4 | – | – | none known | |
| 13 | Gain | 2 | 2q31.1 | – | 12837 | 30 | 33 | 29 | HOXD13 |
| 12 | Loss | 2 | 2q35 | – | 2774 | 9 | 17 | 36 | FEV |
| Gain | 2 | 2q35 | – | – | 4 | 17 | 14 | FEV | |
| 12 | Loss | 1 | 1p35.1 | – | 25184 | 4 | 17 | 7 | MARCKSL1 |
| Gain | 1 | 1p35.1 | – | – | 22 | 17 | 21 | MARCKSL1 | |
| 13 | Loss | 8 | 8p12~p11.2 | – | 938 | 17 | 33 | 7 | ADRB3 |
| Gain | 8 | 8p12~p11.2 | – | – | 13 | – | 21 | ADRB3 | |
| 13 | Loss | 14 | 14q32.32.32.33 | – | 1223220 | 4 | – | 14 | >5 genes |
| Gain | 14 | 14q32.32~32.33 | – | – | 26 | 17 | 21 | >5 genes | |
| 12 | Gain | 21 | 21q22.2 | [10] | 423958 | 48 | 17 | – | ETS2, FLJ45139, PSMG1d |
| 11 | Loss | 14 | 14q11.2 | – | 280777 | 17 | 50 | – | none known |
| Gain | 14 | 14q11.2 | – | – | 9 | – | 14 | none known | |
| 11 | Loss | 17 | 17q12 | – | 29229 | 9 | 17 | 21 | MLLT6 |
| Gain | 17 | 17q12 | – | – | 13 | – | 14 | MLLT6 | |
Abbreviations: ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; Chr, chromosome; CNA, copy number aberration; Ref., references.
Loss is defined as copy number ≤ 1.2. Gain is defined as copy number ≥ 2.8.
Based on > 1,000 genes involved in cancer development: oncogenes, tumor suppressors, DNA repair genes, cell growth genes, and metabolism genes.
The PSMG1 gene was previously described as DSCR2.
Table 3.
Mean number recurring copy number aberrations, by leukemia subtype
| CNA,a mean no. | ||||
|---|---|---|---|---|
| Leukemia subtype | Sample size | Loss | Gain | Total CNAs |
| t(12;21) | n = 6 | 8.5 ± 4.8 | 2.2 ± 2.4 | 10.7 ± 3.7 |
| Hyperploid (47–50) | n = 2 | 7.5 ± 6.4 | 3.0 ± 4.2 | 10.5 ± 2.1 |
| Hyperploid ( > 50) | n = 3 | 6.7 ± 6.4 | 18.7 ± 8.5 | 25.3 ± 14.0 |
| Hypoploid ( < 46) | n = 1 | 8.0 ± 0.0 | 20.0 ± 0.0 | 28.0 ± 0.0 |
| Miscellaneousb | n = 10 | 5.4 ± 3.5 | 9.5 ± 6.3 | 14.9 ± 8.1 |
| None | n = 3 | 6.0 ± 1.7 | 8.3 ± 1.5 | 14.3 ± 3.1 |
| Other | n = 4 | 14.3 ± 3.1 | 10.8 ± 8.7 | 16.8 ± 12.9 |
| Pseudodiploid | n = 3 | 4.0 ± 2.6 | 9.0 ± 7.5 | 13.0 ± 5.3 |
| t(9;22) | n = 1 | 4.0 ± 0.0 | 1.0 ± 0.0 | 5.0 ± 0.0 |
| Pre-T ALL | n = 6 | 9.8 ± 5.9 | 5.5 ± 8.8 | 15.3 ± 13.5 |
| Pre-B ALL | n = 23 | 6.6 ± 4.3 | 8.3 ± 7.8 | 14.9 ± 9.1 |
| AML | n = 14 | 6.0 ± 3.3 | 6.5 ± 7.9 | 12.5 ± 9.1 |
Abbreviations: ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; CNA, copy number aberration.
Loss is defined as copy number ≤ 1.2. Gain is defined as copy number ≥ 2.8.
Of the miscellaneous sub types, “none” refers to samples with undetected cytogenetic abnormalities and “other” includes samples with less commonly reported cytogenetic abnormalities.
3.1. Clinical cytogenetics comparison
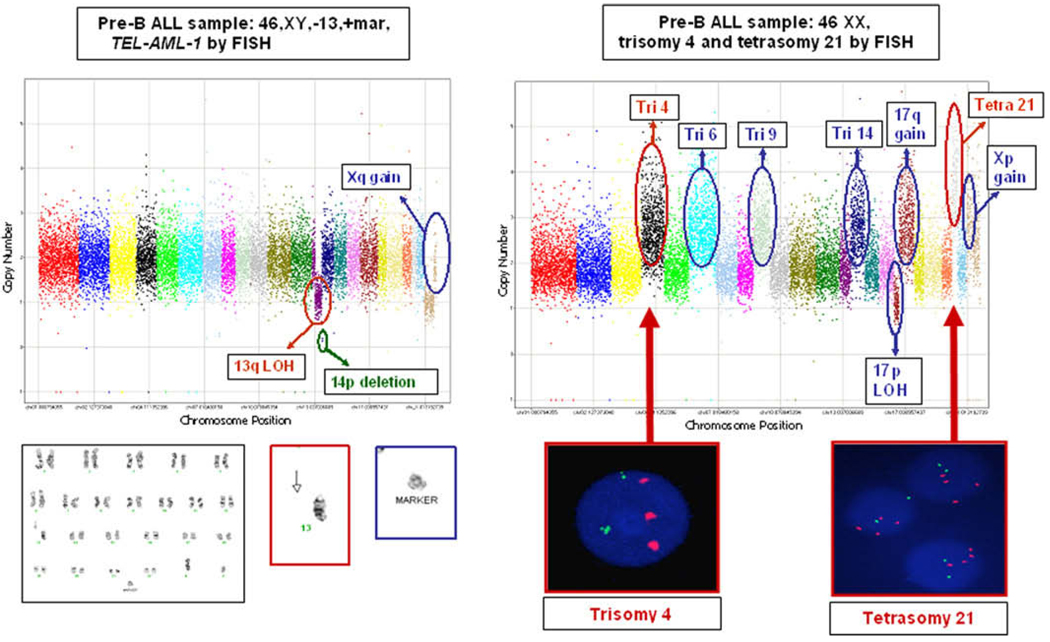
Of the 43 leukemia samples with available karyotype data, , 42 (98%) matched the chromosomal gains and losses identified by MIPs. Three of three samples with trisomy or tetrasomy reported by FISH (trisomies 4 and 10, tetrasomy 21) matched the whole-chromosome amplification reported by MIPs (100%). Additionally, nearly every sample reported a multitude of micro-CNAs and macro-CNAs not reported by karyotype and FISH in the clinical cytogenetics (Figs. 1A and 1B). As expected, MIPs did not detect copy-neutral, balanced translocations reported by FISH. In some cases, unidentified chromosomal material (i.e., marker chromosomes) on the karyotype analysis could be identified as originating from a specific region, as suggested by MIP amplification in that specific location; for example, the sample with extra, unidentified chromosomal material on karyotype and gain in Xq by MIPs most likely has a marker for this extra region of Xq (Fig. 1A).
Fig. 1.
Molecular inversion probe analysis of pre-B ALL samples with customized 24 k cancer panel reveals additional allelic imbalance, compared with conventional karyotyping or fluorescence in situ hybridization (FISH) analysis. Chromosomes 1–22 and X are sequentially labeled in color, with copy number (linear scale) along the y-axis. (A) Karyotype 46,XY,−13, +mar; ETV6/RUNX1 fusion detected with TEL-AML1 FISH probe. The MIPs identified further allelic imbalance information (circled in green and blue) not described in the clinical karyotype. The chromosome 13 deletion (circled in red) was described as an entire deletion in the karyotype (red box, below graph), but was identified by MIPs as an isolated 13q deletion (13p remains intact). The unidentified marker (blue box, below graph) likely represents gain in Xq. (B) Karyotype 46,XX; trisomy 4 and tetrasomy 21 detected with FISH analysis. The MIPs identified further allelic imbalance information (circled in blue) not detected with karyotyping or FISH analysis and validate FISH findings (red boxes, below graph) of trisomy 4 and tetrasomy 21 (circled in red). Abbreviations: LOH, loss of heterozygosity; Tetra, tetrasomy; Tri, trisomy.
3.2. Deletion of PAX5
This childhood leukemia cohort exhibited PAX5 deletions in 4 of 29 pre-B and pre-T ALL samples (14%), confirmed by real time (RT) PCR. One sample had a deletion only in PAX5 exon 8, two samples had deletions only in PAX5 exons 3 and 6, and the one remaining sample had deletions in all three measured PAX5 exons (i.e., 3, 6, and 8). MIP copy number compared with PCR copy number values are displayed in Supplemental Table 5.
3.3. Deletion of 9p21 (CDKN2A)
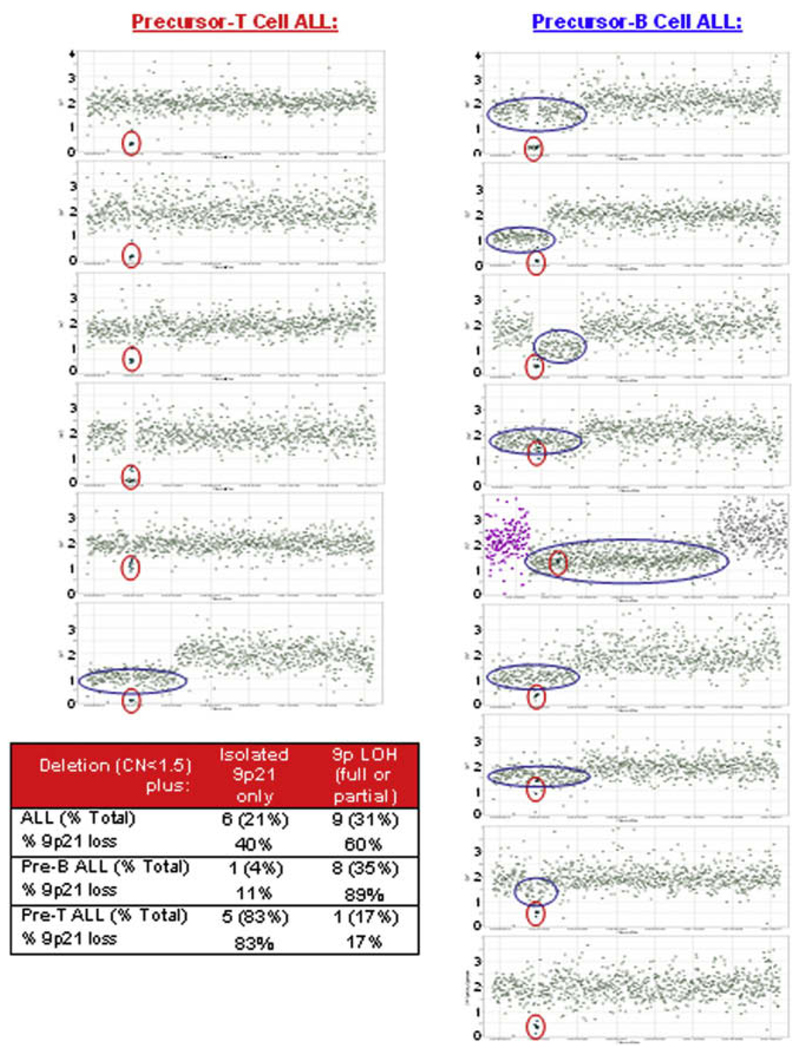
Using a definition of homozygous deletions (CN < 0.5) and hemizygous deletions (CN = 0.8–1.5), 13.8% hemizygous and 37.9% homozygous 9p21 deletions were identified in 29 pediatric ALL samples (precursor B- and T-ALL). The majority of 9p21 deletions were homozygous (11 of 15, 73.3%), and also involved either upstream or downstream hemizygous loss of adjacent 9p regions (9 of 15, 60%). The majority of samples with extended 9p hemizygous loss were from patients with pre-B ALL. Of the six remaining samples with isolated 9p21 deletion and no other associated 9p hemizygous deletion, 5 of 6 (83.3%) were in pre-T ALL samples (Fig. 2). The length of the 9p hemizygous deletions varied considerably from several thousand base pairs to the entire 9p region, with one sample containing deletion of the entire chromosome. The 9p21 deletion consistently encompassed 9 probes, 15 kb in length (chr9:21962445–21977472; NCBI Build 35.1). Fully 100% of the CDKN2A homozygous and hemizygous deletions were confirmed by RT-PCR (Supplemental Table 6).
Fig. 2.
Isolated 9p21.3 (CDKN2A) deletion associated with precursor T-cell ALL and full or partial 9p loss of heterozygosity (LOH) associated with precursor B-cell ALL. The x-axis shows 890 probes ordered by location on chromosome 9, from the short arm (left) to the long arm (right), with linear copy number (CN) along the y-axis. Every sample with 9p21.3 deletion (circled in red) is shown. Areas of LOH are circled in blue. Note that even in samples with 9p LOH (copy number 1), homozygous CDKN2A deletions (copy number < 0.5) are still present. The inset table summarizes findings.
3.4. Clinical associations for ALL
Clinical associations were determined to be significant, with an false discovery rate of < 0.05. The clinical features significantly associated with gene-specific CNAs included age at presentation (pre-B and pre-T ALL), leukemia subtype (pre-B vs. pre-T ALL), relapse (pre-B and pre-T ALL), and central nervous system disease (CNS 1 vs. CNS 2,3) (Table 4).
Table 4.
Clinical correlation with recurring copy number aberrations in childhood leukemia
| Mean CNAa | Region | Cancer genesb | P-value | |
|---|---|---|---|---|
| By age, yr (pre-B ALL + pre-T ALL) | ||||
| <10yr (n = 24) | ≥10yr (n = 5) | |||
| 3.14 | 2.17 | 21q22.2 | ETS2, FLJ45139, PSMG1c | 0.00893 |
| 1.34 | 0.51 | 9p21 | CDKN2A | 0.00338 |
| By leukemia type (pre-B ALL vs. pre-T ALL) | ||||
| Pre-B (n = 23) | Pre-T (n = 6) | |||
| 1.41 | 0.36 | 9p21 | CDKN2A | 0.00037 |
| Relapse? (pre-B ALL + pre-T ALL) | ||||
| No (n = 21) | Yes (n = 6) | |||
| 2.13 | 3.26 | 6p22.1 | none known | 0.01255 |
| CNS disease type (pre-B ALL + pre-T ALL) | ||||
| CNS 1 (n = 17) | CNS 2, 3 (n = 12) | |||
| 2.12 | 2.88 | 4p16.3 | TACC3 | 0.01068 |
| 1.74 | 1.11 | 7q21.13 | FZD1 | 0.00151 |
| 2.26 | 2.72 | 14q11.2~q32.33 | >20 genes | 0.01060 |
Significant gain or loss is highlighted in bold italic type.
Based on > 1,000 genes involved in cancer development: oncogenes, tumor suppressors, DNA repair genes, cell growth genes, and metabolism genes.
The PSMG1 gene was previously described as DSCR2.
4. Discussion
Originally designed for large-scale SNP genotyping, MIP has recently been adapted to detect allelic CNAs [17]. Although MIPs have detected CNAs in other types of cancer [18,20–22], the present study represents, to our knowledge, the first report of MIPs being used on leukemia samples. This technology can identify sequence allele and sequence-specific deletions and amplifications, gene recombination events, and uniparental disomy on minute amounts of genomic DNA template (in the nanogram range), including from paraffin-embedded samples [18]. The MIP cancer panel used for this study comprises 24,037 SNPs from intragenic sequences of > 1,000 genes involved in cancer development (oncogenes, tumor suppressors, DNA repair genes, cell growth genes, and metabolism genes). The MIP cancer panel contains a mean spacing of 100 kb between probes for the entire genome; however, each cancer gene is represented by at least 3–6 probes, which often translates to < 5 kb of sequential coverage within the targeted cancer genes. The traditional 500 k SNP arrays offer denser whole-genome coverage on average (5 kb coverage), but these platforms contain SNPs optimized for microarray hybridization performance and adequate coverage for genome-wide association studies of human disease. Thus, SNP arrays often have very large gaps in specific gene coverage. The targeted MIP approach can be considered a complementary candidate gene approach, for identifying gene-specific CNAs that may be missed by these other methods of varying resolution. Several studies already have taken advantage of this targeted MIP approach to identify CNAs in cancer genes in colon, breast, and ovarian cancer [18,20–22].
4.1. Comparison with other SNP arrays
With the MIP cancer panel, 69 recurring regions of CNAs were identified in 45 pediatric leukemia samples (including pre-B, pre-T, and AML), which is a number similar to data from other published SNP array studies. Notably, Mullighan et al. [10] reported 63 recurring CNAs in 242 childhood ALL samples; Kuiper et al. [11] reported 31 recurring CNAs in 40 childhood ALL samples; and Kawamata et al. [12] reported 23 recurring CNAs (with many more nonrecurring regions identified) in 399 childhood ALL samples.
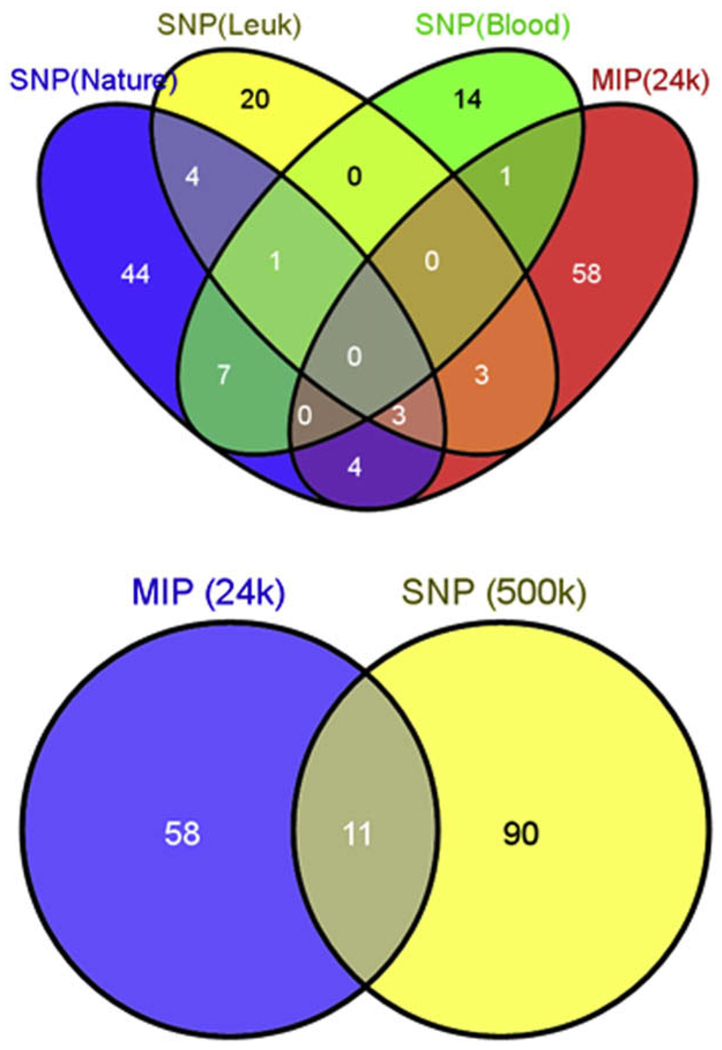
Combining the primary and supplemental data from these three SNP array studies [10–12], we found that 15 common regions of recurring CNAs were reported in at least two of the three publication: 1q, 20q, 3p14.2, 5q33.3, 6q23.3, 7 p, 9p13.2, 9p21.3, 9q, 10 p, 11q23.3, 12p13.2, 12q21.33, 13q14.2, and 17q11.2 (Supplemental Fig. 1). Our MIP analysis identified 11 CNA regions identical to findings from at least one of the three SNP studies: 1p22.2, 2p21, 7q34, 9p13.2, 9p21.3, 11q13.1, 12q21.33, 14q24.2, 16p13.3, 18q12.2, and 21q22.2 (Figs. 3A and 3B). The number of CNA regions in common increases to 28 when larger ranges of overlap are considered in addition to identically reported but limited chromosome locations; for example, the MIP panel identified a 7,673-bp copy number change in 17q21.32 (ITGA2B) and Kawamata et al. [12] reported a gain in a much larger region spanning 17q21.2~q25.3).
Fig. 3.
Venn diagram of recurring copy number aberrations (CNAs) detected by single-nucleotide polymorphism arrays vs. molecular inversion probe arrays (SNP vs. MIP). Top: Comparison of recurring CNAs reported in supplemental data of previously published Affymetrix SNP 500 k arrays on childhood leukemia cohorts vs. those detected by MIP analysis. Bottom: Recurring CNAs in the SNP studies vs. the MIP platform overall. Note equivalent overlap between platforms in the two diagrams. Sources: The SNP(Nature) data are from Mullighan et al., 2007 [10], published in Nature; the SNP(Leuk) data are from Kuiper et al., 2007 [11], published in Leukemia; and the SNP(Blood) data are from Kawamata et al., 2008 [12], published in Blood. MIP (24 k) indicates MIP cancer panel data with 24,000 probes; SNP (500 k) indicates data from the three combined Affymetrix 500 k studies. Figures created using: Oliveros, JC. (2007) VENNY. An interactive tool comparing lists with Venn Diagrams. http://bioinfogp.cnb.csic.es/tools/venny/index.html.
The present MIP analysis validates these overlapping regions of CNAs in childhood leukemia reported by other genome-wide studies, in addition to describing new CNAs in cancer-related genes not previously identified. Given our targeted candidate gene approach and the potential gaps in coverage of the 500 k platforms, it is not surprising that we detected CNAs in regions that were not previously reported (and not covered by the 500 k platform). However, the concordance of similar findings between all three data sets of the identical Affymetrix 500 k SNP platforms is unexpectedly low (much less than 50%). Some of the differences in recurring CNAs between different platforms (or even among identical platforms) may be due to differences in analysis methods or even to differences in the pediatric patients included in each study. This highlights the need for larger studies with alternative methods (such as MIPs) to help complement current childhood leukemia studies and to validate the few loci found in common using the same platforms.
One example of sample selection affecting differences in results between microarray platforms may be the relatively large number of amplifications that we found in our pre-BALL study cohort, compared with previous genome-wide SNP studies. The increased average number of gains in our group of patients may be explained by the number of high-hyperdiploid samples included in our study (13% of our pre-B ALL samples). The inherently large number of gains in the high-hyperdiploid group of patients (average of 19 gains per sample) increases the overall average of pre-B ALL gains and thereby skews our results. As more subtypes are included in future analyses,we expect to find greater CNA overlap with previously published studies and overall decreased number of gains, with smaller confidence intervals. Again, this emphasizes the need for more studies with large patient sizes in order to determine the true rate and location of CNAs in childhood leukemia.
4.2. Deletion of CDKN2A
The CDKN2A locus at 9p21 encodes p16INK4A and p14ARF, which act as tumor suppressors. The p14ARF protein functions as a stabilizer of the tumor suppressor protein p53 and can sequester MDM1, a protein responsible for p53 degradation. The p16INK4A protein is a cyclin-dependent kinase inhibitor that works upstream of the retinoblastoma (RB) protein to control cell cycle arrest. Thus, the deletion or mutation of CDKN2A leads to the removal of proteins essential to controlling tumor suppression. The clinical significance of CDKN2A deletion in childhood leukemia remains both conflicting and controversial [13,16,25]. In fact, the current Children’s Oncology Group risk stratification system does not include 9p21 deletions (p16INK4A) when assigning treatment regimens to children with ALL. However, the frequent finding of CDKN2A deletions in repeated CNA studies of childhood leukemia argues in favor its importance. In 2008, Yang et al. [26] reported significantly shorter remission times in children with relapsed ALL and CDKN2A deletions at diagnosis, and Mullighan et al. [27] reported that nearly 30% of ALL patients acquired new CNAs in CDKN2A at relapse.
Homozygous and hemizygous deletions of 9p21 have been described in pediatric ALL, using a variety of methods. The reported prevalence of 9p21 deletion varies by ALL subtype (5–34% in precursor-B ALL and 60–80% in T-cell ALL), which is consistent with our MIP data (35% in precursor B-ALL and 100% in precursor T-cell ALL). The reported prevalence of hemizygous (6–13%) vs. homozygous (14–35%) 9p21 deletions in pediatric ALL is also consistent with our MIP data (18.5% hemizygous deletions vs. 40.7% homozygous deletions) (Supplemental Table 6). Most importantly, we discovered a unique pattern of 9p loss of heterozygosity deletions (hemizygous) encompassing the 9p21 deletion in the majority of precursor B-ALL samples but rarely seen in the precursorT-ALL samples. The clinical significance of this flanking 9pCNA, which we found in most of our precursor B-ALL samples with CDKN2A deletions, is unknown; however, the isolated deletion of CDKN2A with no other surrounding loss of heterozygosity in almost exclusively precursor T-ALL may reflect lineage-specific leukemogenesis, or perhaps the inherently poor treatment response of precursor T-ALL. Bungaro et al. [28] recently reported their finding of SMAD1 downregulation in CDKN2A homozygous deleted pre-B ALL cases, compared with nondeleted cases, which may also contribute to the clinical behavior of patients harboring CDKN2A loss in their leukemic blasts.
4.3. Recurring CNAs
The allelic quantitation of intragenic SNPs in our MIP cancer panel allowed for precise identification of very small regions of CNAs. The smallest recurring micro-scale copy number changes was just 270 bp in length and was defined by 3 sequential probes within chromosome 14q24.2, whereas the largest recurring CNA was 170 Mb in length and defined by 1,466 individual probes that encompassed all of chromosome 6 (i.e., 6p25.3~q27). Notably, nearly two thirds of recurring CNAs identified by the MIP cancer panel occurred in all three subtypes of leukemia: precursor B-cell, precursor T-cell, and AML. In 21 regions, the CNA was found in the ALL samples but not in the AML samples. There were no regions unique to AML alone in our analysis. This overlap of common recurring CNAs among different leukemia subtypes may reflect a common mechanism of leukemogenesis between both lymphoblastic and myeloid leukemias. Furthermore, these common CNAs may explain the efficacy of some of the same therapeutic agents used to treat both types of leukemia. Finally, common CNAs found in both ALL and AML may represent potential shared targets for future drug development. These findings of recurring CNAs will need to be replicated in larger studies.
4.4. Clinical associations
Four clinical characteristics in our analysis were significantly associated with CNAs in specific genes. The clinical features associated with CNAs in our sample set included age from 1 to 10 years old versus ≥ 10 years old (part of the U.S. National Cancer Institute [NCI]/ Rome risk stratification criteria), leukemia subtype, relapse status, and central nervous system disease. Among these statistically significant clinical correlations, children ≥ 10 years of age (i.e., children in the higher risk NCI/Rome group) were more likely to possess a homozygous CDKN2A deletion in our study cohort. Patients with precursor T-cell ALL are often older, are more likely to have the same homozygous CDKN2A deletion, and require more intensive chemotherapy to achieve remission. However, our findings of increasedCDKN2Adeletions in older ALL patients cannot be explained by subtype alone, because 3 of these 5 older children (60%) in our cohort had precursor B-cell leukemia. Perhaps the increased prevalence of homozygous CDKN2A deletions that we observed in older children relates to its known increase in the traditionally older precursor T-cell ALL patients, and the older age in both these groups somehow contributes to their increased risk of treatment failure. This association may help to explain some of the biological connections among age, phenotype, and outcome. A much larger study reported the same significant association between CDKN2A deletions in children ≥10 years old with precursor B-cell ALL from a cohort of 864 children [29]. Further investigation may offer more biological explanations and so help to determine the currently disputed prognostic significance of 9p21 deletions in childhood ALL.
Not surprisingly, we also found a statistically significant association between this same homozygous CDKN2A deletion and leukemia subtype (when not accounting for age). As previously discussed, 9p21 deletions have been described for many years as associated with precursor T-cell ALL. In our study cohort, 100% of the precursor T-ALL samples had CDKN2A deletions.
5. Conclusions
Here we have reported initial use of novel MIP technology in a cohort of 45 pediatric leukemia patients, which identified unique patterns of 9 p loss of heterozygosity in precursor B-cell versus precursor T-cell ALL samples. Findings were validated using reported clinical cytogenetics for each sample and RT-PCR on CDKN2A and PAX5 deletions (two of the most common deletions in ALL). Both novel CNAs and previously reported CNAs were detected, and equal overlap was demonstrated in recurring loci between MIPs and other SNP 500 k microarray platforms. The MIP analysis also detected specific regions of amplification or deletion with significant clinical correlations, including an increase in CDKN2A deletions in older children. The present application of MIP technology is based on a small number of leukemia samples and will need to be replicated on a larger sample size. We are currently expanding our analysis to include other leukemia cohorts and are adding more loci to the MIP cancer panel.
Supplementary Material
Acknowledgments
We thank Athena Cherry and Charles Bangs in the Department of Pathology and Cytogenetics at Stanford University School of Medicine for their contribution of the clinical cytogenetics results.
This work was supported in part by theWalter V. and Idun Berry Fellowship Foundation (J.D.S.), the Harriet H. Samuelsson Foundation (J.D.S.), the Children’s Fund/Lucile Packard Foundation for Children’s Health (J.D.S.), the American Society of Hematology Scholar Awards Program (J.D.S.), the CureSearch Research Fellowship Award Program (J.D.S.), the For Julie Foundation (N.J.L.), and National Institutes of Health grants CA96879 (H.P.J.) and 2P01HG000205 (R.W.D., H.P.J., K.W.).
Footnotes
Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.cancergencyto.2009.03.005.
References
- 1.Ries LAG, Smith MA, Gurney JG, Linet M, Tamra T, Young JL, Bunin GR, editors. Cancer incidence and survival among children and adolescents: United States SEER Program 1975–1995. Bethesda, MD: National Institutes of Health; 1999. [Google Scholar]
- 2.Pui CH, Evans WE. Treatment of acute lymphoblastic leukemia. N Engl J Med. 2006;354:166–178. doi: 10.1056/NEJMra052603. [DOI] [PubMed] [Google Scholar]
- 3.Schultz KR, Pullen DJ, Sather HN, Shuster JJ, Devidas M, Borowitz MJ, Carroll AJ, Heerema NA, Rubnitz JE, Loh ML, Raetz EA, Winick NJ, Hunger SP, Carroll WL, Gaynon PS, Camitta BM. Risk- and response-based classification of childhood B-precursor acute lymphoblastic leukemia: a combined analysis of prognostic markers from the Pediatric Oncology Group (POG) and Children’s Cancer Group (CCG) Blood. 2007;109:926–935. doi: 10.1182/blood-2006-01-024729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lange BJ, Smith FO, Feusner J, Barnard DR, Dinndorf P, Feig S, Heerema NA, Arndt C, Arceci RJ, Seibel N, Weiman M, Dusenbery K, Shannon K, Luna-Fineman S, Gerbing RB, Alonzo TA. Outcomes in CCG-2961, a Children’s Oncology Group Phase 3 Trial for untreated pediatric acute myeloid leukemia: a report from the Children’s Oncology Group. Blood. 2008;111:1044–1053. doi: 10.1182/blood-2007-04-084293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaynon PS. Childhood acute lymphoblastic leukaemia and relapse. Br J Haematol. 2005;131:579–587. doi: 10.1111/j.1365-2141.2005.05773.x. [DOI] [PubMed] [Google Scholar]
- 6.Meshinchi S, Arceci RJ. Prognostic factors and risk-based therapy in pediatric acute myeloid leukemia. Oncologist. 2007;12:341–355. doi: 10.1634/theoncologist.12-3-341. [DOI] [PubMed] [Google Scholar]
- 7.Carroll WL, Bhojwani D, Min DJ, Moskowitz N, Raetz EA. Childhood acute lymphoblastic leukemia in the age of genomics. Pediatr Blood Cancer. 2006;46:570–578. doi: 10.1002/pbc.20722. [DOI] [PubMed] [Google Scholar]
- 8.Jarosová M, Holzerová M, Jedlicková K, Mihál V, Zuna J, Starý J, Pospísilová D, Zemanová Z, Trka J, Blazek J, Pikalová Z, Indrák K. Importance of using comparative genomic hybridization to improve detection of chromosomal changes in childhood acute lymphoblastic leukemia. Cancer Genet Cytogenet. 2000;123:114–122. doi: 10.1016/s0165-4608(00)00310-1. [DOI] [PubMed] [Google Scholar]
- 9.Strefford JC, Worley H, Barber K, Wright S, Stewart AR, Robinson HM, Bettney G, van Delft FW, Atherton MG, Davies T, Griffiths M, Hing S, Ross FM, Talley P, Saha V, Moorman AV, Harrison CJ. Genome complexity in acute lymphoblastic leukemia is revealed by array-based comparative genomic hybridization. Oncogene. 2007;26:4306–4318. doi: 10.1038/sj.onc.1210190. [DOI] [PubMed] [Google Scholar]
- 10.Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, Girtman K, Mathew S, Ma J, Pounds SB, Su X, Pui CH, Relling MV, Evans WE, Shurtleff SA, Downing JR. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 11.Kuiper RP, Schoenmakers EF, van Reijmersdal SV, Hehir-Kwa JY, van Kessel AG, van Leeuwen FN, Hoogerbrugge PM. High-resolution genomic profiling of childhood ALL reveals novel recurrent genetic lesions affecting pathways involved in lymphocyte differentiation and cell cycle progression. Leukemia. 2007;21:1258–1266. doi: 10.1038/sj.leu.2404691. [DOI] [PubMed] [Google Scholar]
- 12.Kawamata N, Ogawa S, Zimmermann M, Kato M, Sanada M, Hemminki K, Yamatomo G, Nannya Y, Koehler R, Flohr T, Miller CW, Harbott J, Ludwig WD, Stanulla M, Schrappe M, Bartram CR, Koeffler HP. Molecular allelokaryotyping of pediatric acute lymphoblastic leukemias by high-resolution single nucleotide polymorphism oligonucleotide genomic microarray. Blood. 2008;111:776–784. doi: 10.1182/blood-2007-05-088310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okuda T, Shurtleff SA, Valentine MB, Raimondi SC, Head DR, Behm F, Curcio-Brint AM, Liu Q, Pui CH, Sherr CJ. Frequent deletion of p16INK4a/MTS1 and p15INK4b/MTS2 in pediatric acute lymphoblastic leukemia. Blood. 1995;85:2321–2330. [PubMed] [Google Scholar]
- 14.Carter TL, Watt PM, Kumar R, Burton PR, Reaman GH, Sather HN, Baker DL, Kees UR. Hemizygous p16INK4A deletion in pediatric acute lymphoblastic leukemia predicts independent risk of relapse. Blood. 2001;97:572–574. doi: 10.1182/blood.v97.2.572. [DOI] [PubMed] [Google Scholar]
- 15.Heerema NA, Sather HN, Sensel MG, Liu-Mares W, Lange BJ, Bostrom BC, Nachman JB, Steinherz PG, Hutchinson R, Gaynon PS, Arthur DC, Uckun FM. Association of chromosome arm 9 p abnormalities with adverse risk in childhood acute lymphoblastic leukemia: a report from the Children’s Cancer Group. Blood. 1999;94:1537–1544. [PubMed] [Google Scholar]
- 16.van Zutven LJ, van Drunen E, de Bont JM, Wattel MM, Den Boer ML, Pieters R, Hagemeijer A, Slater RM, Beverloo HB. CDKN2 deletions have no prognostic value in childhood precursor-B acute lymphoblastic leukaemia. Leukemia. 2005;19:1281–1284. doi: 10.1038/sj.leu.2403769. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, Moorhead M, Karlin-Neumann G, Falkowski M, Chen C, Siddiqui F, Davis RW, Willis TD, Faham M. Allele quantification using molecular inversion probes (MIP) Nucleic Acids Res. 2005;33:e183. doi: 10.1093/nar/gni177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Moorhead M, Karlin-Neumann G, Wang NJ, Ireland J, Lin S, Chen C, Heiser LM, Chin K, Esserman L, Gray JW, Spellman PT, Faham M. Analysis of molecular inversion probe performance for allele copy number determination. Genome Biol. 2007;8:R246. doi: 10.1186/gb-2007-8-11-r246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hardenbol P, Banér J, Jain M, Nilsson M, Namsaraev EA, Karlin-Neumann GA, Fakhrai-Rad H, Ronaghi M, Willis TD, Landegren U, Davis RW. Multiplexed genotyping with sequence-tagged molecular inversion probes. Nat Biotechnol. 2003;21:673–678. doi: 10.1038/nbt821. [DOI] [PubMed] [Google Scholar]
- 20.Brown LA, Kalloger SE, Miller MA, Shih Ie M, McKinney SE, Santos JL, Swenerton K, Spellman PT, Gray J, Gilks CB, Huntsman DG. Amplification of 11q13 in ovarian carcinoma. Genes Chromosomes Cancer. 2008;47:481–489. doi: 10.1002/gcc.20549. [DOI] [PubMed] [Google Scholar]
- 21.Press JZ, De Luca A, Boyd N, Young S, Troussard A, Ridge Y, Kaurah P, Kalloger SE, Blood KA, Smith M, Spellman PT, Wang Y, Miller DM, Horsman D, Faham M, Gilks CB, Gray J, Huntsman DG. Ovarian carcinomas with genetic and epigenetic BRCA1 loss have distinct molecular abnormalities. BMC Cancer. 2008;8:17. doi: 10.1186/1471-2407-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ji H, Kumm J, Zhang M, Farnam K, Salari K, Faham M, Ford JM, Davis RW. Molecular inversion probe analysis of gene copy alterations reveals distinct categories of colorectal carcinoma. Cancer Res. 2006;66:7910–7919. doi: 10.1158/0008-5472.CAN-06-0595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 24.Grant GR, Liu J, Stoeckert CJ., Jr A practical false discovery rate approach to identifying patterns of differential expression in microarray data. Bioinformatics. 2005;21:2684–2690. doi: 10.1093/bioinformatics/bti407. [DOI] [PubMed] [Google Scholar]
- 25.Heyman M, Rasool O, Borgonovo Brandter L, Liu Y, Grander D, Soderhall S, Gustavsson G, Einhorn S. Prognostic importance of p15INK4B and p16INK4 gene inactivation in childhood acute lymphocytic leukemia. J Clin Oncol. 1996;14:1512–1520. doi: 10.1200/JCO.1996.14.5.1512. [DOI] [PubMed] [Google Scholar]
- 26.Yang JJ, Bhojwani D, Yang W, Cai X, Stocco G, Crews K, Wang J, Morrison D, Devidas M, Hunger SP, Willman CL, Raetz EA, Pui CH, Evans WE, Relling MV, Carroll WL. Genome-wide copy number profiling reveals molecular evolution from diagnosis to relapse in childhood acute lymphoblastic leukemia. Blood. 2008;112:4178–4183. doi: 10.1182/blood-2008-06-165027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mullighan CG, Phillips LA, Su X, Ma J, Miller CB, Shurtleff SA, Downing JR. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science. 2008;322:1377–1380. doi: 10.1126/science.1164266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bungaro S, Dell’Orto MC, Zangrando A, Basso D, Gorletta T, Lo Nigro L, Leszl A, Young BD, Basso G, Bicciato S, Biondi A, te Kronnie G, Cazzaniga G. Integration of genomic and gene expression data of childhood ALL without known aberrations identifies subgroups with specific genetic hallmarks. Genes Chromosomes Cancer. 2009;48:22–38. doi: 10.1002/gcc.20616. [DOI] [PubMed] [Google Scholar]
- 29.Sulong S, Moorman AV, Irving JA, Strefford JC, Konn ZJ, Case MC, Minto L, Barber KE, Parker H, Wright SL, Stewart AR, Bailey S, Bown NP, Hall AG, Harrison CJ. A comprehensive analysis of the CDKN2A gene in childhood acute lymphoblastic leukemia reveals genomic deletion, copy number neutral loss of heterozygosity, and association with specific cytogenetic subgroups. Blood. 2009;113:100–107. doi: 10.1182/blood-2008-07-166801. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.