Abstract
The functional consequences of missense variants are often difficult to predict. This becomes especially relevant when DNA sequence changes are used to determine a diagnosis or prognosis. To analyze the consequences of twelve missense variants in patients with mild forms of ataxia-telangiectasia (A-T), we employed site-directed mutagenesis of ATM cDNA followed by stable transfections into a single A-T cell line to isolate the effects of each allele on the cellular phenotype. After induction of the transfected cells with CdCl2, we monitored for successful ATM transcription and subsequently assessed: 1) intracellular ATM protein levels, 2) ionizing radiation (IR)-induced ATM kinase activity, and 3) cellular radiosensitivity. We then calculated SIFT and PolyPhen scores for the missense changes. Nine variants produced little or no correction of the A-T cellular phenotype and were interpreted to be ATM mutations; SIFT/PolyPhen scores supported this. Three variants corrected the cellular phenotype, suggesting that they represented benign variants or polymorphisms. SIFT and PolyPhen scores supported the functional analyses for one of these variants (c.1709T>C); the other two were predicted to be “not tolerated” (c.6188G>A, and c.6325T>G) and were classified as “operationally neutral”. Genotype/phenotype relationships were compared: three deleterious missense variants were associated with an increased risk of cancer (c.6679C>T, c.7271T>G and c.8494C>T). In situ mutagenesis represents an effective experimental approach for distinguishing deleterious missense mutations from benign or “operationally neutral” missense variants.
Keywords: missense mutations, mutagenesis, ATM, cancer risk
INTRODUCTION
With the growing awareness that a large gene can have hundreds of potential single nucleotide polymorphisms (SNPs) and that ~10% of these will be nonsynonymous missense variants, classifying them into deleterious or benign (or at least “operationally neutral”) is important -- especially when DNA sequencing is part of a diagnostic process. In these studies, we have used DNA variants in the ATM gene as a target of opportunity for comparing computational models to functional studies of stable transfectants.
Loss of ATM function causes the early-onset autosomal recessive disorder, ataxia-telangiectasia (A-T) (MIM# 208900), associated with cerebellar degeneration, hypersensitivity to ionizing radiation (IR), genomic instability, immunodeficiency, and cancer susceptibility (Gatti et al., 1991; Gatti et al., 2001; Perlman et al., 2003). Heterozygotes are also at an increased risk of cancer (Swift et al., 1991; Savitsky et al., 1995; Vorechovsky et al., 1996; Concannon and Gatti, 1997; Gatti et al., 1999; Izatt et al., 1999; Chenevix-Trench et al., 2002; Concannon, 2002; Sommer et al., 2003; Thorstenson et al., 2003; Tamimi et al., 2004; Olsen et al., 2005; Bernstein et al., 2006; Renwick et al., 2006) and have reduced levels of intracellular ATM protein (Chun et al., 2003). Following DNA damage or perturbations of chromatin, ATM autophosphorylates serine residues S367, S1893, and S1981, and activates numerous downstream targets, including p53, CHK1, CHK2, MDM2, BRCA1, NBS1, ATX/SMG1, NFKB, FANCD2, SMC1, RAD17, RAD9, and H2AX (Savitsky et al., 1995; Kim et al., 2002; Bakkenist and Kastan, 2003; Chun et al., 2003; Shiloh, 2003; Abraham, 2004; Ali et al., 2004; Kurz and Lees-Miller, 2004; Kozlov et al., 2006; Linding et al., 2007; Matsuoka et al., 2007). Through these cascading pathways, ATM serine/threonine kinase impacts upon cell cycle checkpoints, oxidative stress, transcription, nonsense mediated decay, apoptosis, and radiosensitivity (Lavin et al., 2006). In addition, recent studies show that ATM is necessary for efficient retroviral infection (Lau et al., 2005; Ariumi and Trono, 2006; Shin et al., 2006).
Approximately 90% of A-T patients are compound heterozygotes, carrying null mutations that result from either splicing aberrations, nonsense mutations, or small frameshifting insertions or deletions (McConville et al., 1996; Stankovic et al., 1998; Sandoval et al., 1999; Mitui et al., 2003; Mitsui et al., 2004; Babaei et al., 2005; Birrell et al., 2005; Cavalieri et al., 2006). In general, null mutations are associated with rapid progression of neurological symptoms and a severe phenotype. In contrast, milder phenotypes have been observed in patients carrying missense mutations, with small but detectable amounts of ATM protein (Gilad et al., 1998; Stankovic et al., 1998; Toyoshima et al., 1998; Sandoval et al., 1999; Becker-Catania et al., 2000; Stewart et al., 2001; Saviozzi et al., 2002). However, it is often difficult to distinguish deleterious missense mutations from benign nonsynonomous polymorphisms (Cooper et al., 2003; Greenblatt et al., 2003; Goldgar et al., 2004; Bao and Cui, 2005; Chan et al., 2007). This becomes clinically relevant when trying to identify ATM mutations in patients with mild symptoms.
Due to the large size of the ATM gene (62 coding exons, 3,056 aa), it has been difficult to manipulate in the laboratory and the instability of full-length cDNA constructs has been a major obstacle to performing ex vivo functional analyses. Two groups have successfully inserted full-length ATM cDNA into Epstein-Barr (EBV)-based vectors (Zhang et al., 1997; Ziv et al., 1997; Scott et al., 2002). Zhang et al. (1997) designated their construct pMAT1. We have used this system to introduce 12 missense changes, found in A-T patients with mild or late-onset symptoms, into pMAT1 and transfected each plasmid into an A-T lymphoblastoid cell line (LCL). After CdCl2 induction, transfected cells were monitored for: 1) ATM transcript, 2) ATM protein expression, 3) ATM kinase function, and 4) radiosensitivity, as a means of evaluating genotype/phenotype associations in these patients. These data also offered a unique opportunity to compare SIFT (sorting intolerant from tolerant) and PolyPhen scores for ATM against the functional and clinical data.
MATERIALS AND METHODS
Cell Culture
Patient blood samples and phenotypes were collected according to approved protocols. Peripheral blood lymphocytes were transformed by EBV. The LCL AT7LA (a.k.a. GM00717A) was used for most transfections; it was derived from an A-T patient with classic phenotype, carrying a homozygous c.1563_1564delAG mutation. It produces no detectable ATM protein by conventional immunoblotting (Chun et al., 2003). The cells were maintained in RPMI 1640 medium with 15% fetal bovine serum (Cyclone, Logan, UT) and 1% penicillin/streptomycin/glutamine (Invitrogen, Carlsbad, CA) in an atmosphere of 5% CO2 at 37°C.
Site-Directed Mutagenesis
To introduce various changes into the full-length ATM cDNA plasmid construct, pMAT1, we used QuikChange™ XL Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) according to the manufacturer’s protocol, with several modifications. Briefly, the PCR amplification mixture contained 80 ng plasmid DNA, 0.2 μM of each primer, 3.75 U PfuTurbo DNA polymerase, 1x reaction buffer, 1 μl of dNTP mix, and 3 μl of QuikSolution from the kit in a total volume of 50 μL. The PCR product was digested with DpnI (20 U/50 μl) for 3 hr at 37°C, ethanol precipitated, and the resuspended pellet was transformed in 45 μl of the XL10-Gold ultracompetent cells supplied with the kit. All constructs were in an EBV-based episomal vector under the control of a cadmium chloride-inducible (CdCl2) metallothionein promoter II (Scott et al., 2002).
Transfection of Human A-T Lymphoblastoid Cells
Ten million AT7LA cells were transfected with 15 μg of either pMAT1 or mutagenized expression construct, using electroporation (250 V, 1,180 μF) in a Cell-Porator (Invitrogen). After electroporation, cells were resuspended in 7 ml growth media and incubated at 37°C and 5% CO2. Selection of resistant cells was performed 72 hr after transfection with 0.2 mg/ml Hygromycin B (Roche Applied Science, Indianapolis, IN). Stably transfected cells were obtained within 3 to 4 weeks after transfection.
Real-Time PCR
The ATM mRNA levels (including mutated endogenous and transfected exogenous) were measured by real-time PCR based on TaqMan Gene Expression Assays (Applied Biosystems, Foster city, CA). The cDNA levels of glyceraldehydes 3-phosphate dehydrogenase (GAPDH) were used to normalize the ATM cDNA levels. Oligonucleotide primers and TaqMan probes for ATM and GAPDH were purchased from Applied Biosystems (ATM ID number: Hs00175892_m1 GAPDH: Hs99999905_m1). Reverse-transcription reactions were catalyzed by Superscript III reverse transcriptase (Invitrogen). PCR was performed in an ABI PRISM 7700 sequence detection system (Applied Biosystems) using an amplification protocol consisting of an initial denaturation and enzyme activation at 95°C for 10 min, followed by 40 cycles at 95°C for 15 sec and 60°C for 1 min. For each sample, two independent RNA extractions were analyzed, with each corresponding cDNA analyzed in duplicate on the same plate. Quantitative real-time PCR results of transcripts were expressed as ATM/GADPH ratios so that data could be combined from different experiments.
Western Immunoblotting
Cells were treated with 7 μM CdCl2 for 17 hr. Induced and uninduced cells were exposed to 2 Gy (for studies of ATM phosphoserine-S1981) and 10 Gy (for phosphorylation of SMC1-S957 and SMC1-S966) and lysed within an hour after irradiation. Nuclear lysates were electrophoresed on 7.5% SDS PAGE, transferred onto polyvinylidene difluoride (PVDF) membrane (BioRad, Hercules, CA), blocked with 10% milk, and incubated for 24 hr at 4°C with antibody to ATM (Novus Biologicals, Littleton, CO), ATM- phosphoS1981 (Rockland Immunochemicals, Gilbertsville, PA), SMC1-phosphoS957 (Novus Biologicals), or SMC1-phosphoS966 (Novus Biologicals). A rabbit-conjugated horseradish peroxidase (HRP) antibody was added at room temperature for 35 min. Enhanced chemiluminescence (ECL; Amersham Pharmacia, Piscataway, NJ) was used to detect the immunoreactive protein.
Colony Survival Assay
For the colony survival assay (CSA), 100 and 200 cells were plated per well in duplicate 96-well plates (Huo et al., 1994; Sun et al., 2002; Mitsui et al., 2004). The plates were irradiated with 1 Gy. After incubation for 10 to 13 days at 37°C, the surviving cells were stained with MTT dye (tetrazolium-based colorimetric assay), followed by another 2 to 4 hr incubation. The wells of each plate were then scored microscopically for formation of viable cell colonies (i.e., clumps of >32 cells), monitoring the colony-forming efficiency (CFE), and calculating the survival fraction (SF). A SF of <21% (13.1±7.2%) has been determined to be the radiosensitive range and >36% (50.1±13.5%) is considered as radionormal (Sun et al., 2002).
Calculation of SIFT and PolyPhen Scores
The effects of missense substitutions on ATM structure and function were evaluated using the programs PolyPhen (www.bork.embl-heidelberg.de/PolyPhen) (Sunyaev et al., 2001) and SIFT (http://blocks.fhcrc.org/sift/SIFT_seq_submit2.html) (Ng and Henikoff, 2001). PolyPhen was accessed through the web interface at http://genetics.bwh.harvard.edu/pph. SIFT scores were calculated using the precomputed BLAST results for ATM from NCBI. Results were filtered for the best hit for each taxon and a minimum hit score of 4,000. The resulting alignment included the sequences listed in Supplementary Table S1 (available online at http://www.interscience.wiley.com/jpages/1059-7794/suppmat).
ESEfinder Estimation of preRNA Binding Sites
The disruption or strengthening of splicing enhancer elements resulting in aberrant splicing has been proposed as an alternative mechanism underlying the molecular pathology of a number of deleterious exonic mutations (Cartegni et al., 2002; Eng et al., 2004). Exonic splicing enhancer (ESE) sequences serve as binding sites for serine/arginine-rich (SR) proteins, which in turn recruit essential components of the spliceosome, including snRNPs U1 and U2AF, processes necessary for splice-site recognition (Kan and Green, 1999; Blencowe, 2000; Graveley, 2000). Using functional Systematic Evolution of Ligands by Exponential enrichment (SELEX), Liu et al. (1998, 2000) identified short degenerate six to eight-nucleotide ESE binding motifs for four SR proteins. In addition, scoring matrices were computed from the frequencies at which a particular nucleotide is found at each position within a motif, facilitating the identification of theoretical ESEs (Liu et al., 1998, 2000; Cartegni et al., 2003). ESEfinder, an online tool utilizing these scoring matrices, is capable of identifying putative ESEs for the SR binding proteins SF2/ASF, SC35, SRp40, and SRp55 (http://rulai.cshl.edu/tools/ESE). The robustness of these predictive scoring matrices continues to be verified, as specific nucleotide variants that reduce motif scores to below threshold values are associated with aberrant splicing (Eng et al., 2004; Coutinho et al., 2005).
Maximun Entropy Scores for Estimation of Splice Site Strengths
In order to estimate the strengths of 5′ and 3′ splice junctions in the ATM gene, splice-site sequence motifs were scored using the splice site-models introduced by Yeo and Burge (2004) and the Maximum Entropy (MaxENT) software (available at: http://genes.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq_acc.html). Briefly, splice-site models that take into account adjacent and non-adjacent dependencies are built under the Maximum Entropy framework using large datasets of human splice-sites. These splice-site models assign a log-odd ratio (MaxENT score) to a 9-mer (5′ splice-site) or a 23-mer (3′ splice-site) sequence. The higher the score, the higher the probability that the sequence is a true splice-site. Also, it can be argued that given two sequences of differing scores, the higher scoring sequence has a higher likelihood of being used (Eng et al., 2004).
Mutation Nomenclature
Mutations were named based on cDNA reference sequence U82828, with 1+ being the A of the initiation start codon (Wildeman et al., 2008).
RESULTS
Selection of ATM Missense Variants
We selected 12 missense variants that were associated with atypical A-T phenotypes and attempted to evaluate the effect of each on the cellular phenotype of a transfected and transduced host A-T cell (AT7LA). This LCL, which was derived from a typical A-T patient carrying a homozygous truncating mutation (c.1563_1564delAG), expresses no detectable ATM protein by immunoblotting of nuclear lysates under conventional conditions. None of the 12 selected missense variants has been observed in an A-T patient with two other deleterious mutations, in healthy controls, or in breast cancer patients, except as noted in the text below.
By site-directed mutagenesis, we introduced each of the changes into a full-length ATM cDNA construct (pMAT1). To check for the integrity of the constructs, we performed BamH1 digests, as well as PCR amplification, of eight overlapping cDNA regions of the ATM gene, and compared the fragment sizes to the expected control pMAT1 fragments. Mutagenized plasmids were also sequenced around the region of the mutagenized site to confirm the presence of the intended cDNA change. Once confirmed, they were used to stably transfect AT7LA cells. Cells with successful integration of the intended construct were selected with Hygromicin B (Roche Applied Science). When sufficient cells were available, they were analyzed for the presence of ATM transcript, ATM protein, ATM functions, and radiosensitivity. Constructs that did not reconstitute ATM functions were re-sequenced across the entire gene to monitor for mutations acquired during the experiments. The data are considered in detail below and are summarized in Table 1.
Table 1.
Analysis of Transfected A-T Host Cells (AT7LA)
| Operational conclusion |
DNA changea | AA change | ATM proteinb |
ATM pS1981 |
SMC1 pS957/S966 |
RS (CSA)c | SIFTd | PolyPhene | Reference patientf | Phenotypeg | Relevant LCL (ATM prot/RS)c,h |
|---|---|---|---|---|---|---|---|---|---|---|---|
| M | c.875C>T | P292L | TD | ND | ND | Sensitive | 0.00 | 3 | AT211LA; AT143LAA | Mild at 20 y/o (AT143LA) | ND/Sensitive; TD/Intermediate |
| P | c.1709T>C | F570S | Normal | Normal | Normal | Normal | 0.40 | 0.371 | Sandoval et al. [1999] | Mild; lived until mid-30s | NA |
| M | c.3248A>G | H1083R | ND | NT | NT | Sensitive | 0.04 | 2.36 | AT83LA | Mild; walked unassisted until 30 y/o | TD/Intermediate |
| M | c.5858C>T | T1953I | TD | ND | ND | Intermediate | 0.00 | 2.05 | AT165LAA | Mild at 35 y/o | TD/Sensitive |
| M | c.6047A>G | D2016G | TD | ND | ND | Sensitive | 0.00 | 2.189 | TAT25H | Typical AT | TD/Sensitive |
| ON | c.6188G>A | G2063E | Normal | Normal | Normal | Normal | 0.00 | 2.274 | TAT41,47,63H | Walked unassisted until 14–22 y/o | TD/sensitive |
| ON | c.6325T>G | W2109G | Normal | Normal | Normal | Normal | 0.00 | 3.677 | AT165LAB | Mild at 35 y/o | TD/sensitive |
| M | c.6679C>T | R2227C | TD | ND | ND | Intermediate | 0.00 | 2.654 | WAR13; AT171LA | Typical AT; ↑cancer riski | TD/sensitive |
| M | c.7271T>G | V2424G | Normal | ND | ND | Sensitive | 0.00 | 2.33 | Stewart et al. [2001] | Mild AT; ↑cancer riski | Normal/NT |
| M | c.7967T>C | L2656P* | TD | ND | ND | Sensitive | 0.00 | 2.051 | Toyoshima et al. [1998] | Typical AT; no immunodeficiency | NA |
| M | c.8030A>G | Y2677C* | ND | NT | NT | Sensitive | 0.00 | 2.758 | Saviozzi et al. [2002] | Late onset; no telangiectasia or immunodeficiency | TD/NA |
| M | c.8494C>T | R2832C* | TD | ND | ND | Intermediate | 0.00 | 2.654 | AT143LAB | Mild at 20 y/o; ↑cancer riski | Red/Intermediate |
DNA numbering is based on cDNA sequence, with +1 corresponding to the A of the ATG translation initiation codon in the reference sequence U82828 (www.hgvs.org/mutnomen). The initiation codon is codon 1.
Detected by immunoblotting
Survival fraction (SF) % ranges: normal, >36%; intermediate, 21–36%; sensitive, <21%.
SIFT: bold score predicts that the variant will be “tolerated”; underlining indicates variance with other data.
PolyPhen: bold score predicts a benign variant; underlining indicates variance with other data.
Superscripts: A, first; B, second allele; H, homozygous.
See Supplementary Materials for clinical summary.
Relevant LCL column gives results of immunoblotting and radiosensitivity (CSA) of an LCL derived from the patient carrying that mutation, if available.
See Supplementary Materials for details.
Changes located in the ATM kinase domain.
ON, “operationally neutral” because functional studies were normal; M, mutation; P, benign polymorphism; ND, non-detectable; TD, trace detected; NT, not tested; NA, not available; RS, radiosensitivity; y/o, years old; Red, reduced.
Validation of ATM Mutagenesis/Transfection Model
To validate the use of site-directed mutagenesis for distinguishing polymorphisms from mutations in the ATM gene, we first tested constructs for two accepted benign variants, c.1744T>C (F582L) and c.2119T>C (S707P) (Johnson et al., 2007), and a known deleterious truncating mutation (c.5908C>T) (not included in Table 1). A-T cells transfected with the former two constructs yielded normal ATM protein levels, normal kinase activity, and corrected radiosensitivity (data not shown). When the same host cell (AT7LA) was transfected with the c.5908C>T construct, no changes in cellular phenotype were noted, i.e., ATM protein was undetectable by immunoblotting, and the cells remained radiosensitive (data not shown).
ATM Protein Expression
Immunoblotting data for ATM protein are shown in Figure 1 and summarized in Table 1 (column “ATM protein”). Untransfected AT7LA showed no detectable ATM protein. After transfection, followed by CdCl2 induction, the wild-type construct (pMAT1) showed a significant amount of ATM protein. This level of ATM was used as a reference “normal” level of ATM protein expression for comparing all other constructs with missense variants. Uninduced cells failed to show ATM protein expression.
Figure 1.
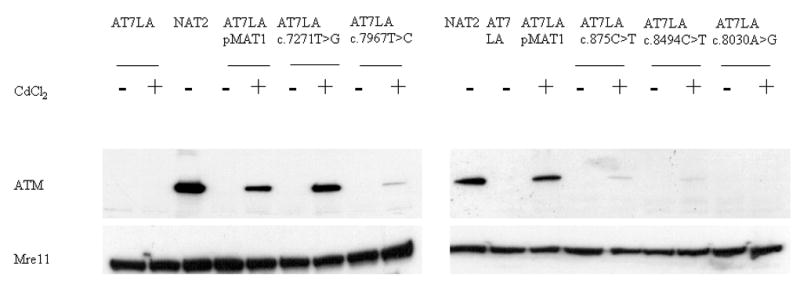
Immunoblots showing ATM expression in AT7LA cells transfected with normal pMAT1 or variants. Constructs were induced with CdCl2. Mre11 was used as a protein loading control. NAT2 is a normal LCL.
Of the 12 constructs tested, four showed levels of ATM protein comparable to AT7LA-pMAT1 (i.e., normal levels): c.1709T>C (F570S), c.6188G>A (G2063E), c.6325T>G (W2109G), and c.7271T>G (V2424G). We interpreted this as evidence that the constructs represented benign polymorphisms or at least “operationally neutral” variants, unless the resulting ATM protein in the transfected cells could be shown to be inactive (kinase-dead), which was the case for the c.7271T>G (V2424G) construct. Six constructs had reduced or trace-detectable amounts of ATM (TD in Table 1): c.875C>T (P292L), c.5858C>T (T1953I), c.6047A>G (D2016G), c.6679C>T (R2227C), c.7967T>C (L2656P), and c.8494C>T (R2832C). These most likely represent “mild” mutations, accounting for the associated mild clinical phenotypes (see Supplementary Materials). Two constructs, c.3248A>G (H1083R) and c.8030A>G (Y2677C), produced non-detectable (ND) levels of ATM (Figure 1; Table 1), suggesting that they were also disease-causing deleterious mutations.
Relative Levels of Induced ATM Transcripts
Expression of the integrated ATM transcripts was monitored for the eight constructs with trace or non-detectable ATM protein levels. Before induction with CdCl2, ATM transcripts were detectable at a basal level, representing endogenous mRNA (Figure 2A, first bar). At 2 hr and 4 hr post-induction, no significant increases of mRNA were observed, whereas at 8 hr and 17 hr post-induction, significant increases of mRNA transcription were detected (Figure 2A), indicating that the promoter was successfully induced by CdCl2. We subsequently chose to monitor ATM mRNA levels at 8 hr post-induction for the eight transfectants with trace or non-detectable ATM protein levels. The relative mRNA levels were normalized to AT7LA-pMAT1 before and after induction (Figure 2B, first and second bars). As compared with these controls, all eight transfectants expressed equal or higher levels of ATM following induction.
Figure 2.
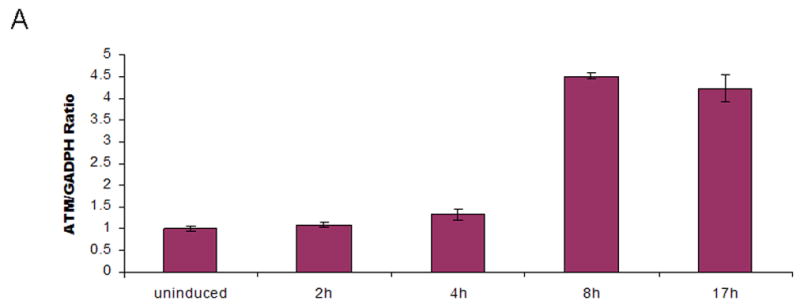
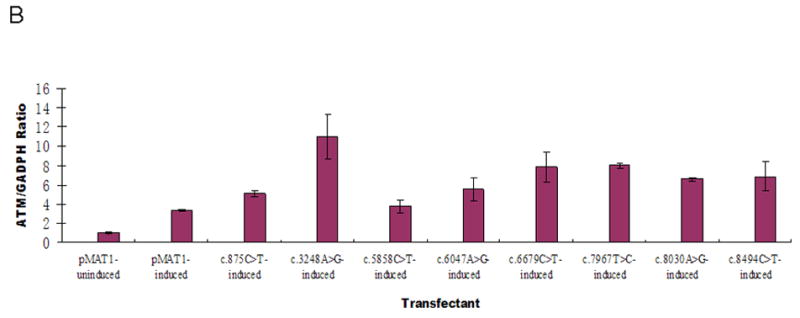
A. AT7LA cells transfected with pMAT1 were induced for 2, 4, 8, and 17h with CdCl2. Significant levels of ATM mRNA expression were detected at 8 and 17h post-induction.
B. ATM mRNA levels at 8h post-induction, detected by quantitative real time PCR. First bar on the left represents endogenous mRNA, without induction by CdCl2. Data were normalized to pMAT1-induced transcription levels (second bar).
ATM Kinase Activity
To test whether expressed ATM protein was functional, the autophosphorylation of ATM-S1981 and the phosphorylation of SMC1 (at S957 or S966) were determined by immunoblot analysis, 1 hr after the cells were irradiated with either 2 Gy or 10 Gy, respectively. Characteristic data are shown in Figures 3A and B; the remaining results are listed in Table 1. The control AT7LA cells transfected with wild-type construct (pMAT1) showed phosphorylation of ATM-S1981 (Figure 3A, lane 4) and SMC1-S966 (Figure 3B, lane 4) after induction of the transfected cells with CdCl2 for 17 hr followed by irradiation.
Figure 3.
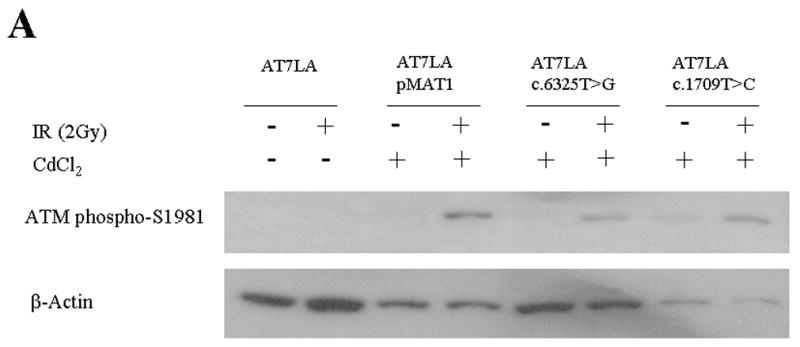
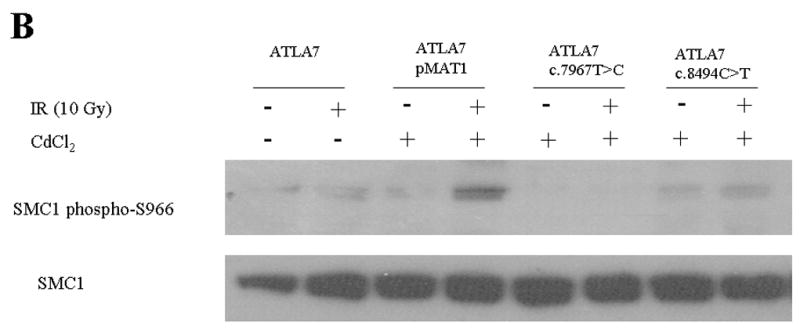
Immunoblot showing radiation-induced phosphorylation. A. ATM-S1981. Stably transfected cells were induced with CdCl2 and irradiated with 2 Gy. β-actin was used as a protein loading control. B. SMC1-S966. Stably transfected cells were induced with CdCl2 and irradiated with 10 Gy. Total SMC1 levels are shown in bottom panel, using an antibody that does not crossreact with phosphorylated SMC1.
Three of the constructs with normal ATM levels showed normal kinase activity for both ATM and SMC1 targets: c.1709T>C (F570S), c.6188G>A (G2063E), and c.6325T>G (W2109G) (Figure 3 and Table 1). The c.5858C>T (T1953I) and c.7271T>G (V2424G) constructs showed trace and normal ATM levels, respectively, but no detectable kinase activity, similar to the LCL derived from the c.7271T>G patient that was described by Stewart et al. (2001).
Colony Survival Assay
Figure 4A shows characteristic results for the radiosensitivity of AT7LA before and after transfection with the wild-type construct (pMAT1), followed by CdCl2 induction and exposure to different doses of IR: only the untransfected AT7LA remained radiosensitive (SF: <10% at 1.0, 1.5, and 2 Gy). The radiosensitivity of each transfectant is shown in Figure 4B and summarized in Table 1 (column RS). The CSA results were concordant with most other functional parameters measured and were consistent with an interpretation of either a deleterious mutation (M) or a variant (operationally neutral, ON; or benign polymorphism, P) (Table 1, first column), with the exception of transfectant c.7271T>G, which was radiosensitive, despite normal levels of ATM protein; however, the protein has little or no ATM kinase (Stewart, et al., 2001). For constructs c.5858C>T, c.6679C>T, and c.8494C>T, the cellular responses to 1.0 Gy were in the intermediate range (22%, 25%, and 22%, respectively). When exposed to 1.5 Gy or 2 Gy, such cells typically fall into the radiosensitive range (Sun et al., 2002).
Figure 4.
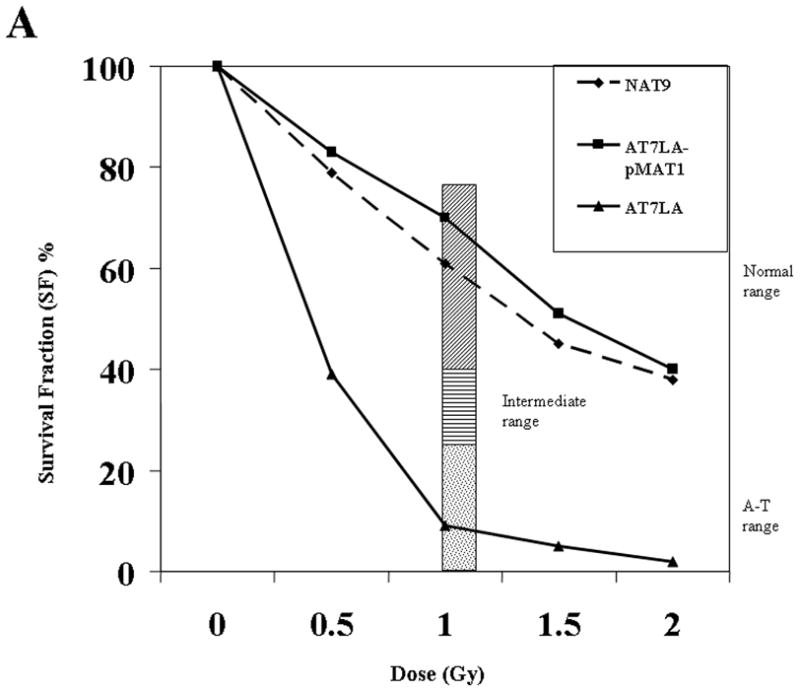
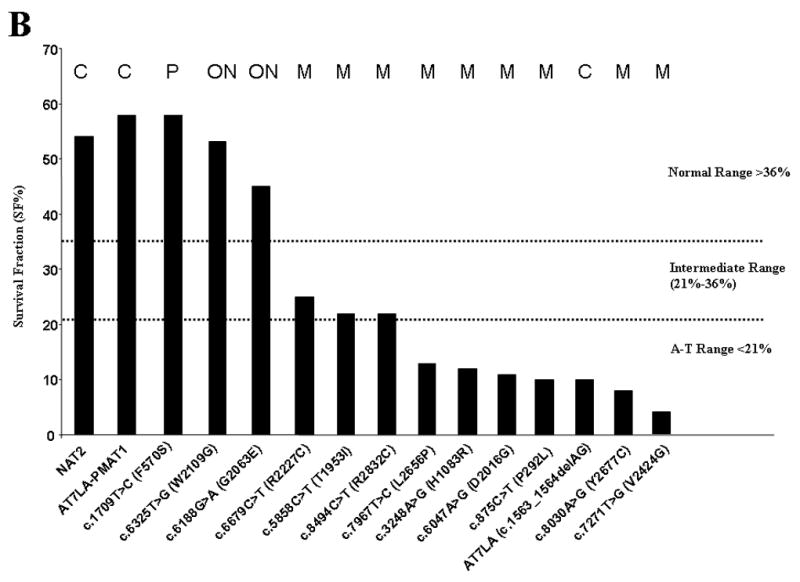
A. The radiosensitivity of AT7LA cells was abrogated by transfection with wild type construct, pMAT1. NAT9: normal; AT7LA: AT cell. Bar: speckled area denotes radiosensitivity range; diagonal-hatching denotes normal range. B. The radiosensitivity of each transfected LCL. Normal, intermediate and A-T ranges are indicated by dotted lines. NAT2 is a control LCL. C, control; M, mutation; ON, ‘operationally neutral’ variant; P, benign polymorphism.
SIFT and PolyPhen Scores
The SIFT and PolyPhen scores correlated with one another for all variants. However, only one variant was predicted to be “tolerated”, c.1709T>C (F570S), despite functional data suggesting that two others (c.6188G>A, and c.6325T>G) were also benign changes. The latter two were, therefore, classified as “operationally neutral” variants (ON in Table 1), until later studies can clarify the consequences of these DNA changes. A renewed effort was made to search for another allele on patients carrying these variants. No additional mutations could be identified. Taken together, the data suggest that these two missense variants (c.6188G>A and c.6325T>G) may lead to splicing errors that cannot be detected when the mutagenized, but correctly spliced, cDNA is inserted into the transfection constructs. Despite this, repeated efforts have failed to identify splicing defects in transcripts from patients carrying these variants (additional in silico analyses of the variants are presented in Supplementary Figures S1 and S2). Alternatively, it is possible that these variants affect functions of the ATM protein that have not yet been identified and therefore cannot be evaluated (see Discussion).
Genotype/Phenotype Comparisons
The cellular and clinical phenotypes for each allele (Table 1) were compared and the missense changes were further interpreted (see Supplementary Materials).
DISCUSSION
We used stable transfectants to evaluate the consequences of clinically relevant missense changes in the ATM gene. We tested constructs for 12 missense variants that had been observed in A-T patients with various atypical phenotypes. In aggregate, the variants categorized herein as mutations by functional studies comprise about one-fifth of all observed missense mutations in A-T patients. As compared to truncating mutations in the ATM gene, deleterious missense mutations appear to be associated with slower progression of neurological symptoms, intermediate ex vivo radiosensitivity responses, and “trace” (vs. “non-detectable”) ATM protein levels (Gilad et al., 1998; Stankovic et al., 1998; Taylor, 1998; Becker-Catania et al., 2000; Sun et al., 2002; Chun et al., 2003). However, unless a cell line is homozygous for a mutation, it is difficult to evaluate the true consequences of any single variant. Furthermore, genetic backgrounds and gene expression profiles differ from patient to patient, as well as from one affected sibling to another, introducing the potential for mutations in other genes to modify the phenotype. Site-directed mutagenesis of ATM cDNA with stable transfection into a single A-T host cell allows the functional consequences of individual variants to be isolated and analyzed on an identical genetic background.
Three constructs corrected the cellular phenotype of AT7LA (c.1709T>C, c.6188G>A, and c.6325T>G) and would be operationally interpreted as benign polymorphisms, based on only the functional studies. This alerted us to re-screen the ATM gene for a second deleterious mutation in families AT165LA, TAT41, TAT47, and TAT63; however, a second mutation was not identified. Since these patients had laboratory-confirmed diagnoses and each had been extensively screened for mutations by most available methods, including protein truncation test (PTT), single-stranded conformation polymorphism (SSCP), denaturing high performance liquid chromatography (dHPLC), multiplex ligation-dependent probe amplification (MLPA), and direct sequencing of cDNA for the entire coding region of the ATM gene (62 exons), it is likely that the missing mutations are hidden by poorly understood mechanisms of mutation, such as sites in the non-coding regions (including miRNA recognition sites), sites of post-translational modification (Vogt et al., 2005), or mitochondrial genes that affect the transfer of certain amino acids (Guan et al., 2006). It is also possible that the second mutation resides in other genes coding for upstream or downstream proteins that interact with ATM. Such candidate proteins include Protein Phosphatase 5 (PP5), recently described as essential to the deactivation of ATM at a site other than serine-1981 (Ali et al., 2004), or Tip60, which acetylates and activates ATM (Eymin et al., 2006). On the other hand, of more than 800 A-T patients genotyped worldwide (http://chromium.liacs.nl/lovd/), most patients have two identified ATM mutations; mutations in other genes have not been described. Nonetheless, this rare event remains a distinct possibility and patients such as TAT 41/47/63 and AT165LA are candidates for having mutations in modifier genes.
Alternatively, because the mutagenized transfected cDNA represents mature mRNA (i.e., it is already correctly spliced in the construct before it is transfected), a missense change that affects splicing would not score as deleterious. We were confronted by this possibility for variants c.6188G>A and c.6325T>G: both had SIFT and PolyPhen scores predicting that they were deleterious changes. (Technically, the PolyPhen actually predicts that these changes would likely damage the protein structure. However, the structure of the ATM protein has not yet been definitely determined). This prediction was inconsistent with the functional data showing that the mutagenized cDNA corrected ATM protein levels and function. Aberrant splicing products corresponding to these variants could not be identified. Additional in silico analyses suggested that c.6188G>A might disrupt an ASF/SF2 SR protein binding motif at the site of the mutation and an adjacent cryptic 5′ splice-site at the same site (see Supplementary Figure S1). Conversely, c.6325T>G creates a SRp55 binding site and a new cryptic 5′ splice site (see Supplementary Figure S2). Despite this, minigene constructs to test these models failed to provide supporting evidence for aberrant splicing (additional details are provided in Supplementary Materials). Once again, these variants were operationally categorized as “neutral” in the absence of evidence that the changes would be deleterious to ATM protein levels or the functions tested.
Variants c.875C>T (P292L) and c.8494C>T (R2832C) were identified in an A-T patient who walked unassisted at 20 years, despite mild ataxia. He maintained high scholastic grades at university level, despite dysarthria. Constructs carrying each of these changes induced expression of only trace levels of ATM protein, with no detectable in vivo p53 kinase function. The presence of low but detectable levels of ATM may partially explain the mild phenotype. The maternal allele, c.8494C>T (R2832C) has been associated with cancer in four A-T patients (all were compound heterozygotes) and with breast cancer in three of four mothers (obligate carriers); despite this, it has not been observed in cohort studies of breast cancer patients (Tamimi et al., 2004; Heikkinen et al., 2005; Olsen et al., 2005; Thompson et al., 2005; Bernstein et al., 2006; Johnson et al., 2007). Taken together, these data suggest that cancer risk and severity of neurological phenotype are independent and result from distinct underlying mechanisms. An extended epidemiological study of cancer risk in c.8494C>T carrier families is warranted. Variant c.875C>T (P292L) was also observed in patient AT211LA, who had typical A-T; the second allele was identified as c.9092_9097delAAGTGA;c.9098A>T (QVN3031L).
The c.7271T>G (V2424G) mutation has been previously described in great detail (Stankovic et al., 1998; Stewart et al., 2001). It was first identified in a family that included the oldest surviving A-T patient in the British Isles (over 50 years old), and a homozygous patient with mild phenotype who bore a normal child (Stankovic et al., 1998). She also had breast cancer, as did multiple members of this family. The c.7271T>G variant has also been studied in screens of breast cancer cohorts (Chenevix-Trench et al., 2002; Concannon, 2002; Bernstein et al., 2003; Thorstenson et al., 2003; Bernstein et al., 2006), and remains as the only ATM mutation that has been consistently associated with breast cancer risk in non-AT cohorts (Gatti et al., 1999; Izatt et al., 1999; Chenevix-Trench et al., 2002; Concannon, 2002; Bernstein et al., 2006). The construct that we transfected into AT7LA cells fully corrected the ATM protein level; despite this, the protein failed to phosphorylate ATM or SMC1 substrates, and the transfected host A-T cells remained radiosensitive. These data validated the site-directed mutagenesis approach for analyzing isolated ATM alleles. Interestingly, the c.7271T>G mutation does not appear to block fertility, despite the marked histological changes observed in gonadal tissues from knockout Atm−/− mice (Barlow et al., 1996).
The missense change c.7967T>C (L2656P) was observed in a patient with no detectable immunodeficiency, with a truncation as the second mutation (Toyoshima et al., 1998). The transfected cells expressed a low but detectable level of ATM, with no phosphorylation of ATM or SMC1 substrates. It is possible that the presence of a reduced level of ATM might have had some protective function and be responsible for the normal immunological profile described in the patient. On the other hand, roughly one-third of A-T patients do not manifest immunodeficiency (Woods and Taylor, 1992; Nowak-Wegrzyn et al., 2004).
In vitro site-directed mutagenesis constitutes a useful, albeit laborious, tool for distinguishing mutations from polymorphisms. This approach allows the successful introduction of a single nucleotide change into a single constant ATM-deficient genetic background and simplifies the causal analysis of phenotypic consequences arising from “variants of unknown significance” (Cooper et al., 2003; Greenblatt et al., 2003; Goldgar et al., 2004; Bao and Cui, 2005; Chan et al., 2007; Du et al., 2007; Lovelock et al., 2007). As a result of performing these analyses, we have improved our understanding of ATM missense variants that are associated with mild A-T phenotypes. Some missense changes have been experimentally validated as a cause of splicing aberrations (Ng and Henikoff, 2001; Eng et al., 2004; Babaei et al., 2005; Du et al., 2007). Taken together, we estimate the frequency of operationally deleterious ATM missense mutations in A-T patients to comprise less than 10% of all known mutations. This is in stark contrast to the much greater frequency of ATM missense variants reported in breast cancer patients, and provides additional support for the hypothesis that missense variants in the ATM gene are associated primarily with a cancer phenotype rather than with a neurological impairment (Vorechovsky et al., 1996; Gatti et al., 1999; Laake et al., 2000; Chenevix-Trench et al., 2002; Concannon, 2002; Bernstein et al., 2003; Buzin et al., 2003; Sommer et al., 2003; Bernstein et al., 2006; Renwick et al., 2006). On the other hand, most missense variants (for all large genes) have not been operationally categorized as “deleterious” or “neutral”, so it is difficult to assess their disease-causing roles.
Supplementary Material
Acknowledgments
This work was supported by grants from NIH (NS35322, NS052528, and AI067769), Joseph Drown Foundation, and the Ataxia-Telangiectasia Medical Research Foundation.
References
- Abraham RT. PI 3-kinase related kinases: ‘big’ players in stress-induced signaling pathways. DNA Repair (Amst) 2004;3(8–9):883–7. doi: 10.1016/j.dnarep.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Ali A, Zhang J, Bao S, Liu I, Otterness D, Dean NM, Abraham RT, Wang XF. Requirement of protein phosphatase 5 in DNA-damage-induced ATM activation. Genes Dev. 2004;18(3):249–54. doi: 10.1101/gad.1176004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariumi Y, Trono D. Ataxia-telangiectasia-mutated (ATM) protein can enhance human immunodeficiency virus type 1 replication by stimulating Rev function. J Virol. 2006;80(5):2445–52. doi: 10.1128/JVI.80.5.2445-2452.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babaei M, Mitui M, Olson ER, Gatti RA. ATM haplotypes and associated mutations in Iranian patients with ataxia-telangiectasia: recurring homozygosity without a founder haplotype. Hum Genet. 2005;117(2–3):101–6. doi: 10.1007/s00439-005-1254-7. [DOI] [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421(6922):499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- Bao L, Cui Y. Prediction of the phenotypic effects of non-synonymous single nucleotide polymorphisms using structural and evolutionary information. Bioinformatics. 2005;21(10):2185–90. doi: 10.1093/bioinformatics/bti365. [DOI] [PubMed] [Google Scholar]
- Barlow C, Hirotsune S, Paylor R, Liyanage M, Eckhaus M, Collins F, Shiloh Y, Crawley JN, Ried T, Tagle D, Wynshaw-Boris A. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell. 1996;86(1):159–71. doi: 10.1016/s0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]
- Becker-Catania SG, Chen G, Hwang MJ, Wang Z, Sun X, Sanal O, Bernatowska-Matuszkiewicz E, Chessa L, Lee EY, Gatti RA. Ataxia-telangiectasia: phenotype/genotype studies of ATM protein expression, mutations, and radiosensitivity. Mol Genet Metab. 2000;70(2):122–33. doi: 10.1006/mgme.2000.2998. [DOI] [PubMed] [Google Scholar]
- Bernstein JL, Bernstein L, Thompson WD, Lynch CF, Malone KE, Teitelbaum SL, Olsen JH, Anton-Culver H, Boice JD, Rosenstein BS, Borresen-Dale AL, Gatti RA, Concannon P, Haile RW. ATM variants 7271T>G and IVS10–6T>G among women with unilateral and bilateral breast cancer. Br J Cancer. 2003;89(8):1513–6. doi: 10.1038/sj.bjc.6601289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein JL, Teraoka S, Southey MC, Jenkins MA, Andrulis IL, Knight JA, John EM, Lapinski R, Wolitzer AL, Whittemore AS, West D, Seminara D, Olson ER, Spurdle AB, Chenevix-Trench G, Giles GG, Hopper JL, Concannon P. Population-based estimates of breast cancer risks associated with ATM gene variants c.7271T>G and c.1066–6T>G (IVS10–6T>G) from the Breast Cancer Family Registry. Hum Mutat. 2006;27(11):1122–8. doi: 10.1002/humu.20415. [DOI] [PubMed] [Google Scholar]
- Birrell GW, Kneebone K, Nefedov M, Nefedova E, Jartsev MN, Mitsui M, Gatti RA, Lavin MF. ATM mutations, haplotype analysis, and immunological status of Russian patients with ataxia telangiectasia. Hum Mutat. 2005;25(6):593. doi: 10.1002/humu.9341. [DOI] [PubMed] [Google Scholar]
- Blencowe BJ. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem Sci. 2000;25(3):106–10. doi: 10.1016/s0968-0004(00)01549-8. [DOI] [PubMed] [Google Scholar]
- Buzin CH, Gatti RA, Nguyen VQ, Wen CY, Mitui M, Sanal O, Chen JS, Nozari G, Mengos A, Li X, Fujimura F, Sommer SS. Comprehensive scanning of the ATM gene with DOVAM-S. Hum Mutat. 2003;21(2):123–31. doi: 10.1002/humu.10158. [DOI] [PubMed] [Google Scholar]
- Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. 2002;3(4):285–98. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31(13):3568–71. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalieri S, Funaro A, Porcedda P, Turinetto V, Migone N, Gatti RA, Brusco A. ATM mutations in Italian families with ataxia telangiectasia include two distinct large genomic deletions. Hum Mutat. 2006;27(10):1061. doi: 10.1002/humu.9454. [DOI] [PubMed] [Google Scholar]
- Chan PA, Duraisamy S, Miller PJ, Newell JA, McBride C, Bond JP, Raevaara T, Ollila S, Nystrom M, Grimm AJ, Christodoulou J, Oetting WS, Greenblatt MS. Interpreting missense variants: comparing computational methods in human disease genes CDKN2A, MLH1, MSH2, MECP2, and tyrosinase (TYR) Hum Mutat. 2007;28(7):683–93. doi: 10.1002/humu.20492. [DOI] [PubMed] [Google Scholar]
- Chenevix-Trench G, Spurdle AB, Gatei M, Kelly H, Marsh A, Chen X, Donn K, Cummings M, Nyholt D, Jenkins MA, Scott C, Pupo GM, Dork T, Bendix R, Kirk J, Tucker K, McCredie MR, Hopper JL, Sambrook J, Mann GJ, Khanna KK. Dominant negative ATM mutations in breast cancer families. J Natl Cancer Inst. 2002;94(3):205–15. doi: 10.1093/jnci/94.3.205. [DOI] [PubMed] [Google Scholar]
- Chun HH, Sun X, Nahas SA, Teraoka S, Lai CH, Concannon P, Gatti RA. Improved diagnostic testing for ataxia-telangiectasia by immunoblotting of nuclear lysates for ATM protein expression. Mol Genet Metab. 2003;80(4):437–43. doi: 10.1016/j.ymgme.2003.09.008. [DOI] [PubMed] [Google Scholar]
- Concannon P. ATM heterozygosity and cancer risk. Nat Genet. 2002;32(1):89–90. doi: 10.1038/ng0902-89. [DOI] [PubMed] [Google Scholar]
- Concannon P, Gatti RA. Diversity of ATM gene mutations detected in patients with ataxia-telangiectasia. Hum Mutat. 1997;10(2):100–7. doi: 10.1002/(SICI)1098-1004(1997)10:2<100::AID-HUMU2>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Cooper GM, Brudno M, Green ED, Batzoglou S, Sidow A. Quantitative estimates of sequence divergence for comparative analyses of mammalian genomes. Genome Res. 2003;13(5):813–20. doi: 10.1101/gr.1064503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho G, Xie J, Du L, Brusco A, Krainer AR, Gatti RA. Functional significance of a deep intronic mutation in the ATM gene and evidence for an alternative exon 28a. Hum Mutat. 2005;25(2):118–24. doi: 10.1002/humu.20170. [DOI] [PubMed] [Google Scholar]
- Du L, Pollard JM, Gatti RA. Correction of prototypic ATM splicing mutations and aberrant ATM function with antisense morpholino oligonucleotides. Proc Natl Acad Sci U S A. 2007;104(14):6007–12. doi: 10.1073/pnas.0608616104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng L, Coutinho G, Nahas S, Yeo G, Tanouye R, Babaei M, Dork T, Burge C, Gatti RA. Nonclassical splicing mutations in the coding and noncoding regions of the ATM Gene: maximum entropy estimates of splice junction strengths. Hum Mutat. 2004;23(1):67–76. doi: 10.1002/humu.10295. [DOI] [PubMed] [Google Scholar]
- Eymin B, Claverie P, Salon C, Leduc C, Col E, Brambilla E, Khochbin S, Gazzeri S. p14ARF activates a Tip60-dependent and p53-independent ATM/ATR/CHK pathway in response to genotoxic stress. Mol Cell Biol. 2006;26(11):4339–50. doi: 10.1128/MCB.02240-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti RA, Becker-Catania S, Chun HH, Sun X, Mitui M, Lai CH, Khanlou N, Babaei M, Cheng R, Clark C, Huo Y, Udar NC, Iyer RK. The pathogenesis of ataxia-telangiectasia. Learning from a Rosetta Stone. Clin Rev Allergy Immunol. 2001;20(1):87–108. doi: 10.1385/CRIAI:20:1:87. [DOI] [PubMed] [Google Scholar]
- Gatti RA, Boder E, Vinters HV, Sparkes RS, Norman A, Lange K. Ataxia-telangiectasia: an interdisciplinary approach to pathogenesis. Medicine (Baltimore) 1991;70(2):99–117. [PubMed] [Google Scholar]
- Gatti RA, Tward A, Concannon P. Cancer risk in ATM heterozygotes: a model of phenotypic and mechanistic differences between missense and truncating mutations. Mol Genet Metab. 1999;68(4):419–23. doi: 10.1006/mgme.1999.2942. [DOI] [PubMed] [Google Scholar]
- Gilad S, Chessa L, Khosravi R, Russell P, Galanty Y, Piane M, Gatti RA, Jorgensen TJ, Shiloh Y, Bar-Shira A. Genotype-phenotype relationships in ataxia-telangiectasia and variants. Am J Hum Genet. 1998;62(3):551–61. doi: 10.1086/301755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldgar DE, Easton DF, Deffenbaugh AM, Monteiro AN, Tavtigian SV, Couch FJ. Integrated evaluation of DNA sequence variants of unknown clinical significance: application to BRCA1 and BRCA2. Am J Hum Genet. 2004;75(4):535–44. doi: 10.1086/424388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR. Sorting out the complexity of SR protein functions. Rna. 2000;6(9):1197–211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt MS, Beaudet JG, Gump JR, Godin KS, Trombley L, Koh J, Bond JP. Detailed computational study of p53 and p16: using evolutionary sequence analysis and disease-associated mutations to predict the functional consequences of allelic variants. Oncogene. 2003;22(8):1150–63. doi: 10.1038/sj.onc.1206101. [DOI] [PubMed] [Google Scholar]
- Guan MX, Yan Q, Li X, Bykhovskaya Y, Gallo-Teran J, Hajek P, Umeda N, Zhao H, Garrido G, Mengesha E, Suzuki T, del Castillo I, Peters JL, Li R, Qian Y, Wang X, Ballana E, Shohat M, Lu J, Estivill X, Watanabe K, Fischel-Ghodsian N. Mutation in TRMU related to transfer RNA modification modulates the phenotypic expression of the deafness-associated mitochondrial 12S ribosomal RNA mutations. Am J Hum Genet. 2006;79(2):291–302. doi: 10.1086/506389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikkinen K, Rapakko K, Karppinen SM, Erkko H, Nieminen P, Winqvist R. Association of common ATM polymorphism with bilateral breast cancer. Int J Cancer. 2005;116(1):69–72. doi: 10.1002/ijc.20996. [DOI] [PubMed] [Google Scholar]
- Huo YK, Wang Z, Hong JH, Chessa L, McBride WH, Perlman SL, Gatti RA. Radiosensitivity of ataxia-telangiectasia, X-linked agammaglobulinemia, and related syndromes using a modified colony survival assay. Cancer Res. 1994;54(10):2544–7. [PubMed] [Google Scholar]
- Izatt L, Greenman J, Hodgson S, Ellis D, Watts S, Scott G, Jacobs C, Liebmann R, Zvelebil MJ, Mathew C, Solomon E. Identification of germline missense mutations and rare allelic variants in the ATM gene in early-onset breast cancer. Genes Chromosomes Cancer. 1999;26(4):286–94. [PubMed] [Google Scholar]
- Johnson N, Fletcher O, Palles C, Rudd M, Webb E, Sellick G, dos Santos Silva I, McCormack V, Gibson L, Fraser A, Leonard A, Gilham C, Tavtigian SV, Ashworth A, Houlston R, Peto J. Counting potentially functional variants in BRCA1, BRCA2 and ATM predicts breast cancer susceptibility. Hum Mol Genet. 2007;16(9):1051–7. doi: 10.1093/hmg/ddm050. [DOI] [PubMed] [Google Scholar]
- Kan JL, Green MR. Pre-mRNA splicing of IgM exons M1 and M2 is directed by a juxtaposed splicing enhancer and inhibitor. Genes Dev. 1999;13(4):462–71. doi: 10.1101/gad.13.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim ST, Xu B, Kastan MB. Involvement of the cohesin protein, Smc1, in Atm-dependent and independent responses to DNA damage. Genes Dev. 2002;16(5):560–70. doi: 10.1101/gad.970602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov SV, Graham ME, Peng C, Chen P, Robinson PJ, Lavin MF. Involvement of novel autophosphorylation sites in ATM activation. Embo J. 2006;25(15):3504–14. doi: 10.1038/sj.emboj.7601231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz EU, Lees-Miller SP. DNA damage-induced activation of ATM and ATM-dependent signaling pathways. DNA Repair (Amst) 2004;3(8–9):889–900. doi: 10.1016/j.dnarep.2004.03.029. [DOI] [PubMed] [Google Scholar]
- Laake K, Jansen L, Hahnemann JM, Brondum-Nielsen K, Lonnqvist T, Kaariainen H, Sankila R, Lahdesmaki A, Hammarstrom L, Yuen J, Tretli S, Heiberg A, Olsen JH, Tucker M, Kleinerman R, Borresen-Dale AL. Characterization of ATM mutations in 41 Nordic families with ataxia telangiectasia. Hum Mutat. 2000;16(3):232–46. doi: 10.1002/1098-1004(200009)16:3<232::AID-HUMU6>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Lau A, Swinbank KM, Ahmed PS, Taylor DL, Jackson SP, Smith GC, O’Connor MJ. Suppression of HIV-1 infection by a small molecule inhibitor of the ATM kinase. Nat Cell Biol. 2005;7(5):493–500. doi: 10.1038/ncb1250. [DOI] [PubMed] [Google Scholar]
- Lavin MF, Delia D, Chessa L. ATM and the DNA damage response. Workshop on ataxia-telangiectasia and related syndromes. EMBO Rep. 2006;7(2):154–60. doi: 10.1038/sj.embor.7400629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linding R, Jensen LJ, Ostheimer GJ, van Vugt MA, Jorgensen C, Miron IM, Diella F, Colwill K, Taylor L, Elder K, Metalnikov P, Nguyen V, Pasculescu A, Jin J, Park JG, Samson LD, Woodgett JR, Russell RB, Bork P, Yaffe MB, Pawson T. Systematic discovery of in vivo phosphorylation networks. Cell. 2007;129(7):1415–26. doi: 10.1016/j.cell.2007.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HX, Chew SL, Cartegni L, Zhang MQ, Krainer AR. Exonic splicing enhancer motif recognized by human SC35 under splicing conditions. Mol Cell Biol. 2000;20(3):1063–71. doi: 10.1128/mcb.20.3.1063-1071.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HX, Zhang M, Krainer AR. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev. 1998;12(13):1998–2012. doi: 10.1101/gad.12.13.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovelock PK, Spurdle AB, Mok MT, Farrugia DJ, Lakhani SR, Healey S, Arnold S, Buchanan D, Investigators K, Couch FJ, Henderson BR, Goldgar DE, Tavtigian SV, Chenevix-Trench G, Brown MA. Identification of BRCA1 missense substitutions that confer partial functional activity: potential moderate risk variants? Breast Cancer Res. 2007;9(6):R82. doi: 10.1186/bcr1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316(5828):1160–6. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- McConville CM, Stankovic T, Byrd PJ, McGuire GM, Yao QY, Lennox GG, Taylor MR. Mutations associated with variant phenotypes in ataxia-telangiectasia. Am J Hum Genet. 1996;59(2):320–30. [PMC free article] [PubMed] [Google Scholar]
- Mitsui M, Nahas S, Chun H, Gatti RA. Diagnosis of Ataxia-Telangiectasia: ATM Mutations Associated With Cancer. Cancer Diagnostics: Current and Future Trends. 2004:473–487. [Google Scholar]
- Mitui M, Campbell C, Coutinho G, Sun X, Lai CH, Thorstenson Y, Castellvi-Bel S, Fernandez L, Monros E, Carvalho BT, Porras O, Fontan G, Gatti RA. Independent mutational events are rare in the ATM gene: haplotype prescreening enhances mutation detection rate. Hum Mutat. 2003;22(1):43–50. doi: 10.1002/humu.10232. [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11(5):863–74. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak-Wegrzyn A, Crawford TO, Winkelstein JA, Carson KA, Lederman HM. Immunodeficiency and infections in ataxia-telangiectasia. J Pediatr. 2004;144(4):505–11. doi: 10.1016/j.jpeds.2003.12.046. [DOI] [PubMed] [Google Scholar]
- Olsen JH, Hahnemann JM, Borresen-Dale AL, Tretli S, Kleinerman R, Sankila R, Hammarstrom L, Robsahm TE, Kaariainen H, Bregard A, Brondum-Nielsen K, Yuen J, Tucker M. Breast and other cancers in 1445 blood relatives of 75 Nordic patients with ataxia telangiectasia. Br J Cancer. 2005;93(2):260–5. doi: 10.1038/sj.bjc.6602658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S, Becker-Catania S, Gatti RA. Ataxia-telangiectasia: diagnosis and treatment. Semin Pediatr Neurol. 2003;10(3):173–82. doi: 10.1016/s1071-9091(03)00026-3. [DOI] [PubMed] [Google Scholar]
- Renwick A, Thompson D, Seal S, Kelly P, Chagtai T, Ahmed M, North B, Jayatilake H, Barfoot R, Spanova K, McGuffog L, Evans DG, Eccles D, Easton DF, Stratton MR, Rahman N. ATM mutations that cause ataxia-telangiectasia are breast cancer susceptibility alleles. Nat Genet. 2006;38(8):873–5. doi: 10.1038/ng1837. [DOI] [PubMed] [Google Scholar]
- Sandoval N, Platzer M, Rosenthal A, Dork T, Bendix R, Skawran B, Stuhrmann M, Wegner RD, Sperling K, Banin S, Shiloh Y, Baumer A, Bernthaler U, Sennefelder H, Brohm M, Weber BH, Schindler D. Characterization of ATM gene mutations in 66 ataxia telangiectasia families. Hum Mol Genet. 1999;8(1):69–79. doi: 10.1093/hmg/8.1.69. [DOI] [PubMed] [Google Scholar]
- Saviozzi S, Saluto A, Taylor AM, Last JI, Trebini F, Paradiso MC, Grosso E, Funaro A, Ponzio G, Migone N, Brusco A. A late onset variant of ataxia-telangiectasia with a compound heterozygous genotype, A8030G/7481insA. J Med Genet. 2002;39(1):57–61. doi: 10.1136/jmg.39.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle DA, Smith S, Uziel T, Sfez S, et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268(5218):1749–53. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- Scott SP, Teh A, Peng C, Lavin MF. One-step site-directed mutagenesis of ATM cDNA in large (20kb) plasmid constructs. Hum Mutat. 2002;20(4):323. doi: 10.1002/humu.9068. [DOI] [PubMed] [Google Scholar]
- Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer. 2003;3(3):155–68. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- Shin YC, Nakamura H, Liang X, Feng P, Chang H, Kowalik TF, Jung JU. Inhibition of the ATM/p53 signal transduction pathway by Kaposi’s sarcoma-associated herpesvirus interferon regulatory factor 1. J Virol. 2006;80(5):2257–66. doi: 10.1128/JVI.80.5.2257-2266.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer SS, Jiang Z, Feng J, Buzin CH, Zheng J, Longmate J, Jung M, Moulds J, Dritschilo A. ATM missense mutations are frequent in patients with breast cancer. Cancer Genet Cytogenet. 2003;145(2):115–20. doi: 10.1016/s0165-4608(03)00119-5. [DOI] [PubMed] [Google Scholar]
- Stankovic T, Kidd AM, Sutcliffe A, McGuire GM, Robinson P, Weber P, Bedenham T, Bradwell AR, Easton DF, Lennox GG, Haites N, Byrd PJ, Taylor AM. ATM mutations and phenotypes in ataxia-telangiectasia families in the British Isles: expression of mutant ATM and the risk of leukemia, lymphoma, and breast cancer. Am J Hum Genet. 1998;62(2):334–45. doi: 10.1086/301706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS, Last JI, Stankovic T, Haites N, Kidd AM, Byrd PJ, Taylor AM. Residual ataxia telangiectasia mutated protein function in cells from ataxia telangiectasia patients, with 5762ins137 and 7271T-->G mutations, showing a less severe phenotype. J Biol Chem. 2001;276(32):30133–41. doi: 10.1074/jbc.M103160200. [DOI] [PubMed] [Google Scholar]
- Sun X, Becker-Catania SG, Chun HH, Hwang MJ, Huo Y, Wang Z, Mitui M, Sanal O, Chessa L, Crandall B, Gatti RA. Early diagnosis of ataxia-telangiectasia using radiosensitivity testing. J Pediatr. 2002;140(6):724–31. doi: 10.1067/mpd.2002.123879. [DOI] [PubMed] [Google Scholar]
- Sunyaev S, Ramensky V, Koch I, Lathe W, 3rd, Kondrashov AS, Bork P. Prediction of deleterious human alleles. Hum Mol Genet. 2001;10(6):591–7. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- Swift M, Morrell D, Massey RB, Chase CL. Incidence of cancer in 161 families affected by ataxia-telangiectasia. N Engl J Med. 1991;325(26):1831–6. doi: 10.1056/NEJM199112263252602. [DOI] [PubMed] [Google Scholar]
- Tamimi RM, Hankinson SE, Spiegelman D, Kraft P, Colditz GA, Hunter DJ. Common ataxia telangiectasia mutated haplotypes and risk of breast cancer: a nested case-control study. Breast Cancer Res. 2004;6(4):R416–22. doi: 10.1186/bcr809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AM. What has the cloning of the ATM gene told us about ataxia telangiectasia? Int J Radiat Biol. 1998;73(4):365–71. doi: 10.1080/095530098142185. [DOI] [PubMed] [Google Scholar]
- Thompson D, Antoniou AC, Jenkins M, Marsh A, Chen X, Wayne T, Tesoriero A, Milne R, Spurdle A, Thorstenson Y, Southey M, Giles GG, Khanna KK, Sambrook J, Oefner P, Goldgar D, Hopper JL, Easton D, Chenevix-Trench G. Two ATM variants and breast cancer risk. Hum Mutat. 2005;25(6):594–5. doi: 10.1002/humu.9344. [DOI] [PubMed] [Google Scholar]
- Thorstenson YR, Roxas A, Kroiss R, Jenkins MA, Yu KM, Bachrich T, Muhr D, Wayne TL, Chu G, Davis RW, Wagner TM, Oefner PJ. Contributions of ATM mutations to familial breast and ovarian cancer. Cancer Res. 2003;63(12):3325–33. [PubMed] [Google Scholar]
- Toyoshima M, Hara T, Zhang H, Yamamoto T, Akaboshi S, Nanba E, Ohno K, Hori N, Sato K, Takeshita K. Ataxia-telangiectasia without immunodeficiency: novel point mutations within and adjacent to the phosphatidylinositol 3-kinase-like domain. Am J Med Genet. 1998;75(2):141–4. [PubMed] [Google Scholar]
- Vogt G, Chapgier A, Yang K, Chuzhanova N, Feinberg J, Fieschi C, Boisson-Dupuis S, Alcais A, Filipe-Santos O, Bustamante J, de Beaucoudrey L, Al-Mohsen I, Al-Hajjar S, Al-Ghonaium A, Adimi P, Mirsaeidi M, Khalilzadeh S, Rosenzweig S, de la Calle Martin O, Bauer TR, Puck JM, Ochs HD, Furthner D, Engelhorn C, Belohradsky B, Mansouri D, Holland SM, Schreiber RD, Abel L, Cooper DN, Soudais C, Casanova JL. Gains of glycosylation comprise an unexpectedly large group of pathogenic mutations. Nat Genet. 2005;37(7):692–700. doi: 10.1038/ng1581. [DOI] [PubMed] [Google Scholar]
- Vorechovsky I, Rasio D, Luo L, Monaco C, Hammarstrom L, Webster AD, Zaloudik J, Barbanti-Brodani G, James M, Russo G, et al. The ATM gene and susceptibility to breast cancer: analysis of 38 breast tumors reveals no evidence for mutation. Cancer Res. 1996;56(12):2726–32. [PubMed] [Google Scholar]
- Wildeman M, van Ophuizen E, den Dunnen JT, Taschner PE. Improving sequence variant descriptions in mutation databases and literature using the Mutalyzer sequence variation nomenclature checker. Hum Mutat. 2008;29(1):6–13. doi: 10.1002/humu.20654. [DOI] [PubMed] [Google Scholar]
- Woods CG, Taylor AM. Ataxia telangiectasia in the British Isles: the clinical and laboratory features of 70 affected individuals. Q J Med. 1992;82(298):169–79. [PubMed] [Google Scholar]
- Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11(2–3):377–94. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- Zhang N, Chen P, Khanna KK, Scott S, Gatei M, Kozlov S, Watters D, Spring K, Yen T, Lavin MF. Isolation of full-length ATM cDNA and correction of the ataxia-telangiectasia cellular phenotype. Proc Natl Acad Sci U S A. 1997;94(15):8021–6. doi: 10.1073/pnas.94.15.8021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv Y, Bar-Shira A, Pecker I, Russell P, Jorgensen TJ, Tsarfati I, Shiloh Y. Recombinant ATM protein complements the cellular A-T phenotype. Oncogene. 1997;15(2):159–67. doi: 10.1038/sj.onc.1201319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


