Abstract
Junctional Adhesion Molecule A (JAM-A) is a multifunctional cell surface protein that has multiple evolutionarily conserved structural features. There is now conclusive evidence that discrete structural elements on JAM-A mediate intracellular signaling events that alter cell migration and paracellular permeability. Specifically, self-dimerization between extracellular Ig-like loops and close apposition of PDZ-dependent, JAM-A-associated intracellular scaffold proteins such as Afadin and guanine-nucleotide exchange factors mediate activation of Rap1 and modulation of epithelial cell migration by effects on β1 integrin. While the same JAM-A structural features also modulate migration of other cell types and paracellular permeability in epithelia/endothelia additional signaling proteins/mechanisms are likely involved. Recent insights into JAM-A outside-in signaling events that regulate these cellular functions are discussed.
II. Introduction
JAM-A, the founding member of a class of proteins termed Junctional Adhesion Molecules, is a cell-surface protein with a high degree of conservation in vertebrates. The consistent nature of the JAM-A sequence indicates that its structure has an important physiologic role. Indeed, JAM-A is known to modulate many cellular functions including migration, polarity, paracellular permeability and proliferation (1–15). This review will focus on relationships between JAM-A structure and intracellular signaling events that lead to the regulation of cell migration and paracellular permeability. For a more general review of the JAM protein family, the reader is referred to Mandel el al., Naik et al., and Weber et al. (16–18).
III. JAM-A Expression and Structure
JAM-A protein has a broad tissue distribution that is reflected by its expression in leukocytes, epithelia, endothelia and other cell types. In leukocytes, JAM-A is expressed on the cell surface where it redistributes in a polarized fashion during cell migration (19). In endothelia and epithelia, JAM-A localizes at cell-cell contacts with the tight junction proteins, occludin and claudins (7). By immunoelectron microscopic analyses of freeze fracture replicas, JAM-A does not appear to be directly incorporated into occludin and claudin –containing epithelial tight junction strands but localizes adjacent to them (20).
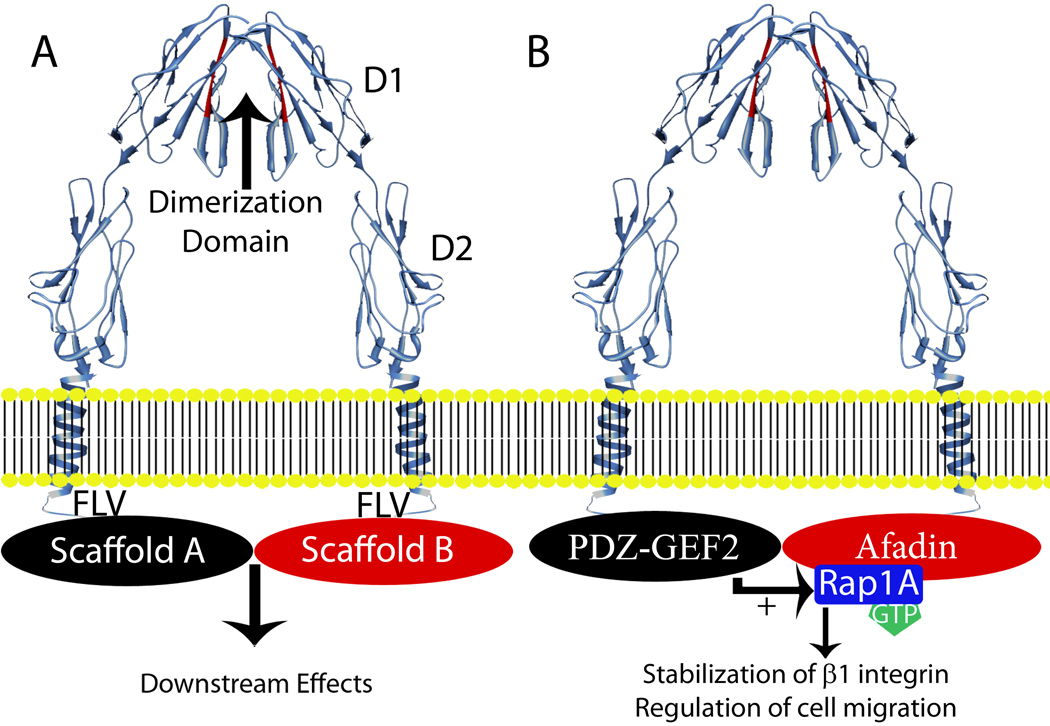
The extracellular domain of JAM-A contains a membrane distal variable immunoglobulin-like loop (D1) and a membrane proximal constant immunoglobulin-like loop (D2). The crystal structure of JAM-A predicts homodimerization through ionic and hydrophobic interactions in D1 (Figure 1A) (21). The short cytoplasmic domain of JAM-A terminates with a PSD-95/Discs-large/ZO-1 (PDZ) binding motif (Figure 1A). The PDZ binding motif of JAM-A facilitates intracellular interactions with scaffold proteins (22). These domains and motifs are conserved in all vertebrates where the sequence for JAM-A is known. Additionally, these domains are found in other JAM-related proteins such as the coxsackie and adenovirus receptor, the nectin-like protein family as well as JAM-B and JAM-C.
Figure 1.
A) Structure of JAM-A and general JAM-A signaling model. JAM-A (crystal structure in blue) has two extracellular Ig-like loops. The membrane distal domain, D1, is comprised of a variable immunoglobulin like loop (IgV) and mediates homodimerization through the motif colored in red. The membrane proximal domain, D2, is comprised of a constant immunoglobulin like loop (IgC). The cytoplasmic tail ends with a carboxy-terminal PDZ-binding motif (FLV) that binds to PDZ domain-containing scaffold proteins. Crystal structure information was reported by Prota et al. (21). The alpha helix depicted for the transmembrane domain is based on the protein sequence. Dimerization of JAM-A is mediated through binding interactions with D1 and brings into close apposition two PDZ-binding domains that facilitate the formation of PDZ containing scaffold protein complexes and subsequent signaling events.
B) Model of outside-in signaling through JAM-A that regulates epithelial cell migration. In this model, dimerized JAM-A brings into close apposition Afadin/Rap1A and PDZGEF2 to activate Rap1A. The active GTPase acts to stabilize cell surface β1 integrin protein and regulate cell migration.
IV. Outside-in signaling and JAM-A regulation of epithelial cell migration
The relationship between conserved JAM-A structural motifs and protein function is important and merits further discussion. A link between the PDZ binding motif of JAM-A and cell polarity has been explored. Specifically, JAM-A has been shown to associate with the Par3/Par6/aPKC complex through interactions with Par3 PDZ domains resulting in the localization of the Par3 complex at the tight junction in epithelial cells. The presence of a JAM-A PDZ binding motif is necessary for proper recruitment of Par3/Par6/aPKC to the tight junction and proper cell polarization secondary to aPKC signaling (6,20).
The extracellular domain of JAM-A contains another structural signaling element that is capable of interacting with other JAM-A molecules (homodimerization). Specifically, in the D1 domain there is a highly conserved motif that mediates homodimerization. Interference with homodimerization has been shown to result in a wide variety of functional effects in multiple cell types (1,7,23,24). However, the signaling events responsible for such functional deficits are poorly understood. Thus, it is useful to examine known signaling events from other junctional proteins.
A general model for signaling from another intercellular junction protein was recently proposed for the nectins. The nectins share structural similarities to JAMs and facilitate the formation of adherens junctions in both epithelial and endothelial cells (25). The cytoplasmic domain of nectin proteins contain a PDZ binding motif that interacts with the scaffolding protein Afadin and activates small G proteins. Such nectin-mediated G protein signaling has been reported to regulate actin polymerization through PI3 kinase and positively regulate cell migration in murine ES cells (26). Since nectins form extracellular homophilic binding interactions and also bind to cytoplasmic scaffolding proteins through PDZ binding motifs, they represent potential analogues for JAM-A signaling.
Investigation of JAM-A signaling has revealed importance of both JAM-A dimerization and PDZ binding motifs within the same signaling cascade. In epithelial cells, JAM-A signaling modulates cell migration in a β1 integrin-dependent manner. Important JAM-A signaling motifs have been identified through the overexpression of mutant proteins. Overexpression of JAM-A dimerization-defective mutants or JAM-A PDZ motif deletion mutants have a dominant negative effect resulting in decreased epithelial cell migration. Conversely, overexpression of full length JAM-A or mutants lacking both the dimerization and PDZ binding motifs does not alter cell migration (1). Decreased cell surface levels of β1 integrin protein after disruption of JAM-A function also correlates with diminished numbers of focal adhesions in both endothelial and epithelial cells (FAs) (1,27). Such JAM-A dependent effects on FAs likely account for changes in cell-matrix interactions and cell migration. These observations are consistent with a model in which JAM-A homodimerization on the cell surface results in close apposition of PDZ domain-bound scaffolding proteins to regulate β1 integrin protein levels and cell migration (Figure 1A) (1).
Scaffolding and signaling proteins downstream of JAM-A that regulate epithelial cell migration have recently been identified through siRNA-based forward genetic screening approaches. Reduced epithelial cell migration was observed after downregulation of JAM-A protein expression in epithelial cells. This reduction was secondary to decreased activity of the small GTPase Rap1 in concert with diminished β1 integrin protein levels (2). Interestingly, Rap1 consists of two paralogs, Rap1A and Rap1B and its activation by binding to GTP has been implicated in the conformational activation of β1 integrin protein in leukocytes and endothelial cells (28–30). In leukocytes, active Rap1 promotes cell migration and in endothelial cells it increases angiogenesis (28–30). However, in epithelial cells, activation of Rap1 promotes β1 integrin protein stability (24,28,29). The respective role of Rap1A versus Rap1B in leukocytes and endothelial cells is unclear from these studies. However, in epithelial cells, downregulation of Rap1A, but not Rap1B, results in decreased β1 integrin protein as well as altered cell migration similar to that observed after downregulation of JAM-A. Thus, JAM-A signaling and modulation of cell migration in epithelial cells appears dependent upon Rap1A activation (2).
There is no experimental evidence demonstrating that Rap1A directly interacts with JAM-A; however, this GTPase has been shown to interact with PDZ containing scaffolding proteins such as Afadin in endothelial cells (31). In epithelial cells, Afadin localizes to both tight and adherens junctions and associates with JAM-A (22,32). Furthermore, Afadin expression is necessary for JAM-A signaling, activation of Rap1A and promoting cell migration (2). Despite this association of Afadin with JAM-A-mediated signaling, Afadin has not been shown to directly activate Rap1A. Such activation has been shown to be dependent on Guanine Nucleotide Exchange Factors (GEFs). Epithelial cells express a GEF termed PDZ-GEF2 that has been shown to activate Rap1A and contains a PDZ domain capable of interacting with the cytoplasmic tail of JAM-A. Consistent with these observations, it was recently shown that JAM-A expression is necessary for co-immunoprecipitation of PDZ-GEF2 with Afadin. Furthermore, it has been observed that downregulation of PDZ-GEF2 has the same negative effects on β1 integrin protein levels and epithelial cell migration as does loss of JAM-A, Afadin or Rap1A (2). Together, the above findings suggest that cis-dimerization of JAM-A promotes close apposition of PDZ containing scaffold proteins including Afadin and PDZ-GEF2 that facilitates activation of signaling mediators such as Rap1A in order to modulate β1 integrin-dependent cell migration (Figure 1B).
V. JAM-A and migration of leukocytes and endothelial cells
JAM-A has been shown to regulate migration in a variety of cells other than epithelia. There is strong evidence for role of JAM-A in migration of endothelial cells and leukocytes. For example, growth factor-induced migration of endothelial cells and angiogenesis by bFGF are decreased in the absence of JAM-A (5,33–37). In addition, while there are no reports of a potential relationship between JAM-A and another endothelial growth factor VEGF, it is worth noting that treatment of endothelial cells with antibodies to the closely related protein JAM-C results in decreased angiogenesis in the presence of high levels of VEGF (38).
JAM-A also plays a role in regulation of leukocyte transendothelial migration. In one of the early reports on JAM-A, it was shown that a JAM-A dimerization inhibiting antibody inhibited trans-endothelial migration of monocytes in vitro (7). Mechanisms for the inhibitory effects of certain JAM-A mAbs on leukocyte migration include direct blockage of adhesion receptor-ligand interactions and functional impairment due to alterations in intracellular signaling. Evidence exists for both of the above proposed mechanisms. A role for JAM-A as a leukocyte adhesion receptor has been highlighted in studies demonstrating that the D2 Ig domain of JAM-A can bind to the leukocyte integrin LFA-1 to modulate transendothelial migration. (39,40). However, these observations have not been universally reproduced in other studies where monoclonal and polyclonal JAM-A antibodies failed to demonstrate inhibition of leukocyte transepithlelial migration in vitro and a showed a lack of colocalization of endothelial JAM-A with LFA-1 during the transmigration process (41,42). Additional observations supporting both mechanisms arise from a recent report demonstrating cytokine-dependent cleavage of the extracellular region of JAM-A that resulted in reduced neutrophil transmigration (43). This finding raises several possibilities: first, JAM-A cleavage may simply remove receptor from the cell surface and thus alter receptor-ligand binding; second, soluble JAM-A released after cleavage may directly compete with LFA-1 for binding to intact JAM-A; third, cleaved receptor may fail to signal due to loss of dimerization domains; fourth, cleaved receptor may disrupt dimerization-mediated signaling through homophilic interactions with intact JAM-A. These possibilities remain to be experimentally defined. Clearly much work remains to be done on the role of JAM-A as an adhesion receptor for leukocytes.
As alluded to above, JAM-A dependent effects on leukocyte migration also appear to involve leukocyte intracellular signaling events. For example, JAM-A deficient neutrophils show diminished transmigration due to defective cell polarization (44). Consistent with this observation, there is evidence that JAM-A signaling through Par3/Par6/aPKC may determine neutrophil polarity responses necessary for transmigration (20). Additional evidence of JAM-A signaling as a key regulator of leukocyte migration comes from a recent study suggesting that JAM-A is necessary for internalization and recycling of β1 integrins during neutrophil uropod extension and transendothelial migration (19). Similar to observations in epithelial cells, JAM-A deficient neutrophils also have decreased levels of active Rap1 (19). While these findings point to similarities between JAM-A signaling in leukocytes and epithelial cells, other observations point to differences that are likely cell-type specific. In particular, Rap1 signaling in leukocytes has been reported to lead to altered trafficking of β1 integrin, while in epithelial cells Rap1 signaling appears to stabilize β1 integrin protein expression levels (1,19).
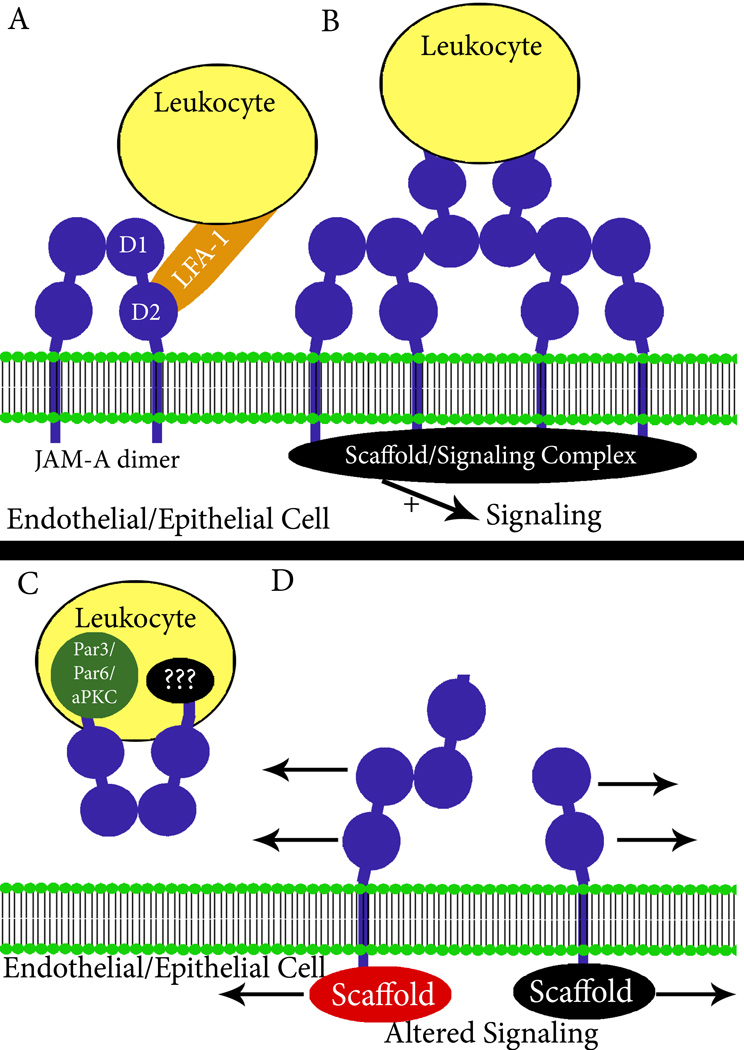
Despite the cell-type specificity of of these JAM-A-dependent functions in leukocytes, it is likely that cis and trans- mediated homodimerization plays a key role in downstream signaling. Based on results from studies with epithelial cells and structural reports indicating that JAM-A likely binds homotypically between cells in trans, one can envision potent transmigration-dependent signals that are generated as leukocytes migrate across intercellular junctions (2,45). Trans-interactions between leukocyte and endothelial or epithelial JAM-A would result in localized oligomerization and the formation of a larger scaffolding/signaling complex at the sites of cell-cell contacts, either between cells of the same type or between epithelial/endothelial cells and leukocytes (Figure 2B). In epithelia and endothelia, it is reasonable to assume that signaling events from such trans interactions between cells could also serve as a crucial regulator of both angiogenesis and restitution. Furthermore, signaling could be interrupted in vivo by any process that disrupts JAM-A dimerization, including receptor cleavage and generation of soluble JAM-A. Interestingly, it was recently reported that the extracellular domain of JAM-A is cleaved by ADAM proteases under inflammatory conditions in endothelial cells resulting in locally increased concentrations of soluble protein (43). Consistent with the model presented in figure 2, cleavage of JAM-A would be expected to alter signaling by removing the dimerization domain. Similarly, a functionally active soluble form of JAM-A could interfere with signaling by disrupting dimerization at cell-cell contacts. Figure 2 summarizes potential mechanisms of JAM-A regulation of leukocyte migration.
Figure 2.
Mechanisms of JAM-A mediated leukocyte transmigration. Potential mechanisms for the effects of JAM-A (blue) on leukocyte (yellow) migration include: A) adhesion receptor-ligand interactions, B) alterations in endothelial/epithelial intracellular signaling due to trans-interactions with leukocytes, C) alterations in JAM-A leukocyte signaling and cell polarity through Par3/Par6/aPKC (green), and D) interference with JAM-A signaling by soluble JAM-A (sJAM-A, in blue). Not shown is intracellular leukocyte signaling due to trans-interactions as highlighted in 2B for endothelial/epithelial cells.
VI. Potential mechanisms of JAM-A signaling that regulate paracellular permeability
It is well documented that JAM-A regulates paracellular permeability of endothelia and epithelia and the functional consequences of such would naturally include effects on leukocyte transmigration (8,10,15,23). Indeed, decreased epithelial JAM-A expression is noted in chronic intestinal inflammatory states such as ulcerative colitis and Crohn’s disease that are characterized by enhanced permeability and mucosal destruction due, in part, by massive leukocyte transmigration (15). However, unlike the abundant evidence supporting mechanisms JAM-A-mediated regulation of epithelial and leukocyte cell migration, the signaling mechanism(s) leading to JAM-A effects on permeability are not well understood. However, clues as to how JAM-A might influence permeability came from a recent study by Laukoetter et al. where it was shown that enhanced intestinal epithelial permeability after loss of JAM-A was associated with increased expression of claudins 10 and 15 both in vitro and in vivo (10). Claudin protein family members are localized to TJ strands (46) and have been shown to form ion-selective pores between epithelial and endothelial cells to regulate paracellular permeability (reviewed in (47,48)). Given that multiple claudins have been shown to be expressed in the same cell and that specific combinations of claudins can pair to form “tight” or “leaky” TJs, these observations provide a plausible mechanism for the observed effects of JAM-A on permeability to small molecules and ions (8,15,49). However, alterations in claudin expression may not account for enhanced permeability to larger solutes and proteins (10–50kD) observed after loss of JAM-A and this requires further investigation (8,50).
The signaling mechanisms by which JAM-A regulates claudin expression and cell permeability are not understood but appear dependent on JAM-A dimerization. In epithelia, overexpression of a JAM-A dimerization-defective mutant or treatment of mice with a dimerization inhibiting JAM-A antibody has been shown to result in enhanced paracellular permeability (8,15). Based on the model presented in figure 1B and likelihood of trans interactions between JAM-A expressing epithelial/endothelial cells analogous to that depicted with leukocytes in figure 2B, it is tempting to speculate that closely apposed scaffolding proteins similarly activate signaling cascades to regulate claudin protein expression and permeability. In support of this, we have observed in unpublished studies that siRNA-mediated downregulation of epithelial Afadin expression in vitro results in enhanced epithelial permeability and altered claudin expression similar to that observed with loss of JAM-A. Thus, with the models presented in figure 1 and figure 2, it is possible to envision how JAM-JAM interactions not only regulate permeability, but how such interactions between leukocytes and endothelial/epithelial cells might result in transient and focal signaling events to facilitate transmigration through alterations in paracellular permeability.
VII. Conclusions
JAM-A signaling regulates a diverse set of complex cellular functions that include migration and paracellular permeability. Multiple regions of the JAM-A protein are highly conserved throughout vertebrate evolution, including dimerization and PDZ binding motifs, indicating their importance in the function of JAM-A. A model of JAMA function has been proposed that highlights critical dependence on these regions of JAM-A for recruitment of scaffold proteins that signal through the small G protein Rap1 to stabilize β1 integrin protein and promote cell migration (Figure 1B).
It is likely that additional JAM-A signaling pathways will be uncovered that are cell-type or function-specific, although there are likely similar elements. A better understanding of JAM-A signaling may allow for the development of specific therapeutics that target the dimerization domain which is accessible from the luminal aspect of the bloodstream and gastrointestinal tract. Small molecules that can manipulate dimerization might thus be exploited to inhibit pathologic inflammation or cell migration associated with tumor progression.
Acknowledgements
We thank Manirath Khounlotham, Eric Peatman, Christopher Capaldo and Asma Nusrat for useful comments. This study was supported by National Institutes of Health grants R01-DK61379 and R01-DK079392.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Outstanding Interest:
(2) Severson et al. This report connects the function of dimerized JAM-A to interactions with the PDZ domain containing scaffold proteins Afadin and PDZ-GEF2. Data is shown to support how such interactions promote activation of the small G protein Rap1A and stabilize cell surface levels of β1 integrin that ultimately modulates epithelial cell migration.
(43) Koenen et al. This study demomnstrates that the extracellular domain of JAM-A is cleaved under inflammatory conditions. It is shown that cleaved JAM-A released under inflammatory conditions can decrease neutrophil transmigration.,
(10) Laukoetter et al. This is the first study demonstrating that JAM-A deficient mice have increased intestinal epithelial permeability and enhanced susceptibility to experimentally induced colitis. It was shown that the enhanced permeability associated with JAM-A deficiency correlates with altered epithelial claudin expression.
Special Interest:
(21) Prota et al. This study reported the human crystal structure of the extracelluar domain of JAM-A provided a structural basis for the further study of JAM-A function. The structural information provided predicted cis-homodimerization of JAM-A.
(15) Vetrano et al. This study reported enhanced permeability in the intestinal mucosa of JAM-A knockout mice that is independent of vascular endothelial cells. In addition, it was shown that treatment of wild type mice with a JAM-A dimerization blocking antibody also results in enhanced intestinal mucosal permeability.
(19) Cera et al. This study compared neutrophils derived from JAM-A knockout and control mice and showed that JAM-A null neutrophils have impaired uropod retraction and β1 integrin internalization. Additionally, it was shown that Rap1 activation was decreased in JAM-A null neutrophils.
References
- 1.Severson EA, Jiang L, Ivanov AI, Mandell KJ, Nusrat A, Parkos CA. Mol Biol Cell. 2008;19:1862–1872. doi: 10.1091/mbc.E07-09-0869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Severson EA, Lee WY, Capaldo CT, Nusrat A, Parkos CA. Mol Biol Cell. 2009 doi: 10.1091/mbc.E08-10-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Naik MU, Naik TU, Suckow AT, Duncan MK, Naik UP. Cancer Res. 2008;68:2194–2203. doi: 10.1158/0008-5472.CAN-07-3057. [DOI] [PubMed] [Google Scholar]
- 4.Naik MU, Naik UP. J Cell Sci. 2006;119:490–499. doi: 10.1242/jcs.02771. [DOI] [PubMed] [Google Scholar]
- 5.Naik MU, Vuppalanchi D, Naik UP. Arterioscler Thromb Vasc Biol. 2003;23:2165–2171. doi: 10.1161/01.ATV.0000093982.84451.87. [DOI] [PubMed] [Google Scholar]
- 6.Ebnet K, Suzuki A, Horikoshi Y, Hirose T, Meyer Zu Brickwedde MK, Ohno S, Vestweber D. Embo J. 2001;20:3738–3748. doi: 10.1093/emboj/20.14.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin-Padura I, Lostaglio S, Schneemann M, Williams L, Romano M, Fruscella P, Panzeri C, Stoppacciaro A, Ruco L, Villa A, Simmons D, Dejana E. J Cell Biol. 1998;142:117–127. doi: 10.1083/jcb.142.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mandell KJ, Babbin BA, Nusrat A, Parkos CA. J Biol Chem. 2005;280:11665–11674. doi: 10.1074/jbc.M412650200. [DOI] [PubMed] [Google Scholar]
- 9.Ozaki H, Ishii K, Horiuchi H, Arai H, Kawamoto T, Okawa K, Iwamatsu A, Kita T. J Immunol. 1999;163:553–557. [PubMed] [Google Scholar]
- 10.Laukoetter MG, Nava P, Lee WY, Severson EA, Capaldo CT, Babbin BA, Williams IR, Koval M, Peatman E, Campbell JA, Dermody TS, Nusrat A, Parkos CA. J Exp Med. 2007;204:3067–3076. doi: 10.1084/jem.20071416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooke VG, Naik MU, Naik UP. Arterioscler Thromb Vasc Biol. 2006;26:2005–2011. doi: 10.1161/01.ATV.0000234923.79173.99. [DOI] [PubMed] [Google Scholar]
- 12.Parris JJ, Cooke VG, Skarnes WC, Duncan MK, Naik UP. Dev Dyn. 2005;233:1517–1524. doi: 10.1002/dvdy.20481. [DOI] [PubMed] [Google Scholar]
- 13.Ong KL, Leung RY, Wong LY, Cherny SS, Sham PC, Lam TH, Lam KS, Cheung BM. J Intern Med. 2008;263:322–332. doi: 10.1111/j.1365-2796.2007.01886.x. [DOI] [PubMed] [Google Scholar]
- 14.Waki H, Liu B, Miyake M, Katahira K, Murphy D, Kasparov S, Paton JF. Hypertension. 2007;49:1321–1327. doi: 10.1161/HYPERTENSIONAHA.106.085589. [DOI] [PubMed] [Google Scholar]
- 15.Vetrano S, Rescigno M, Rosaria Cera M, Correale C, Rumio C, Doni A, Fantini M, Sturm A, Borroni E, Repici A, Locati M, Malesci A, Dejana E, Danese S. Gastroenterology. 2008;135:173–184. doi: 10.1053/j.gastro.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 16.Mandell KJ, Parkos CA. Adv Drug Deliv Rev. 2005;57:857–867. doi: 10.1016/j.addr.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 17.Naik TU, Naik MU, Naik UP. Front Biosci. 2008;13:258–262. doi: 10.2741/2676. [DOI] [PubMed] [Google Scholar]
- 18.Weber C, Fraemohs L, Dejana E. Nat Rev Immunol. 2007;7:467–477. doi: 10.1038/nri2096. [DOI] [PubMed] [Google Scholar]
- 19.Cera MR, Fabbri M, Molendini C, Corada M, Orsenigo F, Rehberg M, Reichel CA, Krombach F, Pardi R, Dejana E. J Cell Sci. 2009;122:268–277. doi: 10.1242/jcs.037127. [DOI] [PubMed] [Google Scholar]
- 20.Itoh M, Sasaki H, Furuse M, Ozaki H, Kita T, Tsukita S. J Cell Biol. 2001;154:491–497. doi: 10.1083/jcb.200103047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prota AE, Campbell JA, Schelling P, Forrest JC, Watson MJ, Peters TR, Aurrand-Lions M, Imhof BA, Dermody TS, Stehle T. Proc Natl Acad Sci U S A. 2003;100:5366–5371. doi: 10.1073/pnas.0937718100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ebnet K, Schulz CU, Meyer Zu Brickwedde MK, Pendl GG, Vestweber D. J Biol Chem. 2000;275:27979–27988. doi: 10.1074/jbc.M002363200. [DOI] [PubMed] [Google Scholar]
- 23.Liu Y, Nusrat A, Schnell FJ, Reaves TA, Walsh S, Pochet M, Parkos CA. J Cell Sci. 2000;113(Pt 13):2363–2374. doi: 10.1242/jcs.113.13.2363. [DOI] [PubMed] [Google Scholar]
- 24.Mandell KJ, McCall IC, Parkos CA. J Biol Chem. 2004;279:16254–16262. doi: 10.1074/jbc.M309483200. [DOI] [PubMed] [Google Scholar]
- 25.Rikitake Y, Takai Y. Cell Mol Life Sci. 2008;65:253–263. doi: 10.1007/s00018-007-7290-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyoshi J, Takai Y. Am J Nephrol. 2007;27:590–604. doi: 10.1159/000108103. [DOI] [PubMed] [Google Scholar]
- 27.Bazzoni G, Tonetti P, Manzi L, Cera MR, Balconi G, Dejana E. J Cell Sci. 2005;118:623–632. doi: 10.1242/jcs.01661. [DOI] [PubMed] [Google Scholar]
- 28.Arai A, Nosaka Y, Kanda E, Yamamoto K, Miyasaka N, Miura O. J Biol Chem. 2001;276:10453–10462. doi: 10.1074/jbc.M004627200. [DOI] [PubMed] [Google Scholar]
- 29.Carmona G, Gottig S, Orlandi A, Scheele J, Bauerle T, Jugold M, Kiessling F, Henschler R, Zeiher AM, Dimmeler S, Chavakis E. Blood. 2009;113:488–497. doi: 10.1182/blood-2008-02-138438. [DOI] [PubMed] [Google Scholar]
- 30.Enserink JM, Price LS, Methi T, Mahic M, Sonnenberg A, Bos JL, Tasken K. J Biol Chem. 2004;279:44889–44896. doi: 10.1074/jbc.M404599200. [DOI] [PubMed] [Google Scholar]
- 31.Linnemann T, Geyer M, Jaitner BK, Block C, Kalbitzer HR, Wittinghofer A, Herrmann C. J Biol Chem. 1999;274:13556–13562. doi: 10.1074/jbc.274.19.13556. [DOI] [PubMed] [Google Scholar]
- 32.Yamamoto T, Harada N, Kano K, Taya S, Canaani E, Matsuura Y, Mizoguchi A, Ide C, Kaibuchi K. J Cell Biol. 1997;139:785–795. doi: 10.1083/jcb.139.3.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naik MU, Mousa SA, Parkos CA, Naik UP. Blood. 2003;102:2108–2114. doi: 10.1182/blood-2003-04-1114. [DOI] [PubMed] [Google Scholar]
- 34.Vlodavsky I, Fuks Z, Ishai-Michaeli R, Bashkin P, Levi E, Korner G, Bar-Shavit R, Klagsbrun M. J Cell Biochem. 1991;45:167–176. doi: 10.1002/jcb.240450208. [DOI] [PubMed] [Google Scholar]
- 35.Sato Y, Hamanaka R, Ono J, Kuwano M, Rifkin DB, Takaki R. Biochem Biophys Res Commun. 1991;174:1260–1266. doi: 10.1016/0006-291x(91)91557-s. [DOI] [PubMed] [Google Scholar]
- 36.Breen EC. J Cell Biochem. 2007;102:1358–1367. doi: 10.1002/jcb.21579. [DOI] [PubMed] [Google Scholar]
- 37.Banerjee S, Mehta S, Haque I, Sengupta K, Dhar K, Kambhampati S, Van Veldhuizen PJ, Banerjee SK. Biochemistry. 2008;47:3345–3351. doi: 10.1021/bi8000352. [DOI] [PubMed] [Google Scholar]
- 38.Lamagna C, Hodivala-Dilke KM, Imhof BA, Aurrand-Lions M. Cancer Res. 2005;65:5703–5710. doi: 10.1158/0008-5472.CAN-04-4012. [DOI] [PubMed] [Google Scholar]
- 39.Fraemohs L, Koenen RR, Ostermann G, Heinemann B, Weber C. J Immunol. 2004;173:6259–6264. doi: 10.4049/jimmunol.173.10.6259. [DOI] [PubMed] [Google Scholar]
- 40.Wojcikiewicz EP, Koenen RR, Fraemohs L, Minkiewicz J, Azad H, Weber C, Moy VT. Biophys J. 2008 doi: 10.1529/biophysj.108.135491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shaw SK, Perkins BN, Lim YC, Liu Y, Nusrat A, Schnell FJ, Parkos CA, Luscinskas FW. Am J Pathol. 2001;159:2281–2291. doi: 10.1016/s0002-9440(10)63078-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shaw SK, Ma S, Kim MB, Rao RM, Hartman CU, Froio RM, Yang L, Jones T, Liu Y, Nusrat A, Parkos CA, Luscinskas FW. J Exp Med. 2004;200:1571–1580. doi: 10.1084/jem.20040965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koenen RR, Pruessmeyer J, Soehnlein O, Fraemohs L, Zernecke A, Schwarz N, Reiss K, Sarabi A, Lindbom L, Hackeng TM, Weber C, Ludwig A. Blood. 2009;113:4799–4809. doi: 10.1182/blood-2008-04-152330. [DOI] [PubMed] [Google Scholar]
- 44.Cera MR, Del Prete A, Vecchi A, Corada M, Martin-Padura I, Motoike T, Tonetti P, Bazzoni G, Vermi W, Gentili F, Bernasconi S, Sato TN, Mantovani A, Dejana E. J Clin Invest. 2004;114:729–738. doi: 10.1172/JCI21231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kostrewa D, Brockhaus M, D'Arcy A, Dale GE, Nelboeck P, Schmid G, Mueller F, Bazzoni G, Dejana E, Bartfai T, Winkler FK, Hennig M. Embo J. 2001;20:4391–4398. doi: 10.1093/emboj/20.16.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Furuse M, Sasaki H, Fujimoto K, Tsukita S. J Cell Biol. 1998;143:391–401. doi: 10.1083/jcb.143.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koval M. Cell Commun Adhes. 2006;13:127–138. doi: 10.1080/15419060600726209. [DOI] [PubMed] [Google Scholar]
- 48.Van Itallie CM, Anderson JM. Annu Rev Physiol. 2006;68:403–429. doi: 10.1146/annurev.physiol.68.040104.131404. [DOI] [PubMed] [Google Scholar]
- 49.Bruewer M, Hopkins AM, Hobert ME, Nusrat A, Madara JL. Am J Physiol Cell Physiol. 2004;287:C327–C335. doi: 10.1152/ajpcell.00087.2004. [DOI] [PubMed] [Google Scholar]
- 50.Mandell KJ, Holley GP, Parkos CA, Edelhauser HF. Invest Ophthalmol Vis Sci. 2006;47:2408–2416. doi: 10.1167/iovs.05-0745. [DOI] [PubMed] [Google Scholar]