Abstract
Protein kinases are important regulators of intracellular signal transduction pathways and play critical roles in diverse cellular functions. Once a protein kinase is activated, its activity is subsequently downregulated through a variety of mechanisms. Accumulating evidence indicates that the activation of protein kinases commonly initiates their downregulation via the ubiquitin/proteasome pathway. Failure to regulate protein kinase activity or expression levels can cause human diseases.
Keywords: activation, downregulation, phosphorylation, ubiquitin
INTRODUCTION
Protein kinases are important regulators of intracellular signal transduction pathways that mediate the development and regulation of both unicellular and multicellular organisms. They play critical roles in cell growth, division, differentiation, adhesion, motility, and death. More than 500 protein kinases have been identified in the human genome (1). Based on their catalytic specificity, they can be subdivided into tyrosine (Tyr, Y)- and serine (Ser, S)/threonine (Thr, T)-specific kinases. Their activity is normally tightly regulated to mediate their cellular function and physiological responses. Perturbation of protein kinases by mutations, altered expression, or dysregulation can cause many human diseases, including cancer and diabetes (2). Normal cellular protein kinases can be positively upregulated by phosphorylation, dephosphorylation, protein cleavage, translocation, ligand/second-messenger or ion binding, dimerization or oligomerization, activating subunit interaction, and protein-protein or protein-lipid interactions. The activity of protein kinases is also governed by negative feedback systems, which can attenuate or terminate kinase activity and thus their induced downstream signals. Several mechanisms serve to negatively regulate kinase activity, including, in the case of ligand-induced receptor tyrosine kinase signaling, antagonistic ligands, hetero-oligomerization with truncated receptors, phosphorylation, dephosphorylation, endocytosis, protein degradation, and reduction of receptor mRNA, all of which serve as means of downregulation (3).
Regulated proteolysis is a common mechanism for downregulating activated protein kinases. In most cases this involves activation-dependent protein degradation mediated through the ubiquitin (Ub)/proteasome or lysosome pathways, although a few kinases, such as Src, AKT, and FAK, can be downregulated by caspase- or calpain protease–mediated protein cleavage in the process of apoptosis (4–9). The ubiquitin-proteasome system (UPS) controls the levels of many proteins, including protein kinases and other intracellular regulatory proteins, and is essential in eukaryotes. Conjugation of Ub to substrate proteins requires three enzymes: the Ub-activating enzyme (E1), an Ub-conjugating enzyme (E2), and an Ub ligase (E3). E1 activates Ub through the formation of a thiol-ester bond between the C terminus of Ub and the active site cysteine (Cys). The activated Ub is then transesterified to a conserved Cys of an E2. The E3 ligase interacts with both E2 and the substrate and facilitates ubiquitination of the substrate. There are two distinct types of E3 ligases: the enzymatic HECT (homologous to E6-AP C terminus) domain E3s and the non-HECT domain E3s. HECT domain E3s form a thiolester with Ub, which can then be transferred directly to the substrate. Thenon-HECT domain E3s, containing a RING, variant RING, or a U box, do not form a thioester with Ub but function as adaptors to facilitate interaction between substrates and a charged E2 (10). E3s have a key role in defining the substrate specificity of ubiquitination. The polyubiquitin chain on substrates is formed through isopeptide linkages between the ε-amino group of a lysine (Lys, K) in one Ub and the C terminus of the next Ub. Seven lysines in Ub can be used for Ub-chain formation. Polyubiquitin chains linked through K48 and, in some cases, K11 are recognized by the proteasome as a signal for degradation (11–13). Polyubiquitin chains linked through other lysines serve other functions. For example, TRAF6, a RING finger E3 ligase, acting together with E2 Ubc13/Uev1A, induces K63 polyubiquitination. TRAF6 polyubiquitination does not result in degradation, but rather is recognized by K63 linkage-specific binding proteins leading to activation of the TAK1 kinase and its downstream targets MKK6 and IκB kinase (IKK) (14, 15).
There are at least 22 known classes of Ub-binding domains (UBDs) [e.g., Ub-associated (UBA) domain) and Ub-interacting motif (UIM)] (16–18). Of 12 human AMP-activated protein kinase (AMPK)-related kinases, 9, including Par-1 paralogs/microtubule-affinityregulating kinases 1–4, possess a UBA domain, which binds intramolecularly to the K29/33 Ub chains formed on AMPK family members. This binding causes dissociation of the UBA domain from the catalytic domain, thus reducing kinase activity, which requires UBA/catalytic domain interaction (19).
Besides Ub, 13 other small Ub-like (UBL) proteins (e.g., SUMO1-3 and NEDD8/Rub1) are structurally similar to Ub, and are conjugated to substrate proteins or lipids via their C termini, thereby regulating different cellular responses (16). In addition, UBLs can be found as an integral UBL domain (ULD) within the larger context of a protein (20). IKKα, IKKβ, IKKi/IKKε, and IKK-related kinase TANK-binding kinase 1 (TBK1/NAK/T2K) contain ULDs. The ULD of TBK1, IKKi, or IKKε is required for kinase domain activation, interaction with kinase substrates, and subsequent degradation of substrates (21, 22).
Many Ser/Thr- and Tyr-specific protein kinases can be degraded through the UPS, and in most cases initiation of the degradation process is triggered upon activation of the protein kinase. The downregulation of activated protein kinases through degradation, and, in some cases, through degradation of regulatory subunits provides negative-feedback regulation of kinase activity, which is essential for the precise control of cellular functions. Not surprisingly, the phosphorylation and dephosphorylation events that regulate protein kinase activity are also involved in their degradation. Activation of the kinase catalytic domain, which commonly occurs through phosphorylation of the activation loop, results in a conformational change that reorients the catalytic center and thereby allows phosphate transfer from ATP to substrate. The activated conformation of a protein kinase is a prerequisite for initiating degradation (23), but its subcellular localization often determines how it is processed for degradation. For instance, in contrast to nonreceptor tyrosine kinases, which are downregulated by the classical UPS-mediated mechanisms, most activated receptor tyrosine kinases are ubiquitinated and directed to the lysosome for degradation. This review focuses on the activation-dependent and UPS-mediated degradation of protein kinases of both the nonreceptor (Table S1: Follow the Supplemental Material link from the Annual Reviews home page at http://www.annualreviews.org) and receptor types (Table S2), including those mediated by protein kinase-specific chaperones.
NONRECEPTOR PROTEIN KINASES
Eukaryotic protein kinases can be divided into two main types: receptor protein kinases and nonreceptor protein kinases. Receptor protein kinases are type I transmembrane proteins with an N-terminal extracellular domain, which can in principle bind activating ligands, a single transmembrane domain, and a C-terminal cytoplasmic domain that includes the catalytic domain. Nonreceptor protein kinases lack a transmembrane domain; most are soluble intracellular proteins, but in some cases, they associate with membranes via a membrane-targeting posttranslational modification, such as an N-terminal myristoyl group, and can act as the catalytic subunit for receptors that lack their own catalytic domain. Eukaryotic protein kinases are specific for either serine/threonine or tyrosine residues in their target protein. Receptor and nonreceptor protein kinases of both amino acid specificities are known (2, 3).
Nonreceptor Serine/Threonine Kinases
Nonreceptor serine/threonine kinases are the largest group of eukaryotic protein kinases comprising 376 out of the total of 518 protein kinases in humans (1). Examples are protein kinase C (PKC), the MAP kinases, and AKT.
Protein kinase C
PKC isoforms are Ser/Thr kinases involved in signal transduction pathways that govern a wide range of physiological processes, including differentiation, proliferation, gene expression, membrane transport, and organization of cytoskeletal and extracellular matrix proteins. Some PKC isoforms are targets of tumor-promoting phorbol esters such as 12-O-tetradecanoylphorbol-13-acetate (TPA). Initially, phorbol ester treatment activates conventional (α, βI, βII, and γ) and novel PKC isoforms (δ, ε, η, and θ), but then, upon prolonged exposure, causes their depletion by proteolytic degradation. Downregulation of PKCδ is responsible for the tumor-promoting effect of phorbol esters (24). The degradation of PKC isoforms induced by diacylglycerol (DAG), a physiological activator of PKC, TPA, or bryostatin-1 is mediated through the UPS. That PKC ubiquitination and degradation is prevented by inhibition of PKC activation demonstrated for the first time that a Ser/Thr protein kinase exhibits activation-dependent protein degradation (25), which acts as a feedback regulatory mechanism. Ubiquitination of several PKC isoforms may be mediated by common E2s and E3s, which can differ according to the nature of the upstream signal. For instance, an active PKCβII mutant interacts with and is ubiquitinated in vitro by E3 complexes composed of RING finger protein HOIL-1L and HOIL-1L–interacting protein, and HOIL-1 deficiency largely inhibits TPA–induced PKCα degradation (26). In contrast, RING finger protein that interacts with C kinase (RINCK) interacts with the C1A domain of PKCβII and ubiquitinates PKCβII. RINCK depletion increases expression of PKCα, PKCδ, and PKCζ in a cell line-specific manner, but has no effect on TPA-mediated downregulation of PKC (27).
In contrast to conventional and novel PKCs, PKCζ, an atypical PKC (aPKC), which is not responsive to TPA or DAG, can be activated by caspase cleavage upon treatment with tumor necrosis factor (TNF)-α (28). Although the physiological significance of PKCζ downregulation is unclear, the released PKCζ kinase domain, which has significantly increased kinase activity, undergoes UPS-dependent degradation (28). The von Hippel-Lindau (VHL) protein regulates ubiquitination of PKCζII, a rapidly and constitutively degraded variant of PKCζ. VHL, together with elongins B/C, Cul-2, and the RING finger-containing protein Rbx1, forms a complex (VCB-Cul2) that functions as an E3 Ub ligase in which VHL recognizes the N-terminal PB1 domain of PKCζII. VHL also regulates ubiquitination of serumactivated PKCλ/ι, another aPKC (29).
Exactly how PKC kinase activation leads to ubiquitination and degradation is not fully understood. In its inactive state, PKC is auto-inhibited as a result of the binding of an N-terminal pseudosubstrate site to the catalytic domain, which prevents the catalytic domain from binding and phosphorylating substrates. A constitutively active form of PKCη, in which mutation of its pseudosubstrate site releases intramolecular inhibition, is rapidly degraded. In contrast, a kinase-dead variant of the PKCη pseudosubstrate-site mutant, which should adopt an active conformation, is resistant to degradation. However, this mutant, but not wild-type (WT) PKCη, which has an inactive conformation, is degraded when co-overexpressed with a constitutively active PKCη mutant (23). These results suggest that the active conformation of PKCη is required but not sufficient for its degradation. Additional modifications, such as auto-/transphosphorylation and/or transphosphorylation of substrates, dephosphorylation, and correct subcellular localization may be required to bring PKC together with the ubiquitination machinery. The importance of phosphorylation is evidenced by the correlation between enhanced PKCδ degradation and PKCδ hyperphosphorylation at T505 in the activation loop and autophosphorylation at S643 induced by treatment with calyculin A, a protein phosphatase PP1/PP2A inhibitor, TPA, or serum. Degradation of the PKCδ T505A mutant is slower than that of its WT counterpart (30). Furthermore, autophosphorylation of PKC βII is required for its ubiquitination by the HOIL-1L/HOIL-1L–interacting protein complex (26). Transphosphorylation can also be involved in PKC ubiquitination. Expression of c-Src or v-Src results in phosphorylation of PKCδ at Y311 and degradation, and mutation of Y311 prevents Src-induced phosphorylation and degradation of PKCδ (31). These results imply that phosphorylated residues on PKCδ may in general provide a docking site for recruiting E3 ubiquitination machinery.
MAP kinase pathways
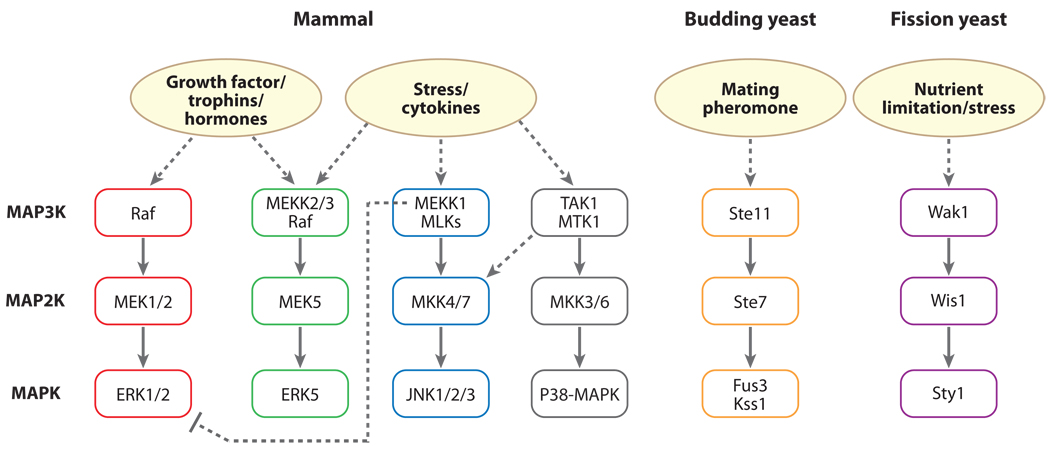
Mitogen-activated protein kinases (MAPKs) transduce signals from the cell surface to the nucleus. MAPK activity is regulated through three-tiered cascades composed of a MAPK, a MAP2K [also named MEK (MAPK/ERK kinase) and MKK (MAPK kinase)], and a MAP3K [also named MAPK kinase kinase or MEKK (MEK kinase)] (Figure 1). When triggered by a mitogenic stimulus, the extracellular signal-regulated kinases 1 and 2 (ERK1/ERK2) MAPKs are phosphorylated and activated by MEK1/2. In response to epidermal growth factor (EGF) or serum treatment, this activated state is only transient, because ERK1/2 can be dephosphorylated byMAPKphosphatases, whose expression is induced downstream of ERK activation. In contrast, a hyperosmotic stimulus results in sustained activation of ERK1/2, which is eventually downregulated by ERK1/2 protein degradation mediated by an upstream MAP3K, MEKK1, which in addition to its Ser/Thr kinase catalytic domain has a variant RING domain [this was initially classified as a plant homeodomain (PHD)], with Ub ligase activity, that promotes ubiquitination of ERK as well as c-Jun and Fra2 (32, 33). Abrogation of MEKK1 and ERK1/2 interaction through mutation of the ERK1/2 docking site reduces sorbitol-induced ERK1/2 degradation and thereby osmotic stress-induced apoptosis. MEKK1 activity is also required for JNK1-dependent activation of the HECT domain E3 Itch and subsequent JunB and c-Jun degradation in T cells (33). MEKK1 autoubiquitinates, and this depends on its kinase activity and RING integrity (33). In addition to autoubiquitination, the C-terminal fragment of MEKK1 containing the kinase domain interacts with and is ubiquitinated by the RING finger protein Deltex, which functions as a suppressor of T-cell activation through selective degradation of MEKK1 (34).
Figure 1.
Conventional (ERK1/2, JNK, and p38 MAP kinase) and ERK5 MAP kinase signaling pathways in mammalian cells and in yeast. Only representative signaling molecules and one of the multiple MAPK pathways for budding or fission yeast are shown. Solid arrows: main pathways; dashed arrows: branched pathways and indirect activation.
MEKK2, another member of the MEKK MAP3K family, interacts with and is ubiquitinated and degraded by Smurf1, a C2-WW-HECT domain E3 ligase. Loss of Smurf1 leads to accumulation of phosphorylated MEKK2 and activation of the downstream JNK signaling cascade, which subsequently enhances osteoblast activity and increases bone mass (35). The c-Raf-1 MAP3K is degraded by the UPS when cell adhesion is disrupted, and the c-Mos MAP3K, which is a regulator of oocyte maturation, is primarily ubiquitinated at K34 and degraded soon after fertilization or activation of Xenopus eggs (36, 37).
ERK7, despite being expressed at mRNA levels comparable to those of ERK2, is present at considerably lower levels in tissues and cultured cells than ERK2; in part this is because, ERK7 is constitutively active, and is rapidly degraded by a Skp1-Cullin-F box (SCF) complex (38). The 20 residues at the N terminus of the kinase domain are a primary determinant of ERK7 degradation (38). Like ERK7, ERK3 is also an unstable protein and is constitutively degraded by the UPS; this is independent of its kinase activity but dependent on two regions in the N-terminal lobe of its kinase domain. ERK3 accumulates during cell differentiation, and expression of stabilized forms of ERK3, in which part of its N-terminal lobe is replaced by the corresponding ERK1-derived sequences, causes G1 arrest (33).
The prototype MAP3K, Ste11 in Saccharomyces cerevisiae, mediates mating and high-osmolarity glycerol and filamentous growth responses. Ste11 phosphorylates and activates Ste7 (MKK), which, in turn, phosphorylates and activates two MAPKs, Fus3 and Kss1. Ste11, Ste7, Fus3, and Kss1 form a complex with the scaffold protein Ste5 in the mating pathway (39). Pheromone stimulation results in UPS-dependent degradation of Ste11 and Ste7. This requires Doa4 (Ubp4p) and Ubp3, respectively (40, 41), which are deubiquitinases (DUBs) that remove and recycle Ub from ubiquitinated proteins before degradation. Ste5, which is a scaffold that assembles the kinase components of this pathway, has a RING finger domain, and this might play a role in the ubiquitination of Ste11 or Ste7. In Drosophila, the expression of Wallenda, a MAP3K homologous to vertebrate DLK and LZK, is regulated by a RING finger protein, highwire (42).
Thus, activated MAPK signaling can be downregulated by UPS-mediated protein degradation at several levels.
Phosphatidylinositol 3-kinase, AKT, and mTOR
Phosphatidylinositol 3-kinase (PI3-K) is a lipid kinase consisting of an 85-kDa regulatory subunit bound to a 110-kDa catalytic subunit. PI3-K catalyzes the production of phosphatidylinositol-3,4,5-trisphosphate and thereby contributes to the activation of various signaling components involved in the regulation of gene expression and cell survival. Tyrosine-phosphorylated c-Cbl, whose identification as the first RING-type E3 ligase led to the idea that other RING finger-containing proteins may have the same function (43, 44) (Figure 2), interacts with, ubiquitinates, and degrades the p85 subunit in multiple experimental systems (45, 46). In addition, Cbl-b has been shown to downregulate bone formation through suppression of insulin-like growth factor (IGF)-1 signaling, including degradation of PI3-K and AKT in osteoblasts (47). p85 was also shown to be ubiquitinated by Cbl-b in a T-cell system, dependent on an interaction between a distal C-terminal proline-rich region in Cbl-b and the c-Src homology (SH)3 domain of p85. In this case, however, the ubiquitination of p85, instead of leading to p85 degradation, inhibits the recruitment of p85 to CD28 and to T-cell antigen receptor (TCR)ζ and CD28 (45).
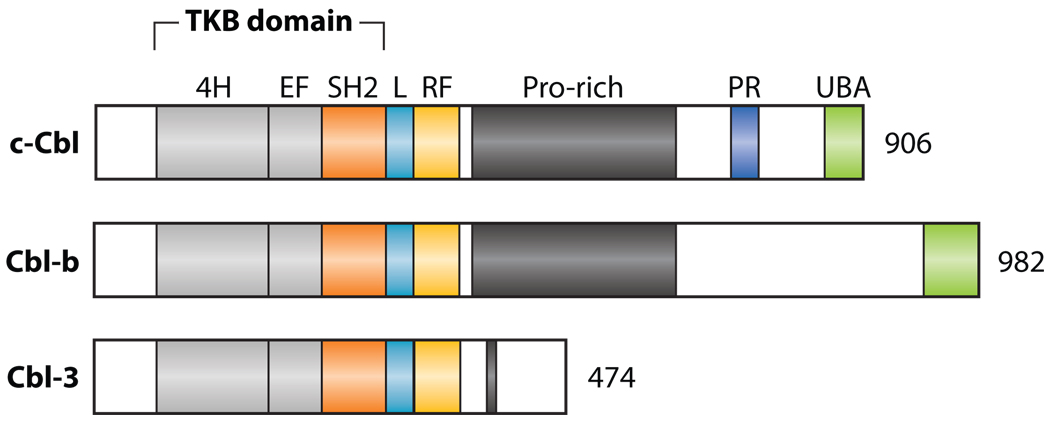
Figure 2.
Cbl protein family in mammals [c-Cbl, Cbl-b, and Cbl-3 (also known as Cbl-c)]. The Cbl proteins contain, from N to C terminus, a TKB domain, a linker region (L), RING finger domain (RF), Pro-rich regions, poly-Pro-Arg motif (PR), and UBA domain. The TKB domain consists of a four-helix bundle (4H), EF hand (EF), and a variant Src homology region 2 (SH2) domain. Cbl-3 lacks the PR motif and UBA domain.
AKT/protein kinase B is one of the major downstream targets of PI3-K. Platelet-derived growth factor (PDGF) and IGF-1 activateAKT in vascular smooth muscle cells accompanied by rapid degradation of AKT mediated by the UPS. Downregulation of AKT requires PI3-K activity but not intrinsic AKT activity (48). Deprivation of vascular endothelial growth factor (VEGF), inhibition of mTOR, or treatment with TNF-α results in both caspase-and proteasome-dependent AKT degradation (49, 50). Furthermore, AKT is preferentially degraded by the UPS in neuronal dendrites during the establishment of neuronal polarity, which is accelerated by inhibition of AKT activity (51). Cbl-b deficiency stabilizes the expression of AKT in osteoblasts during denervation, supporting a role for Cbl-b in AKT degradation. Undoubtedly, this important and conserved signaling pathway will prove to be regulated through ubiquitination at many levels, and a recent report indicates that mTOR, downstream of Akt, is degraded in a phosphorylation-dependent manner through SCFFbw7-mediated ubiquitination (52).
NIK
NF-κB-inducing kinase (NIK), a highly labile Ser/Thr kinase, is a critical regulator of the noncanonical NF-κB pathway. NIK phosphorylates and activates IKKα, which, in turn, phosphorylates the NF-κB2 gene product p100. This subsequently results in processing of p100 to p52, an important functional member of NF-κB family. NIK is ubiquitinated and degraded by the RING finger E3 ligases, inhibitor of apoptosis (IAP) proteins c-IAP1 and c-IAP2, in a RING finger–dependent manner (53). c-IAP1 and c-IAP2 interact, via their baculovirus IAP repeat (BIR) domains, directly with TRAF2 and are recruited to TNF receptor 1- and 2-associated complexes, where they regulate receptor-mediated apoptosis (54). A c-IAP1 mutant that cannot associate with TRAF2 fails to affect NIK levels, suggesting that TRAF2 provides a critical scaffolding link between the E3 ligase c-IAP1 and its substrate NIK. Treatment with IAP antagonists or the TNF family cytokine TWEAK results in autoubiquitination and rapid proteasomal degradation of c-IAPs and leads to a remarkable increase in the levels of NIK, which initiates p100 processing to NF-κB2 (53).
In addition to regulating NIK stability co-ordinately by c-IAP and TRAF2, TRAF3 also interacts with NIK and targets proteasomal degradation of NIK, whereas noncanonical NF-κB stimuli induce degradation of TRAF3 and elevated NIK expression (55). TRAF3 deficiency results in NIK accumulation and constitutive noncanonical NF-κB activity. The rescue of early postnatal lethality of TRAF3 deficiency in mice by compound loss of the p100 gene further indicates that TRAF3 is a critical negative modulator of the noncanonical NF-κB pathway (56).
Serum- and glucocorticoid-induced protein kinases
Serum- and glucocorticoid-induced protein kinase (SGK, also referred to as SGK1) is a stress-induced Ser/Thr kinase that plays a critical role in insulin signaling, cation transport, and cell survival. SGK is ~50% homologous in the catalytic domain with AKT, cAMP-dependent protein kinase, and PKC, and can be phosphorylated and activated through a PI3-K-dependent signaling pathway. Steady-state SGK is rapidly degraded by the UPS. SGK degradation is independent of the catalytic activity and activation site phosphorylation; it requires a hydrophobic motif (GMVAIL; residues 19–24) and six lysines surrounding the GMVAIL motif. The hydrophobic motif is also necessary for SGK localized to the endoplasmic reticulum (ER), where SGK interacts with and is ubiquitinated by a cochaperone and Ub ligase, C terminus of Hsp70-interacting protein (CHIP) (57, 58). The HECT domain Nedd4-2 E3 is another E3 that acts as a negative regulator of SGK. Moreover, SGK phosphorylation of Nedd4-2 potentiates Nedd4-2-mediated SGK ubiquitination and degradation (59).
Nonreceptor Tyrosine Kinases
Nonreceptor tyrosine kinases play a key role in transducing signals from surface receptors following ligand binding. Attenuation of receptor signaling is critical for precise control of cellular responses. This can be achieved by internalization and lysosomal degradation of the receptor proteins themselves, but there are also several mechanisms for downregulating nonreceptor tyrosine kinases following receptor-dependent activation, including dephosphorylation and UPS-mediated degradation. The Cbl family of RING finger E3 ligases play a particularly important part in ubiquitination of activated nonreceptor tyrosine kinases.
Src
Src family tyrosine kinases (Src, Fyn, Yes, Lyn, Hck, Fgr, Lck, and Blk) are important signal transducers that modulate a wide variety of cellular functions downstream of surface receptors, and their constitutive activation leads to cellular transformation. c-Src is activated by the dephosphorylation of Y529 (Y527 in avian c-Src) at the C terminus and is inactivated through phosphorylation of this residue by the carboxyl-terminal Src kinase (Csk) and Csk-type protein tyrosine kinase. In Csk−/− cells, activated c-Src molecules are ubiquitinated and degraded. In addition, active c-Src Y527Phenylalanine (Phe, F) mutant, but not a kinase-dead c-Src mutant, is ubiquitinated and degraded (60, 61). c-Cbl interacts with the Src SH3 domain through its proline-rich region, and phosphorylated Y419 (avian Y416) of c-Src through its TKB domain, mediating ubiquitination of active c-Src Y757F in vitro (62). Overexpression of c-Cbl or another family member, Cbl-3 induces the ubiquitination and downregulation of v-Src and suppresses v-Src-induced cell transformation (45).
Other Src family members are also regulated by Cbl. Src, Fyn, Yes, Lyn, and spleen tyrosine kinase (Syk) coimmunoprecipate with and phosphorylate c-Cbl, albeit at different levels (45, 63). c-Cbl interacts with the Lck SH3 domain, leading to Lck kinase activity–dependent ubiquitination of Lck. Consistently, c-Cbl–deficient T-cell lines have enhanced levels of active Lck, correlating with positive selection of CD4+ thymocytes and thymocyte maturation, both of which are Lck-dependent events (63). Both active Fyn and Lyn are substrates of Cbls, and increased expression of Fyn and Lyn are detected in c-Cbl−/− cells (63, 64). The expression levels of Lck and Lyn in thymocytes are elevated in mice with a loss-of-function mutation C379Alanine (Ala, A) in the c-Cbl RING finger domain, thus recapitulating the effects on Lck and Lyn in c-Cbl knockout mice (65). A membrane-anchored form of c-Cbl ubiquitinates and degrades Hck, reduces total cellular tyrosine phosphorylation levels, and inhibits Hck-induced cellular transformation (45).
c-Abl
c-Abl is another nonreceptor tyrosine kinase whose activity is tightly regulated. The N-terminal half of c-Abl is closely related in structure to Src family kinases. However, unlike Src, c-Abl is not regulated by an inhibitory phosphate that interacts with the SH2 domain, but rather a myristoyl switch involving the N-terminal myristoyl group that serves the same purpose. c-Abl is activated through autophosphorylation and transphosphorylation events. Active and Tyr-phosphorylated c-Abl is unstable and undergoes UPS-mediated degradation (66). c-Abl can be activated by several factors, including Src family kinases, integrin engagement, DNA damage, growth factors, and oxidative stress (67). The expression of v-Src, activated SrcY527F, or Fyn Y528F, which can phosphorylate and activate c-Abl, results in decreased c-Abl stability. Y245 and Y412 are the two major phosphorylated tyrosines on activated c-Abl, and phosphorylation of these sites significantly increases c-Abl activity in vitro (68). In the inactive Abl structure, the SH3-SH2 domains form a clamp in which the SH2 domain is docked onto the C-lobe of the kinase domain, and the SH3 domain binds to the PXXP motif of the SH2-kinase linker where the second Pro is replaced with Y245. Phosphorylation of Y245 will dislodge the SH3 domain and disrupt the assembled structure, and phosphorylation of Y412 in the activation loop will stabilize a conformation that is compatible with substrate binding and catalysis (69). Increased stability is observed in a double Y245/412F mutant of c-Abl but not in an activated mutant with open conformation in which the intramolecular interaction between the Pro-rich region and the SH3 domain is disrupted. This indicates that phosphorylation on Y245 and Y412 of c-Abl contributes to the conformational change and subsequent c-Abl degradation (66). Distinct from the way in which autophosphorylation regulates c-Abl protein stability, Y261 of Arg (the second member of the Abl family) can be autophosphorylated or transphosphorylated by c-Abl, and this phosphorylation both contributes to Arg kinase activity and blocks c- Arg degradation by the UPS in response to oxidative stress (70). The Arg-binding protein 2 (ArgBP2) adaptor protein, which is a substrate for both c-Abl and Arg, links c-Abl to c-Cbl, facilitates phosphorylation of c-Cbl by c-Abl, and promotes Cbl-directed ubiquitination and degradation of c-Abl (45).
Syk and ZAP-70
Syk and zeta-chain–associated protein kinase 70 (ZAP-70) are twin SH2-domain cytoplasmic tyrosine kinases with a highly conserved structure that are essential elements in cascades coupling immune receptors to intracellular responses. Syk is present in all hematopoietic cells, whereas ZAP-70 expression is restricted to T lymphocytes and natural killer (NK) cells. NK cells have a multimeric receptor complex consisting of the ligand-binding α subunit (CD16), which associates noncovalently with homodimers or heterodimers of the TCR-ζ and -γ chains. CD16 engagement on human NK cells rapidly induces recruitment of both Syk and ZAP-70 to the receptor complex, leading to their phosphorylation and activation, ubiquitination, and degradation (45, 63). Syk downregulation is also observed in human basophils in response to IgE stimulation (71).
Cbl has been implicated in the regulation of Syk and Lyn. The c-Cbl TKB domain binds to phosphorylated Y316 and Y323 of Syk and aspartate (D) 290 and Y292 of ZAP-70 (63). Nevertheless, enhanced phosphorylation and activation—but not protein levels—of ZAP-70 and increased Lck and ZAP-70 association are found upon crosslinking of CD3 and CD4 in thymocytes from c-Cbl C379A knockin mice (a loss-of-function mutation in RING finger) and c-Cbl knockout mice (65). These results suggest that Syk or ZAP-70 ubiquitination by c-Cbl/Cbl-b does not lead to their degradation and that c-Cbl or Cbl-b may function as a regulator of the interaction between Syk/ZAP-70 and a phosphatase or an upstream kinase (45, 63). Besides Cbl, it has been reported that the HECT domain of Nedd4 E3 ligases is required for ubiquitination of Syk and Lyn in cells infected with the latent membrane protein 2A of Epstein-Barr virus (72).
FAK
FAK is a ubiquitously expressed tyrosine kinase that is instrumental in integrating signals from integrins, which recognize extracellular matrix proteins, and receptor tyrosine kinases in processes, such as cell adhesion, survival, proliferation, and motility. Coordinated activation of integrins and the PDGF receptor results in increased expression of the suppressors of cytokine signaling (SOCS)1 (also referred to as JAB and SSI-1) and SOCS3 and SOCS-FAK interaction, which depends on theSH2and kinase inhibitory region (KIR) domains of the SOCS protein and FAK Y397 phosphorylation. SOCS proteins interact with elongins B/C via the SOCS box, and thereby associate with cullin-5 and Rbx1, forming a complex with E3 ligase activity that recognizes target proteins through the SH2 domain of the SOCS subunit. Overexpression of SOCS1 and SOCS3 in fibroblasts inhibits kinase activity and tyrosine phosphorylation of FAK and promotes polyubiquitination and degradation of FAK in a SOCS box-dependent manner (73). Signal-transducing adaptor protein (STAP)2, acting together with c-Cbl, also negatively regulates FAKexpression in T cells. STAP2 associates with c-Cbl while concomitantly interacting with the FERM domain of activated FAK via an SH2-like domain in STAP2 and colocalizing with FAK at focal adhesions. STAP2 deficiency or reduced c-Cbl expression enhances FAK protein levels and cell adhesion, whereas STAP2 overexpression induces UPS-dependent FAK degradation and a decrease in integrin-mediated T-cell adhesion to fibronectin (74).
JAK
Many cytokines, hormones, and growth factors elicit their cellular functions through the Janus family of protein-Tyr kinase (Jak)-STAT pathways. Ligand-induced receptor dimerization results in auto- and transphosphorylation of JAK2 on its activation loop and stimulation of its catalytic activity. JAK2, in turn, phosphorylates and activates STAT family transcription factors (75). Activation of JAK2, either by cytokine stimulation or by oncogenic TEL-JAK2 fusion, induces polyubiquitination and degradation of Tyr-phosphorylated JAK2 (75). Phosphorylation of Y1007 in the JAK2 activation loop, which binds to the SH2 domain and an additional N-terminal 12 amino acids of SOCS1, is required for proteasomal degradation of JAK2. This binding also directly inhibits JAK2 kinase activity, probably through preventing the JAK2 substrates or ATP access to the catalytic pocket (75). Conversely, dephosphorylation of Y1007 by the SH2 domain-containing protein-tyrosine phosphatase (SHP)-2 stabilizes JAK2 (76). The C-terminal SOCS box of SOCS1 interacts with elongins B/C and Cul-2, which function as a Ub ligase together with Rbx1 in an SCF-type E3 complex (75). The functional recruitment of the Cul-2 complex through the SOCS box accelerates UPS-dependent degradation of JAK2 (75).
Cyclin-dependent kinases and other cell-cycle-related protein kinases
In addition to direct downregulation of protein kinases by UPS degradation, activated protein kinases can in some cases be shut down by destruction of their activating regulators or upregulation of their inhibitors. A good example of this type of regulation is the cyclin-dependent kinases (CDKs), which govern cell cycle progression. CDKs are activated by cyclin binding and inhibited by CDK inhibitors (CKIs). Although CDKprotein levels do not change significantly, the dynamic activities of CDKs during the cell cycle are regulated through expression, ubiquitination, and degradation of cyclins and CKIs (which are not covered in this review), both temporally and spatially, in addition to phosphorylation and dephosphorylation. Two major types of E3 ligases are involved in the regulation of cell cycle progression: the SCF complexes, which mainly control the transition from G1/S and G2/M, and the anaphase-promoting complex or cyclosome (APC/C), primarily required for mitotic progression and exit as well as G1/S progression.
SCF complexes
The SCF complexes consist of three invariant subunits—SKP1, CUL1 [cell division cycle (Cdc) 53p in yeast], and the RING finger protein Rbx1/Roc1 (Hrt1 in yeast)—as well as a variable component known as an F-box protein. F-box proteins, of which there are ~70 in humans (77), bind to SKP1 through their F box, and recognize substrates through other domains in the F-box protein (78). Three classes of F-box proteins are defined based on their substrate recognition motifs: FBL [Leucine (Leu)-rich repeat (LRR) domain] and FBW (WD40 repeats), which can recognize target degrons in a phosphorylation-dependent manner through their WD40 or LRR domains, and others, where the nature of the recognition domain and the degron that is recognized has only been identified in a few cases (79). SCF complexes, working in concert with the UBC3/Cdc34 E2 (which binds to Rbx1), control G1-S progression and target G1 cyclins (Cln1/2p in yeast and cyclins D and E in mammals) and CKIs (Sic1 and Far1 in yeast and p27 and p21 in mammals) for degradation (80, 81).
Cyclin D
The activation of CDK4/6 by cyclin D1 binding in response to mitogenic signaling results in phosphorylation of Rb, thereby initiating E2F-mediated gene expression and sequestration of p27, which is required for progression into S phase. Whereas free and nonphosphorylated cyclin D1 can also be ubiquitinated, glycogen synthase kinase (GSK)-3β-mediated phosphorylation of T286 of nuclear cyclin D1 during S phase results in its translocation to the cytoplasm, where it is ubiquitinated and degraded by the proteasome. Binding of cyclin D1 to CDK4 enhances T286 phosphorylation by GSK-3β and ubiquitination of cyclin D1 (82). Pin1, a cis/trans peptidyl-prolyl isomerase that acts only on phosphorylated Ser/Thr-Pro bonds, directly binds to cyclin D1 phosphorylated at Thr286–Pro, increases nuclear cyclin D1 levels, and stabilizes cyclin D1. Cyclin D1 expression is reduced in many tissues in Pin1−/− mice (83). Although whether cyclin D1 is a direct substrate of SCFSKP2 is not yet clear, T286-phosphorylated cyclin D1 is recognized and ubiquitinated during S phase by SCFFbx4/αB-crystallin, in which the heat shock protein αB-crystallin functions in concert with the Fbx4 F-box protein (84). Knockdown of either Fbx4 or αB-crystallin attenuates cyclin D1 ubiquitination, enhances cyclin D1 expression, and accelerates cell-cycle progression from G1 to S phase. In addition, GSK3β phosphorylates Fbx4 at S12, which leads to Fbx4 dimerization and cyclin D1 ubiquitination. The detection of mutations in the N-terminal regulatory domain including the GSK3β phosphorylation site in Fbx4 or reduced expression of Fbx4 and αB-crystallin in a subset of primary human cancers that overexpress cyclin D1 (84, 85) implies that impairment of SCFFbx4/αB-crystallin function is a potential mechanism underlying cyclin D1 overexpression in human cancer.
Cyclin E
Cyclin E accumulates at the G1/S-phase boundary, where it is required for activation of CDK2 and initiation of DNA replication, and is degraded as cells progress through S phase (86, 87). Both free and CDK2–complexed cyclin E are ubiquitinated and proteasomally degraded. Free cyclin E is ubiquitinated in a phosphorylation-independent manner by a CUL3-containing E3 complex (88). In contrast, cyclin E in association with CDK2 is ubiquitinated in a phosphorylation-dependent manner by a CUL1-containing SCF complex (86). The different tissue-expression patterns of upregulated cyclin E protein in CUL1−/− and CUL3−/− mouse embryos suggests that different E3s predominantly control the abundance of cyclin E in different tissues (88, 89). Human cyclin E in complex with CDK2 is phosphorylated on at least four different sites: T62 (murine T74), S372, T380 (murine T393), and S384; T380 and S384 are primarily phosphorylated by GSK3 and CDK2, respectively (86). Phosphorylation of cyclin E on all these residues except S372 is important for its binding to the WD40 domains of the Fbw7 F-box protein and the turnover of cyclin E mediated by SCFFbw7 (86). Cyclin E T393A or T74/393A knockin mice exhibited increased cyclin E stability and proliferative anomalies in hematopoietic and epithelial lineages, consistent with a failure to degrade cyclin E appropriately (86).
The three Fbw7 splice variants, α, β, and γ, form homodimers or heterodimers through the D domain, and Fbw7 dimerization enhances the turnover of a weakly associated cyclin E S384A mutant (86). Among the Fbw7 isoforms, SCFFbw7α serves as an essential cofactor for Pin1-dependent prolyl isomerization of cyclin E, which potentiates the ubiquitination of cyclin E by SCFFbw7γ. Knockout of Fbw7α or Pin1 elevated cyclin E expression (86). Consistently, highly expressed cyclin E accompanied with Fbw7 mutations that result in a nonfunctional polypeptide or protein have been detected in several types of human cancers (86). These findings suggest that deregulation of CDK activity, resulting from defective ubiquitination and degradation of their positive regulators, may contribute to tumor development. Cyclin E is also reportedly regulated by other E3 ligases, including SCFSKP2 (87).
Checkpoint kinase 1
Checkpoint kinase 1 (Chk1) is a major effector of S-phase checkpoint signaling during the cellular response to genotoxic stress and is regulated by SCF complexes. Replicative stress induces polyubiquitination and degradation of Chk1, which is dependent on phosphorylation of Chk1 at S345 by ATR. Cul1 and Cul4A selectively associate with Chk1 following genotoxic stress, which induces S345 phosphorylation. Co-expression of both cullins caused an additive increase in Chk1 ubiquitination, and depletion of both proteins had an additive effect on stabilization of Chk1, suggesting that the activation-dependent ubiquitination of Chk1 is carried out by an SCF containing either Cul1 or Cul4A (90). A Cul1 complex containing the Fbx6 F-box protein may in part be responsible for degradation of activated Chk1.
APC/C
Sharing remarkable structural similarities with the SCF complex, the APC/C is a large multisubunit E3 ligase complex, consisting of catalytic core components [including APC11 (an Rbx1-related RING finger protein) and APC2 (a CUL1-related scaffold protein)] as well as a variable component known as an activator (77, 91). Cdc20 and Cdh1 (also named HCT1), two known activators in mitotically cycling cells, and a meiosis-specific adaptor, Ama1 (in budding yeast), confer substrate specificity in the same way that F-box proteins do in the SCF complex, and recognize substrates containing D box, KEN box, A box, or O box motifs (92). In yeast two E2s act in sequence to promote the two steps in polyubiquitin chain assembly on targets of APC/C: Ubc4 (also known as UbcH5) monoubiquitinates APC/C substrates at multiple lysines, whereas Ubc1 (in budding yeast) or UbcH1/E2-25K (a human homolog of Ubc1) catalyzes K48-linked polyubiquitin-chain assembly on preattached Ubs in a manner dependent on the UBA domain in a Ubc1 (93).
APC/C regulates mitosis and exit from mitosis by ubiquitination and degradation of D box–containing mitotic cyclins (cyclin A and B in mammals) and other mitotic proteins (Figure 3). APCCdc20 is active from mid- to late mitosis, whereas APCCdh1 is activated at late mitosis, remains active through G1 phase, and is extinguished at the G1–S boundary (77). In vertebrates, binding of cyclin A to Cdc2/CDK1 is required for entry into mitosis, and association of cyclin A with CDK2 is required for progression through S phase. The activation of APCCdc20 at the onset of prometaphase occurs through CDK1-mediated APC/C phosphorylation and/or SCFβ-TrCP-mediated degradation of the APC/C inhibitor early mitotic inhibitor 1 (Emi1), following phosphorylation by polo-like kinase (Plk) 1 (94, 95). Further enhancement of APC/C activation can be achieved by UPS degradation of the CDK1 inhibitor p21CIP1 by APC/CCdc20, which forms a positive feedback for activation of CDK1 (96). The interaction of tumor suppressor Ras association domain family 1 (RASSF1A) with Cdc20 is another means of inhibiting APC/C, and the RASSF1A gene is commonly silenced in lung cancer and other sporadic tumors (97). Activation of APC/CCdc20 initiates the degradation of cyclin A, thereby inhibiting CDK1. A nondegradable cyclin A mutant with an N-terminal domain deletion maintains hyperactivated CDK1 and arrests cell cycling at mitosis (82). In contrast to cyclin B1, the proteolysis of cyclin A is not delayed by the spindle assembly checkpoint (98). The mitotic checkpoint complex (MCC) composed of Cdc20, Mad2, Mad3 (BubR1 in metazoans), and Bub3 prevents Cdc20 from activating APC/C, thus blocking anaphase onset. Mad2 stimulates the association between Mad3 and Cdc20 (99). Dissociation of Mad2 and BubR1 from Cdc20 mediated by UbcH10 and p31comet, a protein that binds Cdc20-bound Mad2, promotes Cdc20 to activate APC/C and exit from the mitotic checkpoint. This dissociation process involves Cdc20 multi-ubiquitination by APC/C and is counteracted by the deubiquitinating enzyme Ub-specific protease 44 (USP44) (100).
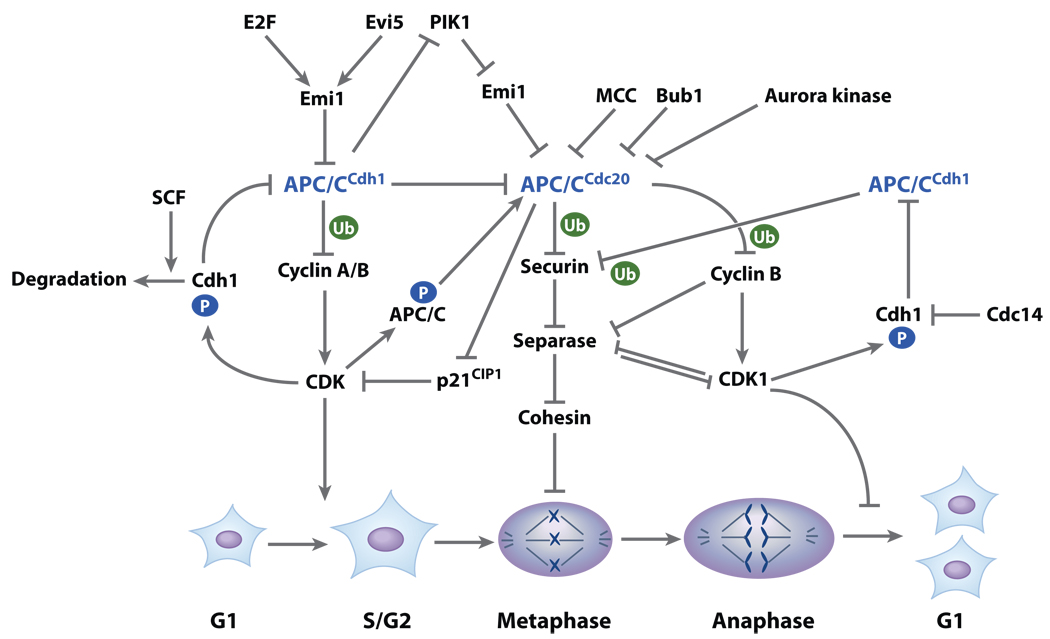
Figure 3.
Function and regulation of the APC/C E3 ligase during somatic cell cycle. At the G1-S transition, accumulated cyclin A and B dependent on the inhibition of APC/CCdc20 by APC/CCdh1 will activate CDKs and promote entry into S phase. In coordination with increased expression of Emi1 regulated by E2F and Evi5, activated CDKs phosphorylate Cdh1, inhibit APC/CCdh1, and cause subsequent accumulation of Plk1. During prometaphase and metaphase, APC/CCdc20 is activated by combinatorial factors including downregulation of APC/CCdh1, CDK1-dependent APC/CCdc20 phosphorylation, dissociation of MCC from APC/CCdc20, degradation of Bub1 and Aurora kinases by APC/CCdh1 in G1 phase, and proteolysis of Emil by Plk1-dependent phosphorylation. Further enhanced activation of APC/CCdc20 can be achieved by degradation of CDK1 inhibitor p21CIP1 by APC/CCdc20. Activated APC/CCdc20 initiates the degradation of cyclin A in prometaphase and cyclin B in metaphase and is involved in destruction of securin, which, in turn, reduces CDK1 activity and results in the dephosphorylation of Cdh1, in conjunction with Cdc14 phosphatase dephosphorylation, and activation of APC/CCdh1. Dephosphorylation of separase together with the APC/CCdh1-dependent proteolysis of securin (Pds1/Cut2) results in the activation of separase, cleavage of cohesin, and anaphase onset. Activated APC/CCdh1 orchestrates the degradation of a wide spectrum of substrates, facilitating mitotic exit and cytokinesis.
APC/CCdc20 and APC/CCdh1 mediate the degradation of cyclin B (Clb2 in yeast) (92, 101). Cyclin B degradation begins in metaphase and inactivates its binding partner CDK1, which allows the removal of inhibitory phosphate residues from Cys protease separase by protein phosphatases. The dephosphorylation of separase together with the APC/C-dependent proteolysis of the separase-binding protein securin (Pds1 in budding yeast/Cut2 in fission yeast) results in activation of separase, which, in turn, cleaves the cohesin protein complex. Dissolving cohesion between sister chromatids allows their separation (77, 92). Cyclin B binds to separase and inhibits its activity. Conversely, separase that is free of securin binds to cyclin B and inhibits CDK1 (102). Expression of a nondegradable cyclin B with a d-box mutation blocks CDK1 inactivation and exit from mitosis (103). The high levels of cyclin B and arrest in metaphase at the two-cell stage of embryos lacking Cdc20 function indicate an essential role of Cdc20 in degradation of cyclin B and in mitosis. Cdc20 and securin double-mutant embryos cannot maintain a metaphase arrest, suggesting that securin participates in preventing mitotic exit (104). Securin is ubiquitinated and regulated by both APC/CCdc20 and APC/CCdh1 (105).
The binding of dephosphorylated Cdh1 to the APC during mitotic exit mediates the destruction of Cdc20 (92). The dephosphorylation of Cdh1 can be a combinatorial consequence of inactivation of CDK1 by APC/CCdc20-mediated degradation of cyclin A and B and removal of inhibitory phosphorylation from Cdh1 by Cdc14 phosphatase (106). At the G1-S transition, Cdh1 is rephosphorylated by CDKs and disassociates from APC, allowing reaccumulation of cyclins until APC/C binds Cdc20 in the subsequent mitosis (92, 103) (Figure 3). Cdh1 can be phosphorylated at different sites in the same domain and inhibited by CDK1 and the CDK-related kinase Ime2. These two kinases overlap functionally as inhibitors of APC/CCdh1 and license chromosome replication during meiosis (107). Aside from inhibition of Cdh1 by rephosphorylation, Emi1 may also contribute to inhibition of APC/CCdh1. The expression of Emi1 is stimulated at the G1-S transition by the E2F transcription factor and stabilized in S/G2 phase by Evi5-mediated antagonism of SCFβ-TrCP-dependent Emi1 ubiquitination and destruction (100). In addition, inactivation of APC/CCdh1 can also be facilitated by degradation of its E2 UbcH10 by APC/CCdh1 (108), autoubiquitination of Cdh1 in G1 and G0 (109), and reduction of Cdh1 at S phase by SCF family-mediated ubiquitination (110).
Other APC/C-regulated protein kinases include Bub1, Aurora A/B, Nek2A, and Plk family members such as Plk1 (vertebrate), Polo kinase (Drosophila), Plx1 (Xenopus), and Cdc5 (budding yeast) (100, 111). Plk1 regulates both the Wee1 kinase and the Cdc25C phosphatase, which, in turn, control CDK1 activity at the G2 to M transition. Plk1 and cyclin B/CDK1 phosphorylate and inhibit Wee1, which inactivates CDK1 by phosphorylation of T15 and T14 on CDK1. Plk1 and cyclin B/CDK1 can constitute another autoactivating feedback loop by phosphorylation and activation of Cdc25C, which dephosphorylates and activates CDK1 (112). Plk1 and Cdc5 accumulate during late G2/M phase and their degradation is initiated at the start of anaphase, which is regulated by APC/CCdh1. Nondegradable Plk1 or Cdc5 causes a reduction in APC/C activity, a defect in the destruction of mitotic cyclins, and a delay in mitotic exit (113, 114). Bub1, which inhibits APC/CCdc20 activity by phosphorylating Cdc20, cooperates with Aurora kinase to promote binding of the mitotic checkpoint complex to the APC/CCdc20 (115, 116). The protein levels and activities of the Bub1 and Aurora kinases peak in mitosis and drop drastically in G1 through the action of APC/CCdh1 (117, 118).
Interplay between SCF complexes and APC/C
Cdh1 is actively proteolyzed during S phase. Depletion of Cul1 or expression of a dominant-negative Cul1 results in Cdh1 accumulation, implicating the SCF family in the degradation of Cdh1 at S phase (110). Conversely, both the SKP2 F-box protein and its cofactor Cks1 are unstable in G1 phase, ubiquitinated and rapidly degraded by APC/CCdh1 upon activation of transforming growth factor (TGF)β/Smad signaling (119–121). In contrast, depletion of Cdh1 in G1 cells stabilizes SKP2 and Cks1, with consequent degradation of p21CIP1 and p27Kip1 and an increased percentage of cells in S phase (119, 120). Phosphorylation of SKP2 by CDK2 and CDK1 stabilizes SKP2 by interfering with its recognition and degradation by APC/CCdh1, which is reversed by dephosphorylation by the mitotic phosphatase Cdc14B during the M to G1 transition (122). Expression of a stable SKP2 mutant that cannot bind APC/CCdh1 induces premature entry into S phase. Thus, the induction of SKP2 and Cks1 degradation in G1 represents a mechanism by which APC/CCdh1 prevents the unscheduled degradation of SCFSKP2–Cks1 substrates and maintains the G1 state (119, 120).
The importance of interplay between SCF complexes and APC/C is also exemplified by the regulation of Wee1. Wee1 family protein kinases that inhibit CDK1 during the G2 phase of the cell cycle are ubiquitinated and downregulated at the onset of mitosis (123). The F-box protein Tome-1 interacts with and ubiquitinates phosphorylated Wee1 in Xenopus egg extracts. Depletion of Tome-1 inhibits Wee1 degradation and mitotic entry; this mitotic entry defect can be rescued by removing Wee1, suggesting that Wee1 is a substrate of SCFTome-1. Tome-1, in turn, is a substrate of APCCdh1 and is degraded during the G1 phase, which allows Wee1 accumulation during interphase (124). In mammalian cells, SCFβ-TrCP, another SCF E3, appears to play a more critical role in Wee1 degradation and entry into mitosis than Tome-1; phosphorylation of S53, S121, and S123 of Wee1 A by Plk1, CK2, and CDK1, respectively, is required for direct binding to β-TrCP, ubiquitination, and degradation (125, 126). Dynamic control of Wee1 and Tome-1 expression by SCF and APC/C complexes contributes to the precise regulation of cyclin B/CDK1 activity and thus mitotic entry and exit.
RECEPTOR PROTEIN KINASES
Ligands bind to and thereby activate receptor protein kinases by inducing dimerization and trans-autophosphorylation, resulting in the activation of a plethora of cellular processes. The receptors must then be inactivated so that the signaling can be switched off. Defects in this inactivation process can lead to cancer. Activation–dependent receptor internalization and ubiquitination-dependent degradation, integrated with feedback from serine/threonine phosphorylation and tyrosine dephosphorylation of receptors, play key roles in switching off ligand-induced activation of receptor protein kinases. Following ligand activation and autophosphorylation, the major mechanism of receptor kinase downregulation depends on receptor ubiquitination, which triggers internalization and trafficking of receptors to the lysosome for degradation rather than for UPS degradation (Table S2). In addition, K63-linked polyubiquitination of the receptors may be required for targeting to lysosomes, as has been reported for EGFR (127). Although the mechanisms for degrading activated nonreceptor and receptor tyrosine kinases are different, the Cbl family E3 ligases play an important role in both systems. Mutations that abrogate interaction with Cbl are a common type of oncogenic mutation in receptor tyrosine kinases. The Nedd4 and Itch family HECT domain E3 ligases, which can associate with membranes via their C2 domains, also regulate multiple receptor kinases.
Receptor Tyrosine Kinases
Receptor tyrosine kinases comprise over half of the 90 tyrosine kinases in the human genome (1). Most of them bind to specific protein ligands, such as growth factors and cytokines, via their extracellular domain, which results in activation of the cytoplasmic catalytic domain upon ligand-mediated oligomerization and phosphorylation of cytoplasmic proteins, thereby transducing extracellular signals across the plasma membrane (3).
EGFR family
The EGF receptor family of tyrosine kinases has four members—EGFR, ErbB2, ErbB3, and ErbB4. EGFR, ErbB3, and ErbB4 bind ligands in the EGF family. EGFR, ErbB2, and ErbB4 are active kinases, whereas ErbB3 lacks catalytic activity. Upon ligand binding, these receptors can heterodimerize, thus generating distinct intracellular signals depending on the combination. For instance, ligand-induced interaction of any of the three active receptors in combination with ErbB3 results in activation of the PI3-K pathway (2).
EGFR ubiquitination by c-Cbl
Ligand-induced downregulation of active EGFR has been extensively studied. EGF induces ubiquitination and downregulation of EGFR. The finding that kinase-dead EGFR is resistant to ubiquitination and is degraded more slowly illustrates the importance of EGFR activation for priming its own degradation (128). Sli-1, the Cbl ortholog in Caenorhabditis elegans, was first identified as a negative regulator of the Let-23 receptor, an EGFR homolog (129). This suggested that c-Cbl could negatively regulate EGFR function. The c-Cbl protein has an N-terminal tyrosine kinase–binding (TKB) domain, a linker region, a RING finger domain, a RING finger C-terminal flank or tail domain, extensive Pro-rich regions, poly-Pro-Arg (PXXXPR) motifs, and a UBA/LZ domain in its C-terminal half. The TKB domain consists of three interacting domains: a four-helix bundle (4H), a Ca2+-binding EF hand, and a variant SH2 domain (130) (Figure 2). Mutation of the conserved Cys (RING) reduces c-Cbl–induced degradation and ubiquitination of EGFR (44), demonstrating an essential role of the c-Cbl RING finger in EGFR degradation. c-Cbl E3 activity is autoinhibited by the TKB domain and linker regions. c-Cbl phosphorylation at Y371 and Y368 may lead to a conformational change that relieves autoinhibition and activates c-Cbl (131). The c-Cbl UBA domain homo- or heterodimerizes with Cbl-b along a large hydrophobic surface formed by helices 2 and 3 and binds Ub at the first helix of the UBA. Cbl-b dimerization is regulated by Ub binding and is required for tyrosine phosphorylation of Cbl-b and ubiquitination of Cbl-b substrates (132, 133).
Upon EGF stimulation, phosphorylated c-Cbl is recruited to he cell surface before the EGFR is internalized (134–136). c-Cbl associates with the EGFR at the plasma membrane and colocalizes in late endosomes and multivesicular bodies (MVBs), suggesting that c-Cbl associates with EGFR throughout the endocytic route (137). The E2 ubiquitin-conjugating enzyme Ubc4/5 that cooperates with c-Cbl is relocated to the plasma membrane, and then to Hrs-positive endosomes, strongly suggesting that EGFR continues to be ubiquitinated after internalization (138). The kinase activity of EGFR and phosphorylation of EGFR at Y1045 that binds to the TKB domain of c-Cbl is necessary for ubiquitination of the EGFR (139, 140). In addition, EGF-induced Y1045 phosphorylation and ubiquitination of EGFR and c-Cbl phosphorylation at Tyr residues require p38 activity (141). The direct link of Y1045 to cancer was evidenced by identification of a truncated EGFR in human glioblastoma that had an intact kinase domain but was missing Y1045 and the internalization signals (142). Mutation of Y1045 in the constitutively active EGFRVIII, which lacks a large part of the extracellular domain and has been detected in various cancer types, inhibits its ubiquitination and downregulation by Cbl-b and enhances its ability to transform cells (143).
Mass spectrometry analysis has identified EGF-induced ubiquitination on six distinct lysines within the EGFR kinase domain, and mutation of Y1045 greatly reduced but did not abolish ubiquitination on these residues (127). The residual ubiquitination could be at-tributable to other E3 ligases or to c-Cbl recruited in a Y1045-independent manner. The latter possibility is supported by the observation that Grb2, which binds directly to phosphorylated Y1068 and Y1086 in activated EGFR, interacts with c-Cbl proline-rich domains via its SH3 domain and recruits Cbl to EGFR. In essence, Cbl may also be recruited to EGFR indirectly by Shc, which forms a complex with Grb2 and binds to phosphosphorylated Y1148 and Y1173 of EGFR (45, 144). Mutation of the six Lys residues in the kinase domain largely prevented EGFR ubiquitination and degradation but not its Tyr phosphorylation and internalization (127). Consistently, c-Cbl−/− cells show a deficiency in EGFR ubiquitination and degradation, but not in its internalization into early endosomes (145). Moreover, a kinase-dead EGFR mutant was resistant to ubiquitination but retained the ability to internalize EGF and escape degradation (146). These results strongly suggest that receptor ubiquitination by Cbl is not required for initial internalization but is required for sorting the receptors at the early endosome and transit to the lysosome for degradation instead of recycling back to the cell membrane. In addition to using Ub to regulate the EGFR, c-Cbl also mediates EGFR modification with NEDD8. EGF stimulates EGFR neddylation, which requires the RING and TKB domains of Cbl, as well as Y1045 of EGFR. EGFR neddylation, in turn, enhances the subsequent ubiquitination and sorting of EGFR for degradation. Multiple Lys residues within the EGFR kinase domain serve as attachment sites both for NEDD8 and Ub (147). These results suggest that EGFR neddylation, in coordination with ubiquitination, accelerates sorting of EGFR for lysosomal degradation.
EGFR can be phosphorylated and inhibited by ligand-induced activation of PKC. PKC-mediated phosphorylation at T654 inhibits ligand-induced phosphorylation, ubiquitination, and degradation of EGFR and diverts internalized EGFR from degradation to the recycling pathway (148, 149). Downregulation of EGFR can be inhibited by the binding of RALT to EGFR and promoted by the binding of the transmembrane protein LRIG1 to ERFR and c-Cbl, and the association between ACK1 and activated EGFR (45, 150, 151).
EGFR regulation by Cbl-b, Cbl-3, and oncogenic Cbls
Cbl-b and Cbl-3, the other two members of the Cbl family, also accelerate EGFR degradation (140). EGFR activation results in coordinated degradation of Cbl-b along with its substrate Shc and Grb2, which requires an intact RING finger and the TKB domain of Cbl-b and binding of Cbl-b to the activated EGFR (152). Notably, activated EGFR interacts with Shc and Grb2 not only at the plasma membrane but also in the endosomal compartment, where EGF induces augmented recruitment of Shc and Grb2. This implies that the three proteins remain associated during the early phase of the endocytic process (153, 154). Nedd4 and Itch, WW domain HECT E3s, also bind Cbl-b and target it for proteasomal degradation. Nedd4 reverses Cbl-b’s effects on EGFR ubiquitination and downregulation of EGFR as well as active c-Src (155), thus providing an additional level of indirect regulation for Cbl substrates. c-Cbl and Cbl-b have important overlapping functions including in ubiquitination and degradation of activated EGFR, and c-Cbl- or Cbl-b-deficient mice are viable and fertile, but the loss of both is embryonically lethal (156, 157).
v-Cbl, a truncated protein consisting of only the first 355 amino acids, and 70Z-Cbl, a deletion mutant lacking 17 internal amino acids that overlap the RING finger, are two RING finger–defective oncogenic forms of c-Cbl (44, 139). However, RING finger mutations in c-Cbl that abolish E3 ligase activity are insufficient for cell transformation. In contrast, mutations within a highly conserved α-helical structure linking the SH2 and RING finger render Cbl proteins oncogenic. The linker helix mutations also cause a loss of ubiquitination and downregulation of EGFR, possibly through disruption of contacts with both the TKB domain of Cbl and the Ubc (158). Thus, in addition to its pivotal role in E3 function, the linker helix of c-Cbl is a critical determinant of Cbl’s ability to transform cells possibly through precisely regulating the position and orientation of the SH2-bound substrate relative to the RING finger-bound E2 enzyme (159). The other possible explanation is that linker helix mutations allow the TKB domain of oncogenic Cbl to bind with higher affinity to targets, like EGFR, than the c-Cbl TKB domain does, and therefore compete more effectively for EGFR binding. As a result, the negative regulation of endogenous c-Cbl on receptor tyrosine kinases is much reduced.
EGFR in the early phases of endocytosis
Whereas the SH2 domain and RING finger are critical for ubiquitination of EGFR, the C terminus including the poly-Pro-Arg motif of c-Cbl functions separately from its Ub ligase activity, interacting with SH3 domains at the N terminus of CIN85 and recruiting the constitutively associated CIN85-endophilin complex to the activated EGFR. Endophilin, which binds to CIN85 via its SH3 domains, modulates plasma membrane invagination during the early steps of endocytosis (144). Sprouty2 forms a tertiary complex with CIN85 and c-Cbl, thereby preventing CIN85-mediated clustering of c-Cbl and blocking EGFR ubiquitination, whereas c-Cbl concurrently ubiquitinates Sprouty2 leading to degradation (144). The interplay between c-Cbl and Sprouty2 acts to fine-tune EGFR regulation. Likewise, the SH3 domain of β-Pix in complex with Cdc42 competes with CIN85 for binding to the poly-Pro-Arg motif of c-Cbl to inhibit c-Cbl-mediated EGFR downregulation. c-Cbl, in turn, promotes ubiquitination and subsequent degradation of β-Pix (45, 160). In addition, ALG-2-interacting protein X (Alix), a CIN85-interacting protein, and the UBA-and SH3-containing protein T-cell Ub Ligand (TULA) also inhibit EGFR ubiquitination by binding to CIN85 or c-Cbl (161).
EGF induces c-Cbl/Cbl-b-mediated monoubiquitination of CIN85 and its homolog CMS (p130Cas ligand with multiple SH3 domains). Impaired monoubiquitination of CIN85 does not affect Cbl-directed EGFR ubiquitination, but blocks EGFR internalization and delays receptor degradation (144). Consistent with the role of monoubiquitination in endocytosis, the endocytic proteins, Eps15, Eps15R, epsins, spartin, HGF-regulated substrate (Hrs) [yeast ortholog vacuolar protein sorting (Vps) 27], Sts1, Sts2, and Ymer are mono- or multiubiquitinated in response to EGF. Eps15 and other two UIM-containing proteins, Eps15R and Epsin, are indispensable for EGFR internalization (162, 163). The RING finger protein Parkin, whose mutation is responsible for a common familial form of Parkinson’s disease, and Nedd4 are involved in the monoubiquitination of Eps15 and Hrs (164). The monoubiquitination of Eps15 depends on the binding of UIMs in Eps15 to the Parkin Ub-like domain. Knockout of Parkin accelerates EGFR endocytosis and degradation, implying that Eps15 ubiquitination may interfere with the ability of the Eps15 UIMs to bind and control the downregulation of ubiquitinated EGFR (165, 166). Monoubiquitination of UBD-containing proteins Eps15, Sts1, Sts2, and Hrs results in inhibitory intramolecular interactions between Ub and their UBD, thereby preventing them from binding in trans to ubiquitinated targets (167). Thus, monoubiquitination of UBD-containing proteins can terminate their interaction with the ubiquitinated receptor complex and allow other UBD-containing proteins to bind the ubiquitinated cargo as they move along the sorting pathway (168).
Among the regulators for EGFR endocytosis, Hrs and signal-transducing adaptor molecule (STAM), each containing a UIM domain as well as a VHS (Vps27, Hrs, and STAM) domain for STAM, associate with each other and localize on the cytoplasmic face of the early endosomal membrane. The Hrs-STAM complex is proposed to be the sorting receptor that recognizes the Ub moieties of activated receptor tyrosine kinases and introduces them into MVBs from early endosomes. Upon EGF stimulation, Hrs is phosphorylated on Y329 and Y334 in a Cbl E3 ligase–dependent manner, and phosphorylation of Hrs is required for the joint degradation of EGFR and Hrs. Depletion or deficiency of Hrs or STAM inhibits receptor tyrosine kinase downregulation in mammalian cells (144, 161).
Both clathrin-dependent and lipid raft-dependent endocytic routes have been proposed for ubiquitinated EGFR molecules (144). EGFR internalized via clathrin-dependent endocytosis is not targeted for degradation, but instead is recycled to the cell surface. By contrast, lipid raft-dependent internalization preferentially commits the receptor to degradation (169).
EGFR in the late phases of endocytosis
Precisely how ubiquitination contributes to receptors being targeted for lysosomal degradation, instead of being recycled back to plasma membrane, is still a puzzle. Recent studies of protein sorting in MVBs in yeast and mammals may provide a link between ubiquitinated protein complexes and their final destination, the lysosome. MVBs are a subset of late endosomes; their typically multivesicular appearance results from the invagination and budding of the limiting (outer) membrane with associated cargo proteins into the compartmental lumen. Fusion of the limiting membrane of the MVB with the lysosomal membrane leads to the lumenal vesicles and their contents being degraded in the lysosome (vacuole in yeast). Membrane proteins, including nonubiquitinated proteins that are excluded from these inner lumenal vesicles and remain in the limiting MVB membrane, can ultimately be transferred to the limiting membrane of the lysosome, recycled back to the plasma membrane, or transported to other sites in the cell. Attachment of a single Ub to non-MVB cargo proteins is sufficient to drive these proteins into the MVB pathway (170). Consistently, ubiquitinated EGFR is sorted with EGF into MVB lumenal vesicles, which are degraded in the lysosome following fusion of the limiting membrane of the MVB with the lysosomal membrane (171). Kinase-dead EGFR mutants, which lack c-Cbl–mediated ubiquitination and degradation, are still able to internalize and reach the limiting MVB membrane, but are excluded from lumenal MVB vesicles and predominantly recycled back to the plasma membrane (139, 140, 146). This suggests that kinase activity and c-Cbl-dependent ubiquitination of EGFR and/or EGFR-associated monoubiquitinated endocytic proteins play a key role for EGFR being delivered to the lumenal MVB vesicles for subsequent degradation.
Active EGFR is dephosphorylated and loses its activity before localizing to late endosomes/ lysosomes (172, 173). Inhibition of proteasome activity or prelysosomal/lysosomal proteolysis reduces EGFR degradation, indicating the involvement of both the proteasome and lysosomes (140). Furthermore, proteasomal inhibitors inhibit both translocation of activated EGFR from the outer limiting membrane to inner membranes of MVBs and the decline in the level of ubiquitinated EGFR, implying that proteasomal activity is required for lysosomal sorting of EGFR and deubiquitination of the EGFR before its lysosomal degradation (134, 174).
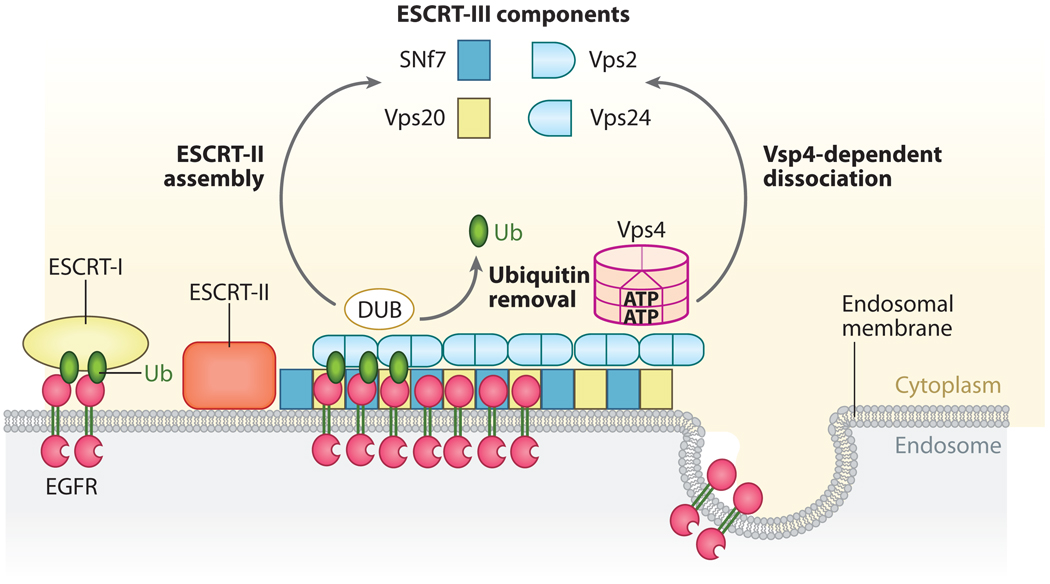
Efficient transport via endosomes relies on endosomal sorting complex required for transport (ESCRT) complexes. In yeast, ESCRT-I, which is recruited transiently to the endosomal membrane from the cytoplasm, is a conserved 350-kDa protein complex composed of Vps23 (mammalian ortholog tumor susceptibility gene Tsg101), Vps28, and Vps37, which are members of the 17 known class E Vps gene products (170). Hrs, in addition to its function in early endosome sorting, binds to Tsg101 via a PSAP motif and mediates the initial recruitment of ESCRT-I to endosomes, thereby indirectly regulating MVB formation. The Ub-conjugating-like domain of Tsg101/Vps23 is required for ESCRT-I to bind Ub in vitro, implying a direct role for Tsg101/ESCRT-1 in the recognition and sorting of ubiquitinated MVB cargo proteins (175). The Golgi-localized, γ-ear-containing, Arfbinding (GGA) protein binds Tsg101 as well as Ub via its VHS and GAT (GGA and TOM) domains and may coordinate with Tsg101 in sorting EGFR. In addition, ubiquitination and degradation of Tsg101 by two E3 ligases, Mahogunin and Tsg101-associated E3 ligase (Tal), contribute to regulation of EGFR degradation (175, 176).
Other ESCRT complexes, such as ESCRT-II (a 155-kDa complex composed of Vps22, Vps25, and Vps36) and ESCRT-III whose endosomal recruitment depends on ESCRT-II, act further downstream to regulate protein sorting in the MVB pathway. ESCRT-III is composed of two functional subcomplexes: a membrane-proximal subcomplex that consists of Snf7/CHMP4a and Vps20/CHMP6, and a peripheral associated subcomplex that consists of Vps2/CHMP2a and Vps24//CHMP3 (177). The Snf7 and Vps20 subcomplex-dependent association of Vps2 and Vps24 with the endosomal membrane results in recruitment of Ub isopeptidases or DUBs as well as Vps4 (in yeast) or SKD1 (in mammals), an AAA-type ATPase that has a role in catalyzing the dissociation of all three ESCRT complexes from endosomes (144, 170, 175). The interaction of CHMP3 with associated molecule with SH3 domain of STAM (AMSH) and recruitment of AMSH to the MVB is necessary for deubiquitination and degradation of EGFR. Besides, AMSH, other DUBs, such as the Ub-specific protease Y (UBPY) [also designated as Ub-specific protease 8 (USP8), an ortholog of the yeast Doa4, and AMSH-like protein (AMSH-LP)] can also remove Ub residues from target proteins before they enter the MVB and thereby recycle Ub to the cytoplasm (161, 175). How these DUBs coordinately deubiquitinate EGFR at the different stage of endocytosis is unclear.
Although further work is needed to define the precise molecular mechanisms of EGFR internalization and degradation, the accumulated evidence suggests the following model of EGF-induced EGFR endocytosis: EGFR that is autophosphorylated at Y1045 within its C-terminal domain recruits and interacts with the TKB domain of c-Cbl. c-Cbl associates with and mediates ubiquitination of the EGFR at several Lys and monoubiquitination of endophilin-bound CIN85 and ubiquitinated EGFR/c-Cbl/CIN85 forms a complex with the UIM-containing endocytotic proteins Eps15, Eps15R, epsins, and Hrs, which are monoubiquitinated by Nedd4 and Parkin, an event that may regulate the assembly/activity of various components of the endocytotic and sorting machinery. EGFR/c-Cbl complexes bound to endocytic proteins travel from early endosomes to late endosomes/MVBs, where Vps23/ESCRT-I recognizes the ubiquitinated EGFR and/or monoubiquitinated CIN85 and/or endocytic proteins. ESCRT-III-recruited DUBs remove the Ub from cargo proteins, thereby resulting in invagination of cargo protein into the lumenal vesicles of MVB. The translocation process of activated EGFR from the outer limiting membrane to the inner membranes of MVBs and deubiquitination of EGFR involves proteasomal activity that targets lumenal vesicles of MVB for fusion with lysosome for final degradation of EGFR (Figure 4 and Figure 5).
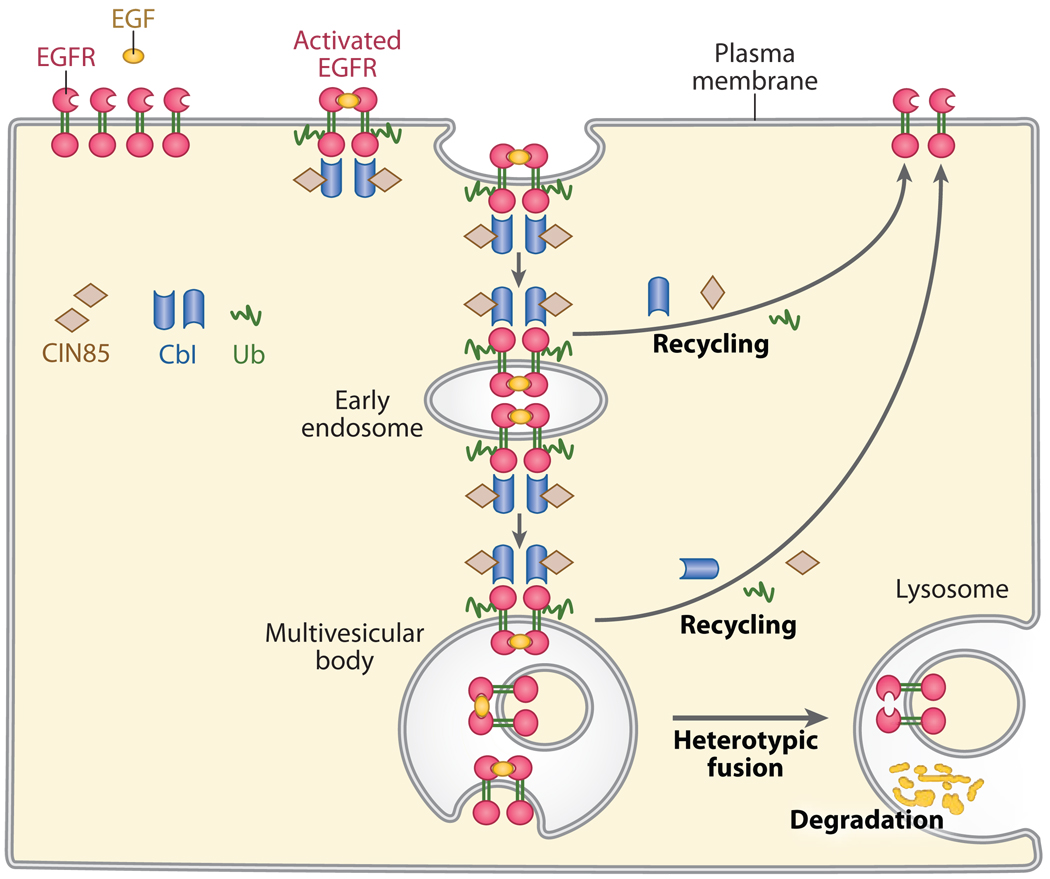
Figure 4.
An EGFR internalization model. Homodimerization and autophosphorylation of EGFR induced by EGF binding recruits and interacts with Cbl/CIN85 complex through the variant SH2 domain of Cbls. Cbls mediate the ubiquitination of EGFR and monoubiquitination of CIN85. EGFR/Cbl/CIN85 with other monoubiquitinated endocytic proteins such as Eps15, Eps15R, epsins and Hrs travel from early endosomes to late endosomes and MVBs. Subsequent invagination of cargo proteins into the lumenal vesicles of MVBs targets MVBs for fusion with lysosomes and then final degradation of EGFR. A certain proportion of EGFR will be deubiquitinated by DUBs and recycled back to the cell membrane.
Figure 5.
A model of ESCRT functioning in EGFR sorting. The UBC-like domain of Vps23, which is a component of ESCRT-I, likely binds to multiubiquitinated EGFR at the membrane of MVBs. ESCRT-II, acting downstream of ESCRT-I, is recruited, and then putatively directs the ESCRT-III complex (which is composed of Vps20, Snf7, Vps2, and Vps24) to the appropriate MVB membrane. ESCRT-III-recruited DUBs remove Ub from cargo proteins, resulting in the invagination of cargo proteins into MVB lumenal vesicles. Vps4 is required for the disassembly and release of the entire MVB sorting machinery, which allows the ESCRT machinery to recycle back into the cytoplasm for further rounds of MVB sorting (170).
Ligand-independent degradation of EGFR
SOCS36E is implicated as a negative regulator of the Drosophila ortholog of EGFR. SOCS5, the mammalian ortholog of SOCS36E, associates with EGFR and promotes EGFR degradation in a ligand- and c-Cbl-independent manner. EGFR degradation requires the SOCS box of SOCS5, which mediates the binding of the elongin B/C complex to recruit an E3 Ub ligase (178). Ligand-independent and constitutive endocytosis of the receptor involves Hrs. Hrs overexpression enhances the ubiquitination of EGFR and leads to translocation of EGFR to Hrs-containing endosomes, both of which depend on the integrity of the lipid-binding motif of Hrs. Hrs itself is polyubiquitinated, which depends on its UIM that is endowed two functions: binding ubiquitinated proteins and recruiting Nedd4 Ub ligase for self-ubiquitination. The polyubiquitination or depletion of Hrs negatively modulates the endocytosis and degradation of EGFR (179).
EGF versus TGFα
Different EGF family ligands do not elicit EGFR downregulation in an identical fashion. EGF induces more sustained phosphorylation and ubiquitination of EGFR and subsequently more efficient degradation than does TGFα, though both ligands recruit c-Cbl to the plasma membrane and cause the same extent of initial ubiquitination and endocytosis of the EGFR. EGFR and c-Cbl colocalize in endosomes after EGF but not TGFα treatment. Moreover, EGF, which is more acidic than TGFα, remains complexed to the EGFR within MVBs, sustaining EGFR kinase activity and triggering lysosomal sorting. In contrast, most TGFα is dissociated in MVBs, allowing the EGFR to recycle to the plasma membrane (134).
ErbB2, ErbB3, and ErbB4
ErbB2 is associated with poor prognosis when overexpressed in cancers. ErbB2 can be activated by EGF through the formation of heterodimers with EGFR or by neuregulin through heterodimerization with ErbB3/ErbB4. ErbB2 ubiquitination and degradation can be induced by EGF, neuregulin 1β (a neuregulin-1 isoform), and tumor-inhibitory ErbB2-specific antibodies (e.g., trastuzumab). The binding of such antibodies to ErbB2 recruits c-Cbl to Y1112 of ErbB2, and a Y1112F mutation retards antibody-induced ErbB2 degradation (144, 161, 180). The involvement of c-Cbl in ligand-induced ErbB family protein degradation requires kinase activity and tyrosine phosphorylation in the C-terminal tails of ErbB proteins, and only the membrane fraction of receptors is targeted by the ligand-dependent mechanism (161). In contrast, the class of ErbB2 tyrosine kinase inhibitors that alkylate a prominent Cys residue uniquely positioned in the nucleotide-binding pocket of ErbB receptors and inhibit kinase activity, and an Hsp90 inhibitor, the ansamycin antibiotic geldanamycin (GA), both induce UPS-mediated degradation of both mature and nascent ErbB2. Moreover, this process appears to be mediated by an E3 ligase, CHIP, in complex with protein chaperones (181).
Nrdp1 (mouse FLRF) possesses an atypical RING finger domain and associates with and stimulates ErbB3 ubiquitination and degradation (161). In the presence of the ErbB3 ligand neuregulin-1, AKT phosphorylates the USP8 DUB at T907 and contributes to USP8 stability, which, in turn, suppresses the ubiquitination and degradation of Nrdp1. The accumulated Nrdp1 promotes ErbB3 ubiquitination (161). Overexpression of Itch does not alter levels of unstimulated EGFR, ErbB-2, or ErbB-3, but it interacts with ErbB4 receptors via its WW domains and promotes its polyubiquitination and degradation (161).
PDGF receptor family
The PDGF family consists of four protein chains that form five biologically active dimers (PDGF-AA, -AB, -BB, -CC, and -DD). Two types of receptors for PDGF, α (which binds PDGFA, -B, and -C chains) and β (which binds PDGF-B and -D chains), have been identified. The PDGFR can be downregulated through reduced mRNA expression, a process regulated by the proto-oncoprotein c-Myc (182) and by the immediate internalization and ubiquitination of the receptor. PDGFRβ was the first receptor tyrosine kinase found to be polyubiquitinated and degraded (183). A kinase-deficient or a C-terminal autophosphorylation site (Y1009/1021F) PDGFRβ mutant exhibits reduced ligand-induced ubiquitination and degradation and amplified mitogenic activity (184). In response to ligand stimulation, the c-Cbl TKB domain interacts with PDGFRα as well as phosphorylated Y1021 of PDGFRβ, a known phospholipase C (PLC)γ1 SH2 domain binding site. The TKB domain as well as an intact RING finger are required for c-Cbl to polyubiquitinate the PDGFR (185). PDGFRα expression is elevated in the mammary glands of c-Cbl−/− mice and c-Cbl deficiency or ablation of the c-Cbl binding site on PDGFRβ enhances PDGF-induced PLCγ1 association with PDGFRβ and impedes receptor sorting to the lysosome (185, 186).
PDGFR, like EGFR, is regulated by proteins associated with receptors and c-Cbl. CMS interacts with both c-Cbl and the small GTPase Rab4 and is involved in the regulation of early endosome morphology. Expression of a truncated form of CMS that retains the ability to interact with Rab4 but not with c-Cbl inhibits ligand-induced PDGFRβ degradation (187). In addition, ligand-induced ubiquitination and degradation of PDGFRβ is inhibited by the interaction of c-Cbl with Alix or the low-density lipoprotein receptor-related protein 1 (LRP1) (45, 188).
Other members of the PDGFR family, the stem cell factor receptor c-Kit, and the CSF-1 receptor CSF-1R/c-Fms behave similarly in response to ligand stimulation. The ubiquitination and degradation of c-Kit is ligand-dependent and requires its intrinsic kinase activity and an involvement of c-Cbl (189, 190). c-Fms in c-Cbl–deficient macrophages, which have higher proliferation rates, is ubiquitinated to a lesser extent, internalized at a slower rate, and is degraded by the lysosomal but not a proteasome-dependent mechanism (191). Direct binding of the c-Cbl TKB domain to phosphorylated Y568 and Y936 of c-Kit or Y973 (mouse; equivalent to Y969 in the human homolog) of c-Fms is required for ligand-induced ubiquitination, internalization, and degradation of c-Kit or c-Fms (189, 192). Notably, Y568 and Y936 in c-Kit and Y973 in c-Fms are absent from oncogenic v-Kit and v-Fms, and deletion of c-Cbl binding sites from c-Kit greatly enhances their transforming ability (192, 193). Consistent with these observations, mutations of the Cbl binding site are frequently observed in c-Fms in human myelodysplasia and acute myeloblastic leukemia, further implicating loss of c-Cbl binding in oncogenic deregulation of c-Fms in human cancer (194, 195).
FGF receptors
In vertebrates, the 24 members of the FGF family have a high affinity for heparan sulfate proteoglycans and require heparan sulfate to activate one of four cell-surface FGFR tyrosine kinases. FGFs have diverse roles in embryonic development, bone morphogenesis, and tissue repair; are important for neuronal signal transduction in the central and peripheral nervous systems; and contribute to the pathogenesis of cancer when inappropriately expressed (196). Mutation at an autophosphorylation site (Y766) of FGFR1, which is essential for interaction with phospholipase C, results in decreased FGFR1 internalization and reduced ligand-induced FGFR1 degradation (197). Activated FGFR phosphorylates FRS2α, a docking protein containing a myristoyl anchor and a phosphotyrosine-binding (PTB) domain at the N terminus that associates with FGFR1 at the juxtamembrane region (amino acids 419–430), whereas the multiple tyrosine phosphorylation sites in the C-terminal tail of FRS2α serve as binding sites for adaptor proteins such as Grb2 and Shp2 (198–200). Grb2 constitutively binds to c-Cbl through both SH3 domains. This complex is recruited to the membrane and directly binds to the phosphorylated FRS2α through the SH2 domain of Grb2. This binding results in ubiquitination of FGFR and FRS2α itself in response to FGF stimulation. Thus, unlike EGFR and PDGFR, which form a direct complex with c-Cbl by way of its TKB domain, FRS2α functions as a link between FGFR and Grb2/c-Cbl complex. This contributes to the dual role of FRS2α for the recruitment of at least two multiprotein complexes, Grb2/c-Cbl and Grb2/SOS, which are responsible for both the activation and attenuation of signals. FGF-induced FGFR downregulation is only partially blocked in FRS2α−/− cells, indicating that attenuation of signaling by FGFR is regulated by redundant or multiple mechanisms (201). Several activating FGFR3 mutations, including G380R in the transmembrane domain as well as K650E and K650M associated with achondroplasia and thanatophoric dysplasia (TD) types II, may disrupt c-Cbl-mediated ubiquitination and degradation by altered conformation of receptor and allow the receptors to escape into a recycling pathway (202).
HGF receptor
Hepatocyte growth factor (HGF) [also known as scatter factor (SF)] binds and activates the widely expressed HGF receptor (HGFR)/Met receptor tyrosine kinase, mediating pleiotropic cellular responses. The human Met family has two members: Met (c-Sea in chickens) and RON (recepteur d’origine nantais) (203). Met is a cell surface–associated 190-kDa disulfide-linked heterodimer consisting of a 50-kDa α chain and a 140-kDa β chain. The α chain is entirely extracellular, and the β chain spans the membrane and possesses intrinsic, ligand-activated tyrosine kinase activity in its intracellular domain. In response to ligand binding, Met undergoes both multiple monoand polyubiquitination and degradation by proteasome- and lysosome-dependent mechanisms, which requires intact tyrosine kinase activity (203). Inhibition of proteasome activity does not affect initial HGF-dependent Met internalization. Instead, it promotes recycling from early endosomes at the expense of sorting to late endosomes, thereby ensuring the rapid return of internalized Met to the cell surface (204). Distinct from these reports, Met has also been shown to be downregulated in a proteasome-independent manner, which requires the formation of K48-linked polyubiquitin chains (205).
c-Cbl/Cbl-b can promote ubiquitination of the Met receptor, and this requires interaction of the c-Cbl/Cbl-b TKB domain with the phosphorylated juxtamembrane Y1003 residue in Met. Met juxtamembrane mutations that lead to decreased Cbl binding and prolonged Met protein stability and signaling have been identified in human lung cancer (206). An oncogenic form of Met, Tpr-Met, generated through chromosomal translocation of Met, and the oncogenic v-Sea receptor, derived from the chicken ortholog of Met, both lack the binding sites for c-Cbl and are not ubiquitinated (203). Although the Met Y1003F mutant is resistant to ubiquitination and degradation and is oncogenically active, it is internalized and undergoes endosomal trafficking with kinetics similar to those of WT Met (203, 207), implying that phosphorylation at Y1003 is critical to targeting Met to components of the lysosomal sorting machinery, whereas phosphorylation of other Tyr residues may be important for the early stages of Met endocytosis. In agreement with this, phosphorylation of Y1349 and Y1356 of Met is essential for ligand-induced Met internalization. Phosphorylated Y1356 is a site for indirect c-Cbl binding via Grb2. Depletion of Grb2 or abrogation of c-Cbl E3 activity blocks Met internalization, which can be rescued by expression of a chimeric protein in which the SH2 domain of Grb2 is tagged at the C terminus c-Cbl. These findings indicate that Cbl E3 ligase activity is critical for Met internalization and suggest a role for Grb2 as an intermediary link between Cbl and this process (203). Met internalization is mediated by clathrin-dependent endocytosis, and expression of a dominant-negative GTPase mutant of dynamin, which inhibits pinching off of invaginating coated pits, reduces the rate of ligand-induced Met receptor downregulation. In addition, as is the case for EGFR, a complex of Met receptor, endophilins, CIN85, Hrs, and Cbl controls the endocytosis of Met (203).
The steady-state level of Met can be regulated by the transmembrane protein LRIG1. Met interacts with the LRIG1 independent of HGF stimulation, and LRIG1 destabilizes Met in a c-Cbl- and proteasome-independent, but lysosome-dependent manner (208). In addition to the regulation of Met by ubiquitination, Met activity is negatively regulated at the transcription level by the RING finger protein Skeletrophin, whose direct relevant substrate is as yet unidentified (209).
Ron is a receptor for macrophage stimulating protein (MSP, also known as HGF-like protein or SF2) that belongs to the plasminogen-prothrombin gene family, which includes plasminogen and HGF among others (203). c-Cbl interacts with Ron at Y1017 (Met Y1003), Y1353 (the multifunctional docking site), and Y1360 (Grb2 binding site) and promotes receptor ubiquitination and internalization in a ligand- and Ron kinase activity-dependent manner. Phosphorylation at Y1017 and Y1353 is required for c-Cbl ubiquitination of Ron, whereas phosphorylation at Y1360 is more important for c-Cbl binding to Ron. In addition, CHIP can act as an E3 ligase for Ron ubiquitination and degradation in response to Hsp90 inhibition by GA (210, 211).
IGF-I receptor
IGFs, which include IGF-I and IGF-II, and their tyrosine kinase receptors (IGF-IR and IGF-IIR) exert potent mitogenic and differentiating effects on most cell types. IGF-IR is a heterotetrameric protein (α2β2) consisting of two identical α extracellular subunits containing a cysteine-rich IGF-binding site and two β transmembrane subunits bearing intrinsic tyrosine kinase activity. IGF-IR activation depends on an ATP-binding site (K1003) and a cluster of three Tyr residues at positions 1131, 1135, and 1136, which are autophosphorylated upon ligand binding (212). Exposure to IGF-I leads to phosphorylation of the IGF-IR β chain, which is subsequently ubiquitinated, internalized, and degraded, in a process mediated by Grb10 that connects IGF-IR to Nedd4; this requires both proteasome and lysosome activity in some cell lines but is dependent only on UPS in other cells (164). The RING finger protein Mdm2, an E3 ligase for p53, associates with and ubiquitinates ligand-unstimulated IGF-IR with involvement of β-arrestin1, the adaptor molecule to bridge Mdm2 and IGF-1R (213, 214). UV radiation-upregulated expression of p53 indirectly induces the expression of IGF-IR by sequestering Mdm2 in the nucleus, thereby reducing Mdm2-mediated IGF-IR degradation. Conversely, inhibition of p53 expression results in Mdm2-dependent ubiquitination and degradation of IGF-IR (213, 214). Also, the heat shock proteins Hsp10 and Hsp60 are involved in regulation of IGF-IR by inhibition of Ub conjugation on IGF-IR. Overexpression of Hsp10 and Hsp60 increases the abundance of IGF-IR by suppression of receptor polyubiquitination, IGF-I-stimulated receptor autophosphorylation, and activation of downstream signaling. Insulin induces expression of Hsp60, and diabetic myocardium is associated with decreased abundance of Hsp60, increased ubiquitination of IGF-1R, and reduced levels of IGF-1R protein (215, 216).
Receptor Serine/Threonine Kinases
In contrast to the large number of receptor tyrosine kinases, there are only 12 receptor serine/ threonine kinases (1). They are all members of a single extended family that binds ligands in the TGFβ family of cytokines and transduces signals that promote growth arrest and differentiation. Signaling by this type of receptor kinase is accomplished through the binding of aTGFβ family member, which induces the formation of a heterodimer containing a type ITGFβ receptor and a type II TGFβ receptor. Within this dimer, the constitutively active type II receptor phosphorylates the inactive type I receptor in the complex, and thereby activates it to phosphorylate downstream cytoplasmic target proteins (3).
TGFβ receptors
Heteromeric complexes of the TGFβ receptor type I, encoded by seven mammalian genes, and theTGFβ receptor type II, encoded by five mammalian genes, are transmembrane Ser/Thr kinase receptors. Twenty-eight members of theTGFβ superfamily, which includes TGFβ isoforms, activins, and bone morphogenetic proteins (BMPs), bind to heteromeric complexes of the receptors, which initiate signals controlling growth, differentiation, and apoptosis of cells and have important functions during embryonic development. Type I receptors, which are transphosphorylated and activated by type II receptors upon ligand binding, mediate specific intracellular signals. Type I receptors phosphorylate receptor-activated Smads (R-Smads including mammalian Smad1, -2, -3, -5, and -8) and initiate Smad signaling facilitated by the Smad anchor protein called Smad anchor for receptor activation (SARA) (217, 218). The inhibitory Smads (I-Smads) Smad6 and Smad7 can negatively regulate TGFβ signaling through association with activated type I receptors, thereby preventing phosphorylation of R-Smads (217, 218). Phosphorylated R-Smads form heteromeric complexes with common-partner Smads (Co-Smads) Smad4 and then translocate into the nucleus, where they modulate transcription either alone or, more commonly, by interacting with DNA-binding partners. Active R-Smads are continuously dephosphorylated in the nucleus, resulting in their dissociation from Smad4. Smad2/3 and Smad4 recycle back to the cytoplasm via a nuclear export receptor, using CRM1-independent and -dependent mechanisms, respectively. The activity of TGFβ receptors is dynamically controlled after ligand stimulation, and if the receptors are still active at the plasma membrane, the R-Smads are rephosphorylated, form complexes with Smad4, and return to the nucleus. If the receptors are no longer active, the R-Smads are retained in the cytoplasm (219, 220). These findings indicate that the Smads are constantly shuttling between the nucleus and the cytoplasm for the duration of active signaling and that the levels of receptors, which are regulated by ubiquitination, may directly determine how long a pool of activated Smads remains in the nucleus, which, in turn, may influence the biological response to TGFβ.
Type I receptor is sumoylated in response to TGFβ and its sumoylation requires the kinase activities of both type I and II receptors (221). Ligand-dependent endocytosis and downregulation of TGFβ receptor complex requires the kinase activity of type II but not type I receptors, even though the kinase activity of both receptors is required for TGFβ-induced signaling (222). This suggests that phosphorylation of the type I receptors or other substrates by type II receptors is necessary for efficient receptor trafficking and downregulation. Upon ligand stimulation, TGFβ receptors are internalized through both clathrin- and caveolae (lipid raft)-dependent mechanisms. Clathrin-dependent endocytosis internalizes receptors into early endosomes, where the Smad2 anchor SARA is enriched. Inhibition of this endocytosis blocks TGFβ-dependent signaling without affecting the half-life of the receptors. In contrast, the lipid raft domain mediates the internalization of Smad7-Smurf2 bound receptor, and interference with lipid raft function stabilizes the receptors and slightly enhances signaling and Smad2 activation. These results support the importance of clathrin-dependent endocytosis for TGFβ signaling through the Smad-dependent pathway, whereas lipid rafts are required for rapid degradation of TGFβ receptors (217).
TGFβ stimulates the expression of Smurf2, a C2-WW-HECT domain Ub ligase, via a PI3-K/AKT-dependent pathway (223). Nuclearly localized Smurfs bind to Smad7 and induce the nuclear export of Smad7, which depends on the nuclear export signal in a C-terminal region of Smurfs and CRM1 (224). The N-terminal conserved 2 (C2) domain of Smurfs associates constitutively with Smad7 primarily through an interaction with a PPXY sequence (PY motif) of Smad7. The recruitment of the complex, in which Smad7 functions as an adaptor, to the activated TGFβ receptor causes degradation of receptors and Smad7 through the catalytic activity of the Smurf2 HECT domain via proteasomal and lysosomal pathways (217, 218). Loss of Smurf1 does not affect Smad-mediated intracellular TGFβ or BMP signaling (35), implying a functional redundancy between endogenous Smurf1 and Smurf2. Acting similarly to Smurfs, the other two C2-WW-HECT E3 ligases, TGIF-interacting Ub ligase 1 (Tiul1)/WWP1 and Nedd4-2, also constitutively associate with Smad7 and induce degradation of the activated type I receptor as well as Smad2, subsequently inhibiting TGFβ and BMP signaling (217, 218).
In contrast to its role in complexing with an E3 ligase for degradation of TGFβ receptor, Smad7 can also act as an adaptor to recruit Ub C-terminal hydrolase 37 (UCH37), a DUB, to the type I TGFβ receptor for deubiquitinating and stabilizing the receptor (217). This finding suggests that competing effects of E3s and DUBs in complex with Smad7 can serve to fine-tune responses to TGFβ under various physiological and pathological conditions. The stability of Smad7 is also regulated by histone acetyltransferase p300-dependent acetylation. Acetylation of Smad7 at K64 and K70, which are also likely ubiquitination sites, or mutation of these Lys residues prevents ubiquitination and degradation of Smad7 mediated by Smurf1. Activation of TGFβ signaling decreases the acetylation of Smad7 and promotes Smad7 degradation (217), implying that acetylation and ubiquitination may compete at the same Lys residues for regulation of smad7.
The TGFβ signaling pathway is regulated at many other levels by ubiquitination, and this provides a good example of the complex manner in which signaling pathways are regulated by ubiquitination. For instance, in addition to degradation of TGFβ receptors and Smad7, Smurfs also exhibit E3 ligase activity toward other Smad proteins. When overexpressed, Smurf2 interacts with and ubiquitinates Smad1 and Smad2, leading to inhibition of their transcriptional activity. Similarly, overexpressed Smurf1 interacts with Smad1 and Smad5 in vitro, mediates their degradation in mammalian cells, and inhibits BMP signaling, thereby affecting embryonic pattern formation in Xenopus and BMP-induced osteogenic conversion (217, 218). Distinct from regulation of Smad2 in response to TGFβ signaling by UPS, Itch promotes Smad2 ubiquitination, facilitates complex formation between TGFβ receptor and Smad2, augments Smad2 phosphorylation, and enhances TGFβ-induced transcription without altering protein levels of Smad2, Smad4, and Smad7. Knockout of Itch results in reduced susceptibility to TGFβ-induced cell growth arrest and decreased phosphorylation of Smad2 (217, 218).
Aside from its role in desensitization of TGF signaling, Smurf2 also positively regulates TGF signaling by its involvement in downregulation of SnoN. SnoN and c-Ski, nuclear corepressor proteins of the Ski family regulating TGFβ-responsive genes, are rapidly ubiquitinated and degraded by Smurf2 in response to TGFβ signaling, which is mediated by Smad2-dependent interaction between Smurf2 and SnoN (217, 218). TGF stimulation also induces the assembly of the complex consisting of Smad2/3, SnoN, and APCCdh1, in which Smad3, and to a lesser extent Smad2, interact with both the APCCdh1 and SnoN. The APCCdh1 regulates ubiquitination and degradation of SnoN with the possible involvement of the E2 UbcH5 (218). Arkadia, a RING finger E3 ligase, is another E3 interacting with SnoN and c-Ski in their free state as well as in the forms bound to Smad proteins that downregulated the levels of SnoN and c-Ski proteins. This regulation requires formation of a complex between Arkadia and phosphorylated Smad2 or Smad3 (225, 226). Arkadia, cooperating with the scaffold protein Axin, is also involved in polyubiquitination and degradation of Smad7 (218). In addition to targeting three major negative regulators of TGFβ signaling—Smad7, SnoN, and c-Ski—Arkadia terminates TGFβ signaling by ubiquitinating and degrading phosphorylated and activated Smad2/3, which may promote a rapid resetting of TGFβ signaling after its activation. Arkadia−/− embryos have a similar phenotype to Smad2−/− embryos and cannot form foregut and prechordal plates (227). Thus, Arkadia fine-tunes TGFβ signaling by controlling both negative and positive regulators of this pathway.
Mass spectrometry analysis has shown Smad4 to be mono- or oligo-ubiquitinated at K507 of the MH2 domain. Mono- or oligoubiquitinated Smad4 has enhanced ability to oligomerize with R-Smads, whereas K507 mutation results in inefficient Smad4/R-Smad hetero-oligomerization and defective transcriptional activity. Thus, oligo-ubiquitination positively regulates Smad4 function (228). The importance of precise Smad4 regulation has been demonstrated in studies of early vertebrate development and tumor formation. TGFβ signals are involved in the formation of mesoderm and endoderm, whereas ectoderm specification requires attenuation of the TGFβ response. This is achieved by Ectodermin (Ecto), a RING finger E3 ligase, which binds and ubiquitinates Smad4, thus restricting the mesoderminducing activity of TGFβ/BMP signals to the mesoderm. Depletion of Ecto in human cells enforces TGFβ-induced cytostasis; overexpression of Ecto or missense mutations in the MH1 domain of Smad4, which leads to the proteasomal degradation of Smad4, has been found in colon cancer (217, 218). Misregulation of Smad4 is also exemplified in other types of tumors. Smad4 interacts with the F-box proteins β-TrCP1 and SKP2 present in the SCFβ-TrCP1 and SCFSKP2 E3 ligases, leading to ubiquitination and degradation. Smad4 mutants derived from pancreatic tumors or acute myelogenous leukemia exhibit increased interaction with β-TrCP1 or SKP2 and significantly elevated UPS-dependent degradation, suggesting that downregulation of Smad4 by mutationinitiated ubiquitination participates in tumor development (217, 218). Downregulation of Smad4 by Smurf1/2 and by Tiul1/WWP1 and Nedd4–2, which requires complex formation with Smad2 or Smad6/7 that act as adaptor proteins, is also observed (217, 218).
Aside from regulation by ubiquitination, endogenous Smad4 is sumoylated at K113 and K159 in the MH1 domain by the conjugating enzyme Ubc9 and by PIAS1 and PIASxβ, members of the PIAS family of SUMO ligases. Five PIAS members have been identified in mammals, specifically PIAS1, -3, -xα, -xβ, and -y. Sumoylation of Smad4 may serve to protect Smad4 from Ub-dependent degradation (229, 230).
In contrast to Smad4, Smad3 as well as Smad7 associate with PIASy, stimulating sumoylation of Smad3 and suppressing TGFβ-mediated Smad3 activation (231). In addition to sumoylation, Smad3 is ubiquitinated by the SCFβ-TrCP1 E3 complex and PRAJA, a RING-H2 protein, in response to TGFβ signaling (217, 218). Basal-expressed Smad3 as well as Smad1 and Smad4 can be directly ubiquitinated and degraded by CHIP independent of TGFβ signaling, whereas Axin facilitates GSK3β-mediated phosphorylation of Smad3, triggering Smad3 ubiquitination and degradation (217, 218). In addition to regulation by the UPS and sumoylation, Smad3 is transcriptionally repressed upon TGFβ1 activation, which may represent another feedback loop controlling TGFβ signaling (232).
In summary, TGFβ signaling is regulated through the UPS pathway by several E3 ligases at multiple levels, initiated from TGFβ receptors, downstream Smads including R-, Co-, and I-Smad, and the Ski nuclear corepressors SnoN and c-Ski. The temporal and spatial control of this signaling has a pivotal role in many cellular functions and in embryonic development. Dysregulation or mutation of TGFβ signaling components such as Smad4 can contribute to cancer formation.
G protein–coupled receptor signaling and GRK2
The GRK2 Ser/Thr kinase is a central regulator of G protein-coupled receptor (GPCR) signaling and a critical regulator of a wide variety of biological processes. GRK2 is a short-lived protein and is degraded by the UPS; this requires GRK2 activity and can be enhanced upon agonist stimulation of GPCRs such as β2-adrenergic receptor (AR), βAR, and CXCR. Interfering with GRK2 degradation potentiates GPCR desensitization. After agonist stimulation, the adaptor molecule β-arrestin binds to GPCRs and mediates GRK2 phosphorylation at Y13/86/92 by recruited c-Src, which facilitates phosphorylation of GRK2 S670 by ERK MAPK. Mutation of these phosphorylation sites retards GRK2 degradation (233, 234). β2AR activation promotes the binding of β-arrestin–mediated interaction between GRK2 and Mdm2. Mdm2-mediated ubiquitination of GRK2 at K19, K21, K30, and K31 is critical for GRK2 degradation. Moreover, IGF-IR activation enhances GRK2 stability by a mechanism involving the PI3-K/AKT pathway and the Mdm2 E3 ligase (235).
PROTEIN KINASE DEGRADATION MEDIATED BY PROTEIN KINASE–SPECIFIC CHAPERONES
Many protein kinases are associated with molecular chaperones, and the levels and activities of these kinases can be downregulated by the regulation of protein kinase/chaperone complexes. Among the many molecular chaperones, Hsp90 and Cdc37 are often observed in complex with signal-regulated protein kinases, and Cdc37 stabilizes client proteins following their interaction with Hsp90 and regulates protein kinase activity (236). Hsp90 associates with at least 105 protein kinases (237). Hsp90 recognizes a common surface in the N-terminal lobe within the kinase domain, and surface features, rather than a linear sequence motif, define the capacity of the Hsp90 chaperone machine to recognize client protein kinases (237, 238).
Heterocomplexes of protein kinases with Hsp90/Cdc37 are assembled by a multiprotein chaperone system comprising Hsp90, Cdc37, Hsp70, Hop, Hsp40, and p23 (239). The Hsp90 family members contain an ATP-binding pocket, and the binding and hydrolysis of ATP are required for the refolding and release of the native protein from the chaperone complex (239). Hsp90 inhibitors, including radicicol, herbimycin-A, GA, and its derivative 17-AAG, bind tightly to the Hsp90 ATP/ADP pocket, prevent ATP binding and the completion of substrate protein refolding, and lead to disruption of protein kinase/chaperones complex. As a result, treatment with these inhibitors leads to UPS-dependent degradation of Hsp90-Cdc37 client proteins, including EGFR, ErbB2, PDGFR, IGF-I R, PI3-K, AKT, Src, Lck, Lyn, Raf1, Bcr-Abl, and CDK4/6 (240).
The expression of the functionally defective mutant Cdc37-1 in S. cerevisiae, the fission yeast Schizosaccharomyces pombe, and Drosophila decreases the levels of the cyclin-dependent protein kinase Cdc28, Spc1 stress-activated protein kinase, and Aurora B, respectively (241). That a transcription factor CCAAT/enhancer binding protein-α (C/EBPα) disrupts a chaperone CDK4/Hsp90-Cdc37 complex via direct interaction with CDK4 and induces UPS-mediated CDK4 degradation (242) further supports the notion that a functional Hsp90-Cdc37 complex is required for the stability and function of target protein kinases.
In addition to the complex formed between protein kinases and Hsp90/Cdc37, the cochaperone CHIP interacts with Hsp70 and Hsp90 through an N-terminal TPR domain and also displays E3 ligase activity mediated by its a C-terminal U box domain (239). Cochaperone Bag family members (BAG1-6 in humans) have a C-terminal BAG domain that associates with the ATPase domain of Hsp70, stimulating ADP-ATP exchange and client protein release (239). Bag1 associates with the 26S proteasome through an N-terminal UBL domain as well as through specific ubiquitin moieties appended to Bag1 by CHIP, in which the individual ubiquitins are linked through K11. The noncanonical polyubiquitin chain does not induce the degradation of Bag1; instead, it stimulates UBL domain-dependent association of Bag1 with the proteasome (243). Bag1 accepts substrates that can be activated by Bag1, such as Raf-1, from Hsp70, and presents Raf-1 to CHIP for ubiquitination (244–246). Thus, Bag1 acts as a physical link between the Hsp70 chaperone system and the proteasome and recruits Hsp70 to the proteasome, resulting in trafficking of ubiquitinated client proteins to proteasomal degradation (239, 244). Similarly, Hsp70- and CHIP-mediated degradation of ErbB2 (181), apoptosis signal-regulating kinase 1 (ASK1) (247), SGK1 (57), and activated dual leucine zipper-bearing kinase (DLK) (248) has been observed.
The ability of Hsp90 inhibitors to downregulate cellular protein kinases has led to their being tested as potential antitumor agents (240). Ron is ubiquitinated in response to GA or 17-AAG by CHIP, which is recruited via Hsp90 and Hsp70. The constitutively active and oncogenic Ron M1254T mutant that escapes c-Cblnegative regulation upon MSP stimulation is recruited into the CHIP-chaperone complex even more efficiently than WT Ron is. Ron M1254T displays increased sensitivity to GA, enhanced interaction with Hsp90, and an increased degradation rate. The transforming potential evoked by Ron M1254T is abrogated upon Hsp90 inhibition (211). These findings imply that Hsp90 antagonists are suitable pharmacological agents for therapy for cancers in which altered Ron signaling is involved.
CONCLUDING REMARKS
The protein kinases encoded by the human genome regulate and maintain physiological responses and normal cellular functions, including cell growth, differentiation, adhesion, motility, and death. Protein kinase activity is regulated positively or negatively by phosphorylation/dephosphorylation, oligomerization, the binding of second messengers, activating subunits and inhibitor proteins, protein cleavage, and intracellular translocation, as well as by expression level. In the majority of cases, protein kinase activity is reversibly modulated through either phosphorylation/dephosphorylation or the binding of small molecule or protein regulators. However, it is increasingly apparent that activation of protein kinases, particularly if it is sustained, can initiate irreversible downregulation by UPS-mediated protein degradation, and that this can be an important mechanism of signal termination.
Several general principles apply to the recognition of activated protein kinases or their regulators for ubiquitination and degradation (249). First, autophosphorylation of a protein kinase can create binding sites for E3 ligases. In the case of receptor tyrosine kinases, ligand engagement results in phosphorylation of receptor tyrosines leading to recruitment of Cbl family E3 ligases either directly or indirectly via Grb2. Second, transphosphorylation of a protein kinase can create a phosphodegron that is recognized by a phospho-dependent SCF ligase (e.g.,Wee1 by Plk1 and CDK1, Chk1 by ATR, and cyclin E by GSK3 and CDK). Third, activation of a protein kinase results in an altered, active conformation that can be recognized by an E3 ligase. For instance, a common surface on the N-terminal lobe of the activated catalytic domain promotes CHIP-mediated ubiquitination (237, 238). In addition, activation may expose a degron outside the catalytic domain in the protein that is otherwise masked in the inactive state (e.g., Chk1). Finally, most protein kinases are targeted by more than one E3 ligase for ubiquitination, with one or more E3 ligases acting to provide constitutive turnover, and other E3 ligases recognizing the activated protein kinase, meaning that multiple E3 ligases often collaborate to terminate kinase action.
Disruption of ubiquitination that regulates protein kinase directly or indirectly and constitutive activation of the protein kinase inevitably lead to human diseases, including cancer. Mutations of protein kinases themselves, such as oncogenic mutants of EGFR and c-Fms, v-Fms, v-Kit, and Tpr–Met, which lack the autophosphorylation site that recruits c-Cbl (195), or the regulatory components of ubiquitination machinery such as oncogenic Cbl, in which a mutation abolishes the E3 Ub ligase activity, result in escape from the surveillance of this Ub-dependent downregulation. Constitutively active kinases can also elicit oncogenic effects through UPS-mediated downregulation of growth-inhibitory proteins and tumor suppressors. For example, BCR-ABL and v-Src tyrosine kinases trigger UPS-dependent destruction of the Abi proteins, a family of Abl-interacting proteins that bind to and antagonize the oncogenic potential of Abl; expression of Abi is lost in cell lines and bone marrow cells from patients with aggressive BCR-ABL-positive leukemias (250).
We can foresee that the accumulating knowledge about the mechanisms of ubiquitination and degradation that regulate protein kinases will permit development of potent and pharmacological modulators of these processes, which may afford a new approach for treating human diseases.
SUMMARY POINTS
The activation of protein kinases commonly initiates their downregulation via the ubiquitin/proteasome pathway.
Autophosphorylation of a protein kinase can create a binding site for an E3 ligase.
Transphosphorylation of a protein kinase can create a phosphodegron that is recognized by a phospho-dependent SCF ligase.
Activation of a protein kinase can result in an altered, active conformation that can be recognized by an E3 ligase.
Protein kinase activation can expose a degron outside the catalytic domain in the protein that is otherwise masked in the inactive state.
Multiple E3 ligases often collaborate to terminate protein kinase action.
Disruption of ubiquitination and degradation of protein kinases leads to human diseases, including cancer.
Supplementary Material
ACKNOWLEDGMENTS
We apologize to the colleagues whose work was not cited or discussed because of the limitation on the number of references. Z.L. is supported by National Cancer Institute grant 5R01CA109035, the Pediatric Brain Tumor Foundation, a Brain Tumor Society Research Grant, Phi Beta Psi Sorority Research Grant, and an institutional research grant from The University of Texas M. D. Anderson Cancer Center. T.H. is supported by National Cancer Institute grants CA80100, CA82683, and CA116402. T.H. is a Frank and Else Schilling American Cancer Society Research Professor.
Glossary
- Protein kinases
enzymes that modify other proteins by chemically adding phosphate groups to them
- Tyr, Y
tyrosine
- Ser, S
serine
- Thr, T
threonine
- phosphorylation
the addition of a phosphate (PO4) group to a protein or other organic molecule
- Ub
ubiquitin
- UPS
ubiquitin-proteasome system
- E3
Ub ligase
- Ubiquitination
the posttranslational modification of a protein by the covalent attachment of one or more ubiquitin monomers
- UBDs
Ub-binding domains
- UBA
Ub-associated domain
- UIM
Ub-interacting motif
- PHD
plant homeodomain
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 2.Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 3.Hunter T. Signaling–2000 and beyond. Cell. 2000;100:113–127. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- 4.Oda A, Druker BJ, Ariyoshi H, Smith M, Salzman EW. pp60src is an endogenous substrate for calpain in human blood platelets. J. Biol. Chem. 1993;268:12603–12608. [PubMed] [Google Scholar]
- 5.Karni R, Levitzki A. pp60(cSrc) is a caspase-3 substrate and is essential for the transformed phenotype of A431 cells. Mol. Cell Biol. Res. Commun. 2000;3:98–104. doi: 10.1006/mcbr.2000.0197. [DOI] [PubMed] [Google Scholar]
- 6.Gervais FG, Thornberry NA, Ruffolo SC, Nicholson DW, Roy S. Caspases cleave focal adhesion kinase during apoptosis to generate a FRNK-like polypeptide. J. Biol. Chem. 1998;273:17102–17108. doi: 10.1074/jbc.273.27.17102. [DOI] [PubMed] [Google Scholar]
- 7.Carragher NO, Fincham VJ, Riley D, Frame MC. Cleavage of focal adhesion kinase by different proteases during SRC-regulated transformation and apoptosis. Distinct roles for calpain and caspases. J. Biol. Chem. 2001;276:4270–4275. doi: 10.1074/jbc.M008972200. [DOI] [PubMed] [Google Scholar]
- 8.Asselin E, Mills GB, Tsang BK. XIAP regulates Akt activity and caspase-3-dependent cleavage during cisplatin-induced apoptosis in human ovarian epithelial cancer cells. Cancer Res. 2001;61:1862–1868. [PubMed] [Google Scholar]
- 9.Bachelder RE, Wendt MA, Fujita N, Tsuruo T, Mercurio AM. The cleavage of Akt/protein kinase B by death receptor signaling is an important event in detachment-induced apoptosis. J. Biol. Chem. 2001;276:34702–34707. doi: 10.1074/jbc.M102806200. [DOI] [PubMed] [Google Scholar]
- 10.Sun Y. Targeting E3 ubiquitin ligases for cancer therapy. Cancer Biol. Ther. 2003;2:623–629. [PubMed] [Google Scholar]
- 11.Pickart CM. Ubiquitin in chains. Trends Biochem. Sci. 2000;25:544–548. doi: 10.1016/s0968-0004(00)01681-9. [DOI] [PubMed] [Google Scholar]
- 12.Koegl M, Hoppe T, Schlenker S, Ulrich HD, Mayer TU, Jentsch S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 1999;96:635–644. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- 13.Jin L, Williamson A, Banerjee S, Philipp I, Rape M. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell. 2008;133:653–665. doi: 10.1016/j.cell.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 15.Deng L, Wang C, Spencer E, Yang L, Braun A, et al. Activation of the IκB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 16.Kirkin V, Dikic I. Role of ubiquitin- and Ubl-binding proteins in cell signaling. Curr. Opin. Cell Biol. 2007;19:199–205. doi: 10.1016/j.ceb.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 17.Hurley JH, Lee S, Prag G. Ubiquitin-binding domains. Biochem. J. 2006;399:361–372. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woelk T, Oldrini B, Maspero E, Confalonieri S, Cavallaro E, et al. Molecular mechanisms of coupled monoubiquitination. Nat. Cell Biol. 2006;8:1246–1254. doi: 10.1038/ncb1484. [DOI] [PubMed] [Google Scholar]
- 19.Al-Hakim AK, Zagorska A, Chapman L, Deak M, Peggie M, Alessi DR. Control of AMPK-related kinases by USP9X and atypical Lys(29)/Lys(33)-linked polyubiquitin chains. Biochem. J. 2008;411:249–260. doi: 10.1042/BJ20080067. [DOI] [PubMed] [Google Scholar]
- 20.Hartmann-Petersen R, Gordon C. Integral UBL domain proteins: a family of proteasome interacting proteins. Semin. Cell Dev. Biol. 2004;15:247–259. doi: 10.1016/j.semcdb.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 21.Ikeda F, Hecker CM, Rozenknop A, Nordmeier RD, Rogov V, et al. Involvement of the ubiquitin-like domain of TBK1/IKK-i kinases in regulation of IFN-inducible genes. EMBO J. 2007;26:3451–3462. doi: 10.1038/sj.emboj.7601773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuranaga E, Kanuka H, Tonoki A, Takemoto K, Tomioka T, et al. Drosophila IKK-related kinase regulates nonapoptotic function of caspases via degradation of IAPs. Cell. 2006;126:583–596. doi: 10.1016/j.cell.2006.05.048. [DOI] [PubMed] [Google Scholar]
- 23.Kang BS, French OG, Sando JJ, Hahn CS. Activation-dependent degradation of protein kinase C eta. Oncogene. 2000;19:4263–4272. doi: 10.1038/sj.onc.1203779. [DOI] [PubMed] [Google Scholar]
- 24.Lu Z, Hornia A, Jiang Y-W, Zang Q, Ohno S, Foster DA. Tumor promotion by depleting cells of protein kinase Cδ. Mol. Cell Biol. 1997;17:3418–3428. doi: 10.1128/mcb.17.6.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu Z, Liu D, Hornia A, Devonish W, Pagano M, Foster DA. Activation of protein kinase C triggers its ubiquitination and degradation. Mol. Cell Biol. 1998;18:839–845. doi: 10.1128/mcb.18.2.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakamura M, Tokunaga F, Sakata S, Iwai K. Mutual regulation of conventional protein kinase C and a ubiquitin ligase complex. Biochem. Biophys. Res. Commun. 2006;351:340–347. doi: 10.1016/j.bbrc.2006.09.163. [DOI] [PubMed] [Google Scholar]
- 27.Chen D, Gould C, Garza R, Gao T, Hampton RY, Newton AC. Amplitude control of protein kinase C by RINCK, a novel E3 ubiquitin ligase. J. Biol. Chem. 2007;282:33776–33787. doi: 10.1074/jbc.M703320200. [DOI] [PubMed] [Google Scholar]
- 28.Smith L, Wang Z, Smith JB. Caspase processing activates atypical protein kinase C zeta by relieving autoinhibition and destabilizes the protein. Biochem. J. 2003;375:663–671. doi: 10.1042/BJ20030926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okuda H, Saitoh K, Hirai S, Iwai K, Takaki Y, et al. The von Hippel-Lindau tumor suppressor protein mediates ubiquitination of activated atypical protein kinase C. J. Biol. Chem. 2001;276:43611–43617. doi: 10.1074/jbc.M107880200. [DOI] [PubMed] [Google Scholar]
- 30.Srivastava J, Procyk KJ, Iturrioz X, Parker PJ. Phosphorylation is required for PMA- and cell-cycle-induced degradation of protein kinase C delta. Biochem. J. 2002;368:349–355. doi: 10.1042/BJ20020737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blake RA, Garcia-Paramio P, Parker PJ, Courtneidge SA. Src promotes PKC δ degradation. Cell Growth Differ. 1999;10:231–241. [PubMed] [Google Scholar]
- 32.Lu Z, Xu S, Joazeiro C, Cobb MH, Hunter T. The PHD domain of MEKK1 acts as an E3 ubiquitin ligase and mediates ubiquitination and degradation of ERK1/2. Mol. Cell. 2002;9:945–956. doi: 10.1016/s1097-2765(02)00519-1. [DOI] [PubMed] [Google Scholar]
- 33.Laine A, Ronai Z. Ubiquitin chains in the ladder of MAPK signaling. Sci. STKE. 2005;2005:re5. doi: 10.1126/stke.2812005re5. [DOI] [PubMed] [Google Scholar]
- 34.Liu WH, Lai MZ. Deltex regulates T-cell activation by targeted degradation of active MEKK1. Mol. Cell Biol. 2005;25:1367–1378. doi: 10.1128/MCB.25.4.1367-1378.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamashita M, Ying SX, Zhang GM, Li C, Cheng SY, et al. Ubiquitin ligase Smurf1 controls osteoblast activity and bone homeostasis by targeting MEKK2 for degradation. Cell. 2005;121:101–113. doi: 10.1016/j.cell.2005.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manenti S, Delmas C, Darbon JM. Cell adhesion protects c-Raf-1 against ubiquitin-dependent degradation by the proteasome. Biochem. Biophys. Res. Commun. 2002;294:976–980. doi: 10.1016/S0006-291X(02)00594-6. [DOI] [PubMed] [Google Scholar]
- 37.Sheng J, Kumagai A, Dunphy WG, Varshavsky A. Dissection of c-MOS degron. EMBO J. 2002;21:6061–6071. doi: 10.1093/emboj/cdf626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuo WL, Duke CJ, Abe MK, Kaplan EL, Gomes S, Rosner MR. ERK7 expression and kinase activity is regulated by the ubiquitin-proteosome pathway. J. Biol. Chem. 2004;279:23073–23081. doi: 10.1074/jbc.M313696200. [DOI] [PubMed] [Google Scholar]
- 39.Dohlman HG, Thorner JW. Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu. Rev. Biochem. 2001;70:703–754. doi: 10.1146/annurev.biochem.70.1.703. [DOI] [PubMed] [Google Scholar]
- 40.Esch RK, Errede B. Pheromone induction promotes Ste11 degradation through aMAPK feedback and ubiquitin-dependent mechanism. Proc. Natl. Acad. Sci. USA. 2002;99:9160–9165. doi: 10.1073/pnas.142034399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y, Dohlman HG. Pheromone-dependent ubiquitination of the mitogen-activated protein kinase kinase Ste7. J. Biol. Chem. 2002;277:15766–15772. doi: 10.1074/jbc.M111733200. [DOI] [PubMed] [Google Scholar]
- 42.Collins CA, Wairkar YP, Johnson SL, DiAntonio A. Highwire restrains synaptic growth by attenuating a MAP kinase signal. Neuron. 2006;51:57–69. doi: 10.1016/j.neuron.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 43.Joazeiro CA, Wing SS, Huang H, Leverson JD, Hunter T, Liu YC. The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- 44.Waterman H, Levkowitz G, Alroy I, Yarden Y. The RING finger of c-Cbl mediates desensitization of the epidermal growth factor receptor. J. Biol. Chem. 1999;274:22151–22154. doi: 10.1074/jbc.274.32.22151. [DOI] [PubMed] [Google Scholar]
- 45.Swaminathan G, Tsygankov AY. The Cbl family proteins: ring leaders in regulation of cell signaling. J. Cell Physiol. 2006;209:21–43. doi: 10.1002/jcp.20694. [DOI] [PubMed] [Google Scholar]
- 46.Guenou H, Kaabeche K, Dufour C, Miraoui H, Marie PJ. Down-regulation of ubiquitin ligase Cbl induced by twist haploinsufficiency in Saethre-Chotzen syndrome results in increased PI3K/Akt signaling and osteoblast proliferation. Am. J. Pathol. 2006;169:1303–1311. doi: 10.2353/ajpath.2006.060102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suzue N, Nikawa T, Onishi Y, Yamada C, Hirasaka K, et al. Ubiquitin ligase Cbl-b downregulates bone formation through suppression of IGF-I signaling in osteoblasts during denervation. J. Bone Miner. Res. 2006;21:722–734. doi: 10.1359/jbmr.060207. [DOI] [PubMed] [Google Scholar]
- 48.Adachi M, Katsumura KR, Fujii K, Kobayashi S, Aoki H, Matsuzaki M. Proteasome-dependent decrease in Akt by growth factors in vascular smooth muscle cells. FEBS Lett. 2003;554:77–80. doi: 10.1016/s0014-5793(03)01109-8. [DOI] [PubMed] [Google Scholar]
- 49.Riesterer O, Zingg D, Hummerjohann J, Bodis S, Pruschy M. Degradation of PKB/Akt protein by inhibition of the VEGF receptor/mTOR pathway in endothelial cells. Oncogene. 2004;23:4624–4635. doi: 10.1038/sj.onc.1207596. [DOI] [PubMed] [Google Scholar]
- 50.Medina EA, Afsari RR, Ravid T, Castillo SS, Erickson KL, Goldkorn T. Tumor necrosis factor-α decreases Akt protein levels in 3T3-L1 adipocytes via the caspase-dependent ubiquitination of Akt. Endocrinology. 2005;146:2726–2735. doi: 10.1210/en.2004-1074. [DOI] [PubMed] [Google Scholar]
- 51.Yan D, Guo L, Wang Y. Requirement of dendritic Akt degradation by the ubiquitin-proteasome system for neuronal polarity. J. Cell Biol. 2006;174:415–424. doi: 10.1083/jcb.200511028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mao JH, Kim IJ, Wu D, Climent J, Kang HC, et al. FBXW7 targets mTOR for degradation and cooperates with PTEN in tumor suppression. Science. 2008;321:1499–1502. doi: 10.1126/science.1162981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Varfolomeev E, Blankenship JW, Wayson SM, Fedorova AV, Kayagaki N, et al. IAP antagonists induce autoubiquitination of c-IAPs, NF-κB activation, and TNFα-dependent apoptosis. Cell. 2007;131:669–681. doi: 10.1016/j.cell.2007.10.030. [DOI] [PubMed] [Google Scholar]
- 54.Rothe M, Pan MG, Henzel WJ, Ayres TM, Goeddel DV. The TNFR2-TRAF signaling complex contains two novel proteins related to baculoviral inhibitor of apoptosis proteins. Cell. 1995;83:1243–1252. doi: 10.1016/0092-8674(95)90149-3. [DOI] [PubMed] [Google Scholar]
- 55.Liao G, Zhang M, Harhaj EW, Sun SC. Regulation of the NF-κB-inducing kinase by tumor necrosis factor receptor-associated factor 3-induced degradation. J. Biol. Chem. 2004;279:26243–26250. doi: 10.1074/jbc.M403286200. [DOI] [PubMed] [Google Scholar]
- 56.He JQ, Zarnegar B, Oganesyan G, Saha SK, Yamazaki S, et al. Rescue of TRAF3-null mice by p100 NF-κB deficiency. J. Exp. Med. 2006;203:2413–2418. doi: 10.1084/jem.20061166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Belova L, Sharma S, Brickley DR, Nicolarsen JR, Patterson C, Conzen SD. Ubiquitin-proteasome degradation of serum- and glucocorticoid-regulated kinase-1 (SGK-1) is mediated by the chaperone-dependent E3 ligase CHIP. Biochem. J. 2006;400:235–244. doi: 10.1042/BJ20060905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bogusz AM, Brickley DR, Pew T, Conzen SD. A novel N-terminal hydrophobic motif mediates constitutive degradation of serum- and glucocorticoid-induced kinase-1 by the ubiquitin-proteasome pathway. FEBS J. 2006;273:2913–2928. doi: 10.1111/j.1742-4658.2006.05304.x. [DOI] [PubMed] [Google Scholar]
- 59.Zhou R, Snyder PM. Nedd4-2 phosphorylation induces serum and glucocorticoid-regulated kinase (SGK) ubiquitination and degradation. J. Biol. Chem. 2005;280:4518–4523. doi: 10.1074/jbc.M411053200. [DOI] [PubMed] [Google Scholar]
- 60.Hakak Y, Martin GS. Ubiquitin-dependent degradation of active Src. Curr. Biol. 1999;9:1039–1042. doi: 10.1016/s0960-9822(99)80453-9. [DOI] [PubMed] [Google Scholar]
- 61.Harris KF, Shoji I, Cooper EM, Kumar S, Oda H, Howley PM. Ubiquitin-mediated degradation of active Src tyrosine kinase. Proc. Natl. Acad. Sci. USA. 1999;96:13738–13743. doi: 10.1073/pnas.96.24.13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sanjay A, Houghton A, Neff L, DiDomenico E, Bardelay C, et al. Cbl associates with Pyk2 and Src to regulate Src kinase activity, αvβ3 integrin-mediated signaling, cell adhesion, and osteoclast motility. J. Cell Biol. 2001;152:181–195. doi: 10.1083/jcb.152.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rao N, Dodge I, Band H. The Cbl family of ubiquitin ligases: critical negative regulators of tyrosine kinase signaling in the immune system. J. Leukoc. Biol. 2002;71:753–763. [PubMed] [Google Scholar]
- 64.Shao Y, Yang C, Elly C, Liu Y-C. Differential regulation of the B cell receptor-mediated signaling by the E3 ubiquitin ligase Cbl. J. Biol. Chem. 2004;279:43646–43653. doi: 10.1074/jbc.M404082200. [DOI] [PubMed] [Google Scholar]
- 65.Thien CB, Blystad FD, Zhan Y, Lew AM, Voigt V, et al. Loss of c-Cbl RING finger function results in high-intensity TCR signaling and thymic deletion. EMBO J. 2005;24:3807–3819. doi: 10.1038/sj.emboj.7600841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Echarri A, Pendergast AM. Activated c-Abl is degraded by the ubiquitin-dependent proteasome pathway. Curr. Biol. 2001;11:1759–1765. doi: 10.1016/s0960-9822(01)00538-3. [DOI] [PubMed] [Google Scholar]
- 67.Smith JM, Mayer BJ. Abl: mechanisms of regulation and activation. Front. Biosci. 2002;7:d31–d42. doi: 10.2741/a767. [DOI] [PubMed] [Google Scholar]
- 68.Brasher BB, Van Etten RA. c-Abl has high intrinsic tyrosine kinase activity that is stimulated by mutation of the Src homology 3 domain and by autophosphorylation at two distinct regulatory tyrosines. J. Biol. Chem. 2000;275:35631–35637. doi: 10.1074/jbc.M005401200. [DOI] [PubMed] [Google Scholar]
- 69.Hantschel O, Superti-Furga G. Regulation of the c-Abl and Bcr-Abl tyrosine kinases. Nat. Rev. Mol. Cell Biol. 2004;5:33–44. doi: 10.1038/nrm1280. [DOI] [PubMed] [Google Scholar]
- 70.Cao C, Li Y, Leng Y, Li P, Ma Q, Kufe D. Ubiquitination and degradation of the Arg tyrosine kinase is regulated by oxidative stress. Oncogene. 2005;24:2433–2440. doi: 10.1038/sj.onc.1208454. [DOI] [PubMed] [Google Scholar]
- 71.MacGlashan D, Jr, Miura K. Loss of syk kinase during IgE-mediated stimulation of human basophils. J. Allergy Clin. Immunol. 2004;114:1317–1324. doi: 10.1016/j.jaci.2004.08.037. [DOI] [PubMed] [Google Scholar]
- 72.Winberg G, Matskova L, Chen F, Plant P, Rotin D, et al. Latent membrane protein 2A of Epstein-Barr virus binds WW domain E3 protein-ubiquitin ligases that ubiquitinate B-cell tyrosine kinases. Mol. Cell Biol. 2000;20:8526–8535. doi: 10.1128/mcb.20.22.8526-8535.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu E, Cote JF, Vuori K. Negative regulation of FAK signaling by SOCS proteins. EMBO J. 2003;22:5036–5046. doi: 10.1093/emboj/cdg503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sekine Y, Tsuji S, Ikeda O, Sugiyma K, Oritani K, et al. Signal-transducing adaptor protein-2 regulates integrin-mediated T cell adhesion through protein degradation of focal adhesion kinase. J. Immunol. 2007;179:2397–2407. doi: 10.4049/jimmunol.179.4.2397. [DOI] [PubMed] [Google Scholar]
- 75.Ungureanu D, Silvennoinen O. SLIM trims STATs: ubiquitin E3 ligases provide insights for specificity in the regulation of cytokine signaling. Sci. STKE. 2005;2005:pe49. doi: 10.1126/stke.3042005pe49. [DOI] [PubMed] [Google Scholar]
- 76.Ali S, Nouhi Z, Chughtai N, Ali S. SHP-2 regulates SOCS-1-mediated Janus kinase-2 ubiquitination/degradation downstream of the prolactin receptor. J. Biol. Chem. 2003;278:52021–52031. doi: 10.1074/jbc.M306758200. [DOI] [PubMed] [Google Scholar]
- 77.Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat. Rev. Cancer. 2006;6:369–381. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- 78.DeSalle LM, Pagano M. Regulation of the G1 to S transition by the ubiquitin pathway. FEBS Lett. 2001;490:179–189. doi: 10.1016/s0014-5793(01)02121-4. [DOI] [PubMed] [Google Scholar]
- 79.Busino L, Bassermann F, Maiolica A, Lee C, Nolan PM, et al. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science. 2007;316:900–904. doi: 10.1126/science.1141194. [DOI] [PubMed] [Google Scholar]
- 80.Harper JW, Burton JL, Solomon MJ. The anaphase-promoting complex: it’s not just for mitosis any more. Genes Dev. 2002;16:2179–2206. doi: 10.1101/gad.1013102. [DOI] [PubMed] [Google Scholar]
- 81.Nakayama KI, Hatakeyama S, Nakayama K. Regulation of the cell cycle at the G1-S transition by proteolysis of cyclin E and p27Kip1. Biochem. Biophys. Res. Commun. 2001;282:853–860. doi: 10.1006/bbrc.2001.4627. [DOI] [PubMed] [Google Scholar]
- 82.Yew PR. Ubiquitin-mediated proteolysis of vertebrate G1- and S-phase regulators. J. Cell Physiol. 2001;187:1–10. doi: 10.1002/1097-4652(2001)9999:9999<1::AID-JCP1049>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 83.Liou YC, Ryo A, Huang HK, Lu PJ, Bronson R, et al. Loss of Pin1 function in the mouse causes phenotypes resembling cyclin D1-null phenotypes. Proc. Natl. Acad. Sci. USA. 2002;99:1335–1340. doi: 10.1073/pnas.032404099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lin DI, Barbash O, Kumar KG, Weber JD, Harper JW, et al. Phosphorylation-dependent ubiquitination of cyclin D1 by the SCFFBX4-αB crystallin complex. Mol. Cell. 2006;24:355–366. doi: 10.1016/j.molcel.2006.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Barbash O, Zamfirova P, Lin DI, Chen X, Yang K, et al. Mutations in Fbx4 inhibit dimerization of the SCFFbx4 ligase and contribute to cyclin D1 overexpression in human cancer. Cancer Cell. 2008;14:68–78. doi: 10.1016/j.ccr.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat. Rev. Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 87.Möröy T, Geisen C. Cyclin E. Int. J. Biochem. Cell Biol. 2004;36:1424–1439. doi: 10.1016/j.biocel.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 88.Singer JD, Gurian-West M, Clurman B, Roberts JM. Cullin-3 targets cyclin E for ubiquitination and controls S phase in mammalian cells. Genes Dev. 1999;13:2375–2387. doi: 10.1101/gad.13.18.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dealy MJ, Nguyen KV, Lo J, Gstaiger M, Krek W, et al. Loss of Cul1 results in early embryonic lethality and dysregulation of cyclin E. Nat. Genet. 1999;23:245–248. doi: 10.1038/13886. [DOI] [PubMed] [Google Scholar]
- 90.Zhang YW, Otterness DM, Chiang GG, Xie W, Liu YC, et al. Genotoxic stress targets human Chk1 for degradation by the ubiquitin-proteasome pathway. Mol. Cell. 2005;19:607–618. doi: 10.1016/j.molcel.2005.07.019. [DOI] [PubMed] [Google Scholar]
- 91.Ohi MD, Feoktistova A, Ren L, Yip C, Cheng Y, et al. Structural organization of the anaphase-promoting complex bound to the mitotic activator Slp1. Mol. Cell. 2007;28:871–885. doi: 10.1016/j.molcel.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- 93.Rodrigo-Brenni MC, Morgan DO. Sequential E2s drive polyubiquitin chain assembly on APC targets. Cell. 2007;130:127–139. doi: 10.1016/j.cell.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 94.Guardavaccaro D, Kudo Y, Boulaire J, Barchi M, Busino L, et al. Control of meiotic and mitotic progression by the F box protein β-Trcp1 in vivo. Dev. Cell. 2003;4:799–812. doi: 10.1016/s1534-5807(03)00154-0. [DOI] [PubMed] [Google Scholar]
- 95.Moshe Y, Boulaire J, Pagano M, Hershko A. Role of Polo-like kinase in the degradation of early mitotic inhibitor 1, a regulator of the anaphase promoting complex/cyclosome. Proc. Natl. Acad. Sci. USA. 2004;101:7937–7942. doi: 10.1073/pnas.0402442101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Amador V, Ge S, Santamaría PG, Guardavaccaro D, Pagano M. APC/CCdc20 controls the ubiquitin-mediated degradation of p21 in prometaphase. Mol. Cell. 2007;27:462–473. doi: 10.1016/j.molcel.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Song MS, Song SJ, Ayad NG, Chang JS, Lee JH, et al. The tumour suppressor RASSF1A regulates mitosis by inhibiting the APC-Cdc20 complex. Nat. Cell Biol. 2004;6:129–137. doi: 10.1038/ncb1091. [DOI] [PubMed] [Google Scholar]
- 98.Geley S, Kramer E, Gieffers C, Gannon J, Peters JM, Hunt T. Anaphase-promoting complex/cyclosome-dependent proteolysis of human cyclin A starts at the beginning of mitosis and is not subject to the spindle assembly checkpoint. J. Cell Biol. 2001;153:137–148. doi: 10.1083/jcb.153.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Burton JL, Solomon MJ. Mad3p, a pseudosubstrate inhibitor of APCCdc20 in the spindle assembly checkpoint. Genes Dev. 2007;21:655–667. doi: 10.1101/gad.1511107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yu H. Cdc20: aWD40 activator for a cell cycle degradation machine. Mol. Cell. 2007;27:3–16. doi: 10.1016/j.molcel.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 101.Fang G, Yu H, Kirschner MW. Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol. Cell. 1998;2:163–171. doi: 10.1016/s1097-2765(00)80126-4. [DOI] [PubMed] [Google Scholar]
- 102.Gorr IH, Boos D, Stemmann O. Mutual inhibition of separase and Cdk1 by two-step complex formation. Mol. Cell. 2005;19:135–141. doi: 10.1016/j.molcel.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 103.Zachariae W, Nasmyth K. Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev. 1999;13:2039–2058. doi: 10.1101/gad.13.16.2039. [DOI] [PubMed] [Google Scholar]
- 104.Li M, York JP, Zhang P. Loss of Cdc20 causes a securin-dependent metaphase arrest in two-cell mouse embryos. Mol. Cell Biol. 2007;27:3481–3488. doi: 10.1128/MCB.02088-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zur A, Brandeis M. Securin degradation is mediated by fzy and fzr, and is required for complete chromatid separation but not for cytokinesis. EMBO J. 2001;20:792–801. doi: 10.1093/emboj/20.4.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pesin JA, Orr-Weaver TL. Regulation of APC/C activators in mitosis and meiosis. Annu. Rev. Cell Dev. Biol. 2008;24:475–499. doi: 10.1146/annurev.cellbio.041408.115949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Holt LJ, Hutti JE, Cantley LC, Morgan DO. Evolution of Ime2 phosphorylation sites on Cdk1 substrates provides a mechanism to limit the effects of the phosphatase Cdc14 in meiosis. Mol. Cell. 2007;25:689–702. doi: 10.1016/j.molcel.2007.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rape M, Kirschner MW. Autonomous regulation of the anaphase-promoting complex couples mitosis to S-phase entry. Nature. 2004;432:588–595. doi: 10.1038/nature03023. [DOI] [PubMed] [Google Scholar]
- 109.Listovsky T, Oren YS, Yudkovsky Y, Mahbubani HM, Weiss AM, et al. Mammalian Cdh1/Fzr mediates its own degradation. EMBO J. 2004;23:1619–1626. doi: 10.1038/sj.emboj.7600149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Benmaamar R, Pagano M. Involvement of the SCF complex in the control of Cdh1 degradation in S-phase. Cell Cycle. 2005;4:1230–1232. doi: 10.4161/cc.4.9.2048. [DOI] [PubMed] [Google Scholar]
- 111.Hames RS, Wattam SL, Yamano H, Bacchieri R, Fry AM. APC/C-mediated destruction of the centrosomal kinase Nek2A occurs in early mitosis and depends upon a cyclin A-type D-box. EMBO J. 2001;20:7117–7127. doi: 10.1093/emboj/20.24.7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Myer DL, Bahassi el M, Stambrook PJ. The Plk3-Cdc25 circuit. Oncogene. 2005;24:299–305. doi: 10.1038/sj.onc.1208278. [DOI] [PubMed] [Google Scholar]
- 113.Lindon C, Pines J. Ordered proteolysis in anaphase inactivates Plk1 to contribute to proper mitotic exit in human cells. J. Cell Biol. 2004;164:233–241. doi: 10.1083/jcb.200309035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Charles JF, Jaspersen SL, Tinker-Kulberg RL, Hwang L, Szidon A, Morgan DO. The Polo-related kinase Cdc5 activates and is destroyed by the mitotic cyclin destruction machinery in S. cerevisiae. Curr. Biol. 1998;8:497–507. doi: 10.1016/s0960-9822(98)70201-5. [DOI] [PubMed] [Google Scholar]
- 115.Morrow CJ, Tighe A, Johnson VL, Scott MI, Ditchfield C, Taylor SS. Bub1 and aurora B cooperate to maintain BubR1-mediated inhibition of APC/CCdc20. J. Cell Sci. 2005;118:3639–3652. doi: 10.1242/jcs.02487. [DOI] [PubMed] [Google Scholar]
- 116.Tang Z, Shu H, Oncel D, Chen S, Yu H. Phosphorylation of Cdc20 by Bub1 provides a catalytic mechanism for APC/C inhibition by the spindle checkpoint. Mol. Cell. 2004;16:387–397. doi: 10.1016/j.molcel.2004.09.031. [DOI] [PubMed] [Google Scholar]
- 117.Stewart S, Fang G. Destruction box-dependent degradation of aurora B is mediated by the anaphase-promoting complex/cyclosome and Cdh1. Cancer Res. 2005;65:8730–8735. doi: 10.1158/0008-5472.CAN-05-1500. [DOI] [PubMed] [Google Scholar]
- 118.Qi W, Yu H. KEN-box-dependent degradation of the Bub1 spindle checkpoint kinase by the anaphase-promoting complex/cyclosome. J. Biol. Chem. 2007;282:3672–3679. doi: 10.1074/jbc.M609376200. [DOI] [PubMed] [Google Scholar]
- 119.Wei W, Ayad NG, Wan Y, Zhang GJ, Kirschner MW, Kaelin WG., Jr Degradation of the SCF component Skp2 in cell-cycle phase G1 by the anaphase-promoting complex. Nature. 2004;428:194–198. doi: 10.1038/nature02381. [DOI] [PubMed] [Google Scholar]
- 120.Bashir T, Dorrello NV, Amador V, Guardavaccaro D, Pagano M. Control of the SCF(Skp2-Cks1) ubiquitin ligase by the APC/C(Cdh1) ubiquitin ligase. Nature. 2004;428:190–193. doi: 10.1038/nature02330. [DOI] [PubMed] [Google Scholar]
- 121.Liu W, Wu G, Li W, Lobur D, Wan Y. Cdh1-anaphase-promoting complex targets Skp2 for destruction in transforming growth factor β-induced growth inhibition. Mol. Cell Biol. 2007;27:2967–2979. doi: 10.1128/MCB.01830-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rodier G, Coulombe P, Tanguay PL, Boutonnet C, Meloche S. Phosphorylation of Skp2 regulated by CDK2 and Cdc14B protects it from degradation by APC(Cdh1) in G1 phase. EMBO J. 2008;27:679–691. doi: 10.1038/emboj.2008.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Watanabe N, Arai H, Nishihara Y, Taniguchi M, Watanabe N, et al. M-phase kinases induce phospho-dependent ubiquitination of somatic Wee1 by SCFβ-TrCP. Proc. Natl. Acad. Sci. USA. 2004;101:4419–4424. doi: 10.1073/pnas.0307700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ayad NG, Rankin S, Murakami M, Jebanathirajah J, Gygi S, Kirschner MW. Tome-1, a trigger of mitotic entry, is degraded during G1 via the APC. Cell. 2003;113:101–113. doi: 10.1016/s0092-8674(03)00232-0. [DOI] [PubMed] [Google Scholar]
- 125.Watanabe N, Arai H, Iwasaki J, Shiina M, Ogata K, et al. Cyclin-dependent kinase (CDK) phosphorylation destabilizes somaticWee1 via multiple pathways. Proc. Natl. Acad. Sci. USA. 2005;102:11663–11668. doi: 10.1073/pnas.0500410102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Asano S, Park JE, Sakchaisri K, Yu LR, Song S, et al. Concerted mechanism of Swe1/Wee1 regulation by multiple kinases in budding yeast. EMBO J. 2005;24:2194–2204. doi: 10.1038/sj.emboj.7600683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Huang F, Kirkpatrick D, Jiang X, Gygi S, Sorkin A. Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol. Cell. 2006;21:737–748. doi: 10.1016/j.molcel.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 128.Galcheva-Gargova Z, Theroux SJ, Davis RJ. The epidermal growth factor receptor is covalently linked to ubiquitin. Oncogene. 1995;11:2649–2655. [PubMed] [Google Scholar]
- 129.Yoon CH, Lee J, Jongeward GD, Sternberg PW. Similarity of sli-1, a regulator of vulval development in C. elegans, to the mammalian proto-oncogene c-cbl. Science. 1995;269:1102–1105. doi: 10.1126/science.7652556. [DOI] [PubMed] [Google Scholar]
- 130.Thien CB, Langdon WY. c-Cbl and Cbl-b ubiquitin ligases: substrate diversity and the negative regulation of signalling responses. Biochem. J. 2005;391:153–166. doi: 10.1042/BJ20050892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kassenbrock CK, Anderson SM. Regulation of ubiquitin protein ligase activity in c-Cbl by phosphorylation-induced conformational change and constitutive activation by tyrosine to glutamate point mutations. J. Biol. Chem. 2004;279:28017–28027. doi: 10.1074/jbc.M404114200. [DOI] [PubMed] [Google Scholar]
- 132.Kozlov G, Peschard P, Zimmerman B, Lin T, Moldoveanu T, et al. Structural basis for UBA-mediated dimerization of c-Cbl ubiquitin ligase. J. Biol. Chem. 2007;282:27547–27555. doi: 10.1074/jbc.M703333200. [DOI] [PubMed] [Google Scholar]
- 133.Peschard P, Kozlov G, Lin T, Mirza IA, Berghuis AM, et al. Structural basis for ubiquitin-mediated dimerization and activation of the ubiquitin protein ligase Cbl-b. Mol. Cell. 2007;27:474–485. doi: 10.1016/j.molcel.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 134.Longva KE, Blystad FD, Stang E, Larsen AM, Johannessen LE, Madshus IH. Ubiquitination and proteasomal activity is required for transport of the EGF receptor to inner membranes of multivesicular bodies. J. Cell Biol. 2002;156:843–854. doi: 10.1083/jcb.200106056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fukazawa T, Miyake S, Band V, Band H. Tyrosine phosphorylation of Cbl upon epidermal growth factor (EGF) stimulation and its association with EGF receptor and downstream signaling proteins. J. Biol. Chem. 1996;271:14554–14559. doi: 10.1074/jbc.271.24.14554. [DOI] [PubMed] [Google Scholar]
- 136.Stang E, Johannessen LE, Knardal SL, Madshus IH. Polyubiquitination of the epidermal growth factor receptor occurs at the plasma membrane upon ligand-induced activation. J. Biol. Chem. 2000;275:13940–13947. doi: 10.1074/jbc.275.18.13940. [DOI] [PubMed] [Google Scholar]
- 137.de Melker AA, Van Der Horst G, Calafat J, Jansen H, Borst J. c-Cbl ubiquitinates the EGF receptor at the plasma membrane and remains receptor associated throughout the endocytic route. J. Cell Sci. 2001;114:2167–2178. doi: 10.1242/jcs.114.11.2167. [DOI] [PubMed] [Google Scholar]
- 138.Umebayashi K, Stenmark H, Yoshimori T. Ubc4/5 and c-Cbl continue to ubiquitinate EGF receptor after internalization to facilitate polyubiquitination and degradation. Mol. Biol. Cell. 2008;19:3454–3462. doi: 10.1091/mbc.E07-10-0988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Levkowitz G, Waterman H, Zamir E, Kam Z, Oved S, et al. c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 1998;12:3663–3674. doi: 10.1101/gad.12.23.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Levkowitz G, Waterman H, Ettenberg SA, Katz M, Tsygankov AY, et al. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol. Cell. 1999;4:1029–1040. doi: 10.1016/s1097-2765(00)80231-2. [DOI] [PubMed] [Google Scholar]
- 141.Frey MR, Dise RS, Edelblum KL, Polk DB. p38 kinase regulates epidermal growth factor receptor downregulation and cellular migration. EMBO J. 2006;25:5683–5692. doi: 10.1038/sj.emboj.7601457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Frederick L, Wang XY, Eley G, James CD. Diversity and frequency of epidermal growth factor receptor mutations in human glioblastomas. Cancer Res. 2000;60:1383–1387. [PubMed] [Google Scholar]
- 143.Davies GC, Ryan PE, Rahman L, Zajac-Kaye M, Lipkowitz S. EGFRvIII undergoes activation-dependent downregulation mediated by the Cbl proteins. Oncogene. 2006;25:6497–6509. doi: 10.1038/sj.onc.1209662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Marmor MD, Yarden Y. Role of protein ubiquitylation in regulating endocytosis of receptor tyrosine kinases. Oncogene. 2004;23:2057–2070. doi: 10.1038/sj.onc.1207390. [DOI] [PubMed] [Google Scholar]
- 145.Duan L, Miura Y, Dimri M, Majumder B, Dodge IL, et al. Cbl-mediated ubiquitinylation is required for lysosomal sorting of epidermal growth factor receptor but is dispensable for endocytosis. J. Biol. Chem. 2003;278:28950–28960. doi: 10.1074/jbc.M304474200. [DOI] [PubMed] [Google Scholar]
- 146.Felder S, Miller K, Moehren G, Ullrich A, Schlessinger J, Hopkins CR. Kinase activity controls the sorting of the epidermal growth factor receptor within the multivesicular body. Cell. 1990;61:623–634. doi: 10.1016/0092-8674(90)90474-s. [DOI] [PubMed] [Google Scholar]
- 147.Oved S, Mosesson Y, Zwang Y, Santonico E, Shtiegman K, et al. Conjugation to Nedd8 instigates ubiquitylation and down-regulation of activated receptor tyrosine kinases. J. Biol. Chem. 2006;281:21640–21651. doi: 10.1074/jbc.M513034200. [DOI] [PubMed] [Google Scholar]
- 148.Thiel KW, Carpenter G. Epidermal growth factor receptor juxtamembrane region regulates allosteric tyrosine kinase activation. Proc. Natl. Acad. Sci. USA. 2007;104:19238–19243. doi: 10.1073/pnas.0703854104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bao J, Alroy I, Waterman H, Schejter ED, Brodie C, et al. Threonine phosphorylation diverts internalized epidermal growth factor receptors from a degradative pathway to the recycling endosome. J. Biol. Chem. 2000;275:26178–26186. doi: 10.1074/jbc.M002367200. [DOI] [PubMed] [Google Scholar]
- 150.Fiorentino L, Pertica C, Fiorini M, Talora C, Crescenzi M, et al. Inhibition of ErbB-2 mitogenic and transforming activity by RALT, a mitogen-induced signal transducer which binds to the ErbB-2 kinase domain. Mol. Cell Biol. 2000;20:7735–7750. doi: 10.1128/mcb.20.20.7735-7750.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Shen F, Lin Q, Gu Y, Childress C, Yang W. Activated Cdc42-associated kinase 1 is a component of EGF receptor signaling complex and regulates EGF receptor degradation. Mol. Biol. Cell. 2007;18:732–742. doi: 10.1091/mbc.E06-02-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ettenberg SA, Magnifico A, Cuello M, Nau MM, Rubinstein YR, et al. Cbl-b-dependent coordinated degradation of the epidermal growth factor receptor signaling complex. J. Biol. Chem. 2001;276:27677–27684. doi: 10.1074/jbc.M102641200. [DOI] [PubMed] [Google Scholar]
- 153.Sorkin A. Internalization of the epidermal growth factor receptor: role in signalling. Biochem. Soc. Trans. 2001;29:480–484. doi: 10.1042/bst0290480. [DOI] [PubMed] [Google Scholar]
- 154.Balbis A, Parmar A, Wang Y, Baquiran G, Posner BI. Compartmentalization of signaling-competent epidermal growth factor receptors in endosomes. Endocrinology. 2007;148:2944–2954. doi: 10.1210/en.2006-1674. [DOI] [PubMed] [Google Scholar]
- 155.Magnifico A, Ettenberg S, Yang C, Mariano J, Tiwari S, et al. WW domain HECT E3s target Cbl RING finger E3s for proteasomal degradation. J. Biol. Chem. 2003;278:43169–43177. doi: 10.1074/jbc.M308009200. [DOI] [PubMed] [Google Scholar]
- 156.Thien CB, Langdon WY. Cbl: many adaptations to regulate protein tyrosine kinases. Nat. Rev. Mol. Cell Biol. 2001;2:294–307. doi: 10.1038/35067100. [DOI] [PubMed] [Google Scholar]
- 157.Naramura M, Jang IK, Kole H, Huang F, Haines D, Gu H. c-Cbl and Cbl-b regulate T cell responsiveness by promoting ligand-induced TCR down-modulation. Nat. Immunol. 2002;3:1192–1199. doi: 10.1038/ni855. [DOI] [PubMed] [Google Scholar]
- 158.Thien CB, Walker F, Langdon WY. RING finger mutations that abolish c-Cbl-directed polyubiquitination and downregulation of the EGF receptor are insufficient for cell transformation. Mol. Cell. 2001;7:355–365. doi: 10.1016/s1097-2765(01)00183-6. [DOI] [PubMed] [Google Scholar]
- 159.Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 160.Schmidt MH, Husnjak K, Szymkiewicz I, Haglund K, Dikic I. Cbl escapes Cdc42-mediated inhibition by downregulation of the adaptor molecule βPix. Oncogene. 2006;25:3071–3078. doi: 10.1038/sj.onc.1209329. [DOI] [PubMed] [Google Scholar]
- 161.Sorkin A, Goh LK. Endocytosis and intracellular trafficking of ErbBs. Exp. Cell Res. 2008;314:3093–3106. doi: 10.1016/j.yexcr.2008.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Sigismund S, Woelk T, Puri C, Maspero E, Tacchetti C, et al. Clathrin-independent endocytosis of ubiquitinated cargos. Proc. Natl. Acad. Sci. USA. 2005;102:2760–2765. doi: 10.1073/pnas.0409817102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Stang E, Blystad FD, Kazazic M, Bertelsen V, Brodahl T, et al. Cbl-dependent ubiquitination is required for progression of EGF receptors into clathrin-coated pits. Mol. Biol. Cell. 2004;15:3591–3604. doi: 10.1091/mbc.E04-01-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Chen C, Matesic LE. The Nedd4-like family of E3 ubiquitin ligases and cancer. Cancer Metastasis Rev. 2007;26:587–604. doi: 10.1007/s10555-007-9091-x. [DOI] [PubMed] [Google Scholar]
- 165.de Melker AA, van der Horst G, Borst J. c-Cbl directs EGF receptors into an endocytic pathway that involves the ubiquitin-interacting motif of Eps15. J. Cell Sci. 2004;117:5001–5012. doi: 10.1242/jcs.01354. [DOI] [PubMed] [Google Scholar]
- 166.Fallon L, Belanger CM, Corera AT, Kontogiannea M, Regan-Klapisz E, et al. A regulated interaction with the UIM protein Eps15 implicates parkin in EGF receptor trafficking and PI(3)K-Akt signalling. Nat. Cell Biol. 2006;8:834–842. doi: 10.1038/ncb1441. [DOI] [PubMed] [Google Scholar]
- 167.Hoeller D, Crosetto N, Blagoev B, Raiborg C, Tikkanen R, et al. Regulation of ubiquitin-binding proteins by monoubiquitination. Nat. Cell Biol. 2006;8:163–169. doi: 10.1038/ncb1354. [DOI] [PubMed] [Google Scholar]
- 168.Polo S, Sigismund S, Faretta M, Guidi M, Capua MR, et al. A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature. 2002;416:451–455. doi: 10.1038/416451a. [DOI] [PubMed] [Google Scholar]
- 169.Sigismund S, Argenzio E, Tosoni D, Cavallaro E, Polo S, Di Fiore PP. Clathrin-mediated internalization is essential for sustained EGFR signaling but dispensable for degradation. Dev. Cell. 2008;15:209–219. doi: 10.1016/j.devcel.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 170.Katzmann DJ, Odorizzi G, Emr SD. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- 171.Futter CE, Pearse A, Hewlett LJ, Hopkins CR. Multivesicular endosomes containing internalized EGF-EGF receptor complexes mature and then fuse directly with lysosomes. J. Cell Biol. 1996;132:1011–1023. doi: 10.1083/jcb.132.6.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Burke P, Schooler K, Wiley HS. Regulation of epidermal growth factor receptor signaling by endocytosis and intracellular trafficking. Mol. Biol. Cell. 2001;12:1897–1910. doi: 10.1091/mbc.12.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Haj FG, Verveer PJ, Squire A, Neel BG, Bastiaens PI. Imaging sites of receptor dephosphorylation by PTP1B on the surface of the endoplasmic reticulum. Science. 2002;295:1708–1711. doi: 10.1126/science.1067566. [DOI] [PubMed] [Google Scholar]
- 174.Alwan HA, van Zoelen EJ, van Leeuwen JE. Ligand-induced lysosomal epidermal growth factor receptor (EGFR) degradation is preceded by proteasome-dependent EGFR deubiquitination. J. Biol. Chem. 2003;278:35781–35790. doi: 10.1074/jbc.M301326200. [DOI] [PubMed] [Google Scholar]
- 175.Kirisits A, Pils D, Krainer M. Epidermal growth factor receptor degradation: an alternative view of oncogenic pathways. Int. J. Biochem. Cell Biol. 2007;39:2173–2182. doi: 10.1016/j.biocel.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 176.Kim BY, Olzmann JA, Barsh GS, Chin LS, Li L. Spongiform neurodegeneration-associated E3 ligase Mahogunin ubiquitylates TSG101 and regulates endosomal trafficking. Mol. Biol. Cell. 2007;18:1129–1142. doi: 10.1091/mbc.E06-09-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD. Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell. 2002;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- 178.Kario E, Marmor MD, Adamsky K, Citri A, Amit I, et al. Suppressors of cytokine signaling 4 and 5 regulate epidermal growth factor receptor signaling. J. Biol. Chem. 2005;280:7038–7048. doi: 10.1074/jbc.M408575200. [DOI] [PubMed] [Google Scholar]
- 179.Jekely G, Rorth P. Hrs mediates downregulation of multiple signalling receptors in Drosophila. EMBO Rep. 2003;4:1163–1168. doi: 10.1038/sj.embor.7400019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Klapper LN, Waterman H, Sela M, Yarden Y. Tumor-inhibitory antibodies to HER-2/ErbB-2 may act by recruiting c-Cbl and enhancing ubiquitination of HER-2. Cancer Res. 2000;60:3384–3388. [PubMed] [Google Scholar]
- 181.Xu W, Marcu M, Yuan X, Mimnaugh E, Patterson C, Neckers L. Chaperone-dependent E3 ubiquitin ligase CHIP mediates a degradative pathway for c-ErbB2/Neu. Proc. Natl. Acad. Sci. USA. 2002;99:12847–12852. doi: 10.1073/pnas.202365899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Oster SK, Marhin WW, Asker C, Facchini LM, Dion PA, et al. Myc is an essential negative regulator of platelet-derived growth factor beta receptor expression. Mol. Cell Biol. 2000;20:6768–6778. doi: 10.1128/mcb.20.18.6768-6778.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Mori S, Heldin CH, Claesson-Welsh L. Ligand-induced polyubiquitination of the platelet-derived growth factor beta-receptor. J. Biol. Chem. 1992;267:6429–6434. [PubMed] [Google Scholar]
- 184.Mori S, Heldin CH, Claesson-Welsh L. Ligand-induced ubiquitination of the platelet-derived growth factor beta-receptor plays a negative regulatory role in its mitogenic signaling. J. Biol. Chem. 1993;268:577–583. [PubMed] [Google Scholar]
- 185.Reddi AL, Ying G, Duan L, Chen G, Dimri M, et al. Binding of Cbl to a phospholipase Cγ1-docking site on platelet-derived growth factor receptor β provides a dual mechanism of negative regulation. J. Biol. Chem. 2007;282:29336–29347. doi: 10.1074/jbc.M701797200. [DOI] [PubMed] [Google Scholar]
- 186.Crowley MR, Bowtell D, Serra R. TGF-β, c-Cbl, and PDGFR-α the in mammary stroma. Dev. Biol. 2005;279:58–72. doi: 10.1016/j.ydbio.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 187.Cormont M, Meton I, Mari M, Monzo P, Keslair F, et al. CD2AP/CMS regulates endosome morphology and traffic to the degradative pathway through its interaction with Rab4 and c-Cbl. Traffic. 2003;4:97–112. doi: 10.1034/j.1600-0854.2003.40205.x. [DOI] [PubMed] [Google Scholar]
- 188.Lennartsson J, Wardega P, Engström U, Hellman U, Heldin CH. Alix facilitates the interaction between c-Cbl and platelet-derived growth factor β-receptor and thereby modulates receptor down-regulation. J. Biol. Chem. 2006;281:39152–39158. doi: 10.1074/jbc.M608489200. [DOI] [PubMed] [Google Scholar]
- 189.Masson K, Heiss E, Band H, Ronnstrand L. Direct binding of Cbl to Tyr568 and Tyr936 of the stem cell factor receptor/c-Kit is required for ligand-induced ubiquitination, internalization and degradation. Biochem. J. 2006;399:59–67. doi: 10.1042/BJ20060464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Zeng S, Xu Z, Lipkowitz S, Longley JB. Regulation of stem cell factor receptor signaling by Cbl family proteins (Cbl-b/c-Cbl) Blood. 2005;105:226–232. doi: 10.1182/blood-2004-05-1768. [DOI] [PubMed] [Google Scholar]
- 191.Lee PS, Wang Y, Dominguez MG, Yeung YG, Murphy MA, et al. The Cbl protooncoprotein stimulates CSF-1 receptor multiubiquitination and endocytosis, and attenuates macrophage proliferation. EMBO J. 1999;18:3616–3628. doi: 10.1093/emboj/18.13.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Wilhelmsen K, Burkhalter S, van der Geer P. C-Cbl binds the CSF-1 receptor at tyrosine 973, a novel phosphorylation site in the receptor’s carboxy-terminus. Oncogene. 2002;21:1079–1089. doi: 10.1038/sj.onc.1205166. [DOI] [PubMed] [Google Scholar]
- 193.Herbst R, Munemitsu S, Ullrich A. Oncogenic activation of v-kit involves deletion of a putative tyrosine-substrate interaction site. Oncogene. 1995;10:369–379. [PubMed] [Google Scholar]
- 194.Ridge SA, Worwood M, Oscier D, Jacobs A, Padua RA. FMS mutations in myelodysplastic, leukemic, and normal subjects. Proc. Natl. Acad. Sci. USA. 1990;87:1377–1380. doi: 10.1073/pnas.87.4.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Peschard P, Park M. Escape from Cbl-mediated downregulation: a recurrent theme for oncogenic deregulation of receptor tyrosine kinases. Cancer Cell. 2003;3:519–523. doi: 10.1016/s1535-6108(03)00136-3. [DOI] [PubMed] [Google Scholar]
- 196.Ornitz DM, Itoh N. Fibroblast growth factors. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-3-reviews3005. reviews3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Sorokin A, Mohammadi M, Huang J, Schlessinger J. Internalization of fibroblast growth factor receptor is inhibited by a point mutation at tyrosine 766. J. Biol. Chem. 1994;269:17056–17061. [PubMed] [Google Scholar]
- 198.Kouhara H, Hadari YR, Spivak-Kroizman T, Schilling J, Bar-Sagi D, et al. A lipid-anchored Grb2-binding protein that links FGF-receptor activation to the Ras/MAPK signaling pathway. Cell. 1997;89:693–702. doi: 10.1016/s0092-8674(00)80252-4. [DOI] [PubMed] [Google Scholar]
- 199.Hadari YR, Kouhara H, Lax I, Schlessinger J. Binding of Shp2 tyrosine phosphatase to FRS2 is essential for fibroblast growth factor-induced PC12 cell differentiation. Mol. Cell Biol. 1998;18:3966–3973. doi: 10.1128/mcb.18.7.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Ong SH, Guy GR, Hadari YR, Laks S, Gotoh N, et al. FRS2 proteins recruit intracellular signaling pathways by binding to diverse targets on fibroblast growth factor and nerve growth factor receptors. Mol. Cell Biol. 2000;20:979–989. doi: 10.1128/mcb.20.3.979-989.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Wong A, Lamothe B, Lee A, Schlessinger J, Lax I, Li A. FRS2α attenuates FGF receptor signaling by Grb2-mediated recruitment of the ubiquitin ligase Cbl. Proc. Natl. Acad. Sci. USA. 2002;99:6684–6689. doi: 10.1073/pnas.052138899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Cho JY, Guo C, Torello M, Lunstrum GP, Iwata T, et al. Defective lysosomal targeting of activated fibroblast growth factor receptor 3 in achondroplasia. Proc. Natl. Acad. Sci. USA. 2004;101:609–614. doi: 10.1073/pnas.2237184100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Peschard P, Park M. From Tpr-Met to Met, tumorigenesis and tubes. Oncogene. 2007;26:1276–1285. doi: 10.1038/sj.onc.1210201. [DOI] [PubMed] [Google Scholar]
- 204.Hammond DE, Carter S, McCullough J, Urbe S, Vande Woude G, Clague MJ. Endosomal dynamics of Met determine signaling output. Mol. Biol. Cell. 2003;14:1346–1354. doi: 10.1091/mbc.E02-09-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Carter S, Urbe S, Clague MJ. The met receptor degradation pathway: requirement for Lys48-linked polyubiquitin independent of proteasome activity. J. Biol. Chem. 2004;279:52835–52839. doi: 10.1074/jbc.M407769200. [DOI] [PubMed] [Google Scholar]
- 206.Kong-Beltran M, Seshagiri S, Zha J, Zhu W, Bhawe K, et al. Somatic mutations lead to an oncogenic deletion of met in lung cancer. Cancer Res. 2006;66:283–289. doi: 10.1158/0008-5472.CAN-05-2749. [DOI] [PubMed] [Google Scholar]
- 207.Abella JV, Peschard P, Naujokas MA, Lin T, Saucier C, et al. Met/Hepatocyte growth factor receptor ubiquitination suppresses transformation and is required for Hrs phosphorylation. Mol. Cell Biol. 2005;25:9632–9645. doi: 10.1128/MCB.25.21.9632-9645.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Shattuck DL, Miller JK, Laederich M, Funes M, Petersen H, et al. LRIG1 is a novel negative regulator of the Met receptor and opposes Met and Her2 synergy. Mol. Cell Biol. 2007;27:1934–1946. doi: 10.1128/MCB.00757-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Takeuchi T, Adachi Y, Sonobe H, Furihata M, Ohtsuki Y. A ubiquitin ligase, skeletrophin, is a negative regulator of melanoma invasion. Oncogene. 2006;25:7059–7069. doi: 10.1038/sj.onc.1209688. [DOI] [PubMed] [Google Scholar]
- 210.Penengo L, Rubin C, Yarden Y, Gaudino G. c-Cbl is a critical modulator of the Ron tyrosine kinase receptor. Oncogene. 2003;22:3669–3679. doi: 10.1038/sj.onc.1206585. [DOI] [PubMed] [Google Scholar]
- 211.Germano S, Barberis D, Santoro MM, Penengo L, Citri A, et al. Geldanamycins trigger a novel Ron degradative pathway, hampering oncogenic signaling. J. Biol. Chem. 2006;281:21710–21719. doi: 10.1074/jbc.M602014200. [DOI] [PubMed] [Google Scholar]
- 212.Laviola L, Natalicchio A, Giorgino F. The IGF-I signaling pathway. Curr. Pharm. Des. 2007;13:663–669. doi: 10.2174/138161207780249146. [DOI] [PubMed] [Google Scholar]
- 213.Girnita L, Girnita A, Larsson O. Mdm2-dependent ubiquitination and degradation of the insulin-like growth factor 1 receptor. Proc. Natl. Acad. Sci. USA. 2003;100:8247–8252. doi: 10.1073/pnas.1431613100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Vasilcanu R, Vasilcanu D, Rosengren L, Natalishvili N, Sehat B, et al. Picropodophyllin induces downregulation of the insulin-like growth factor 1 receptor: potential mechanistic involvement of Mdm2 and β-arrestin1. Oncogene. 2008;27:1629–1638. doi: 10.1038/sj.onc.1210797. [DOI] [PubMed] [Google Scholar]
- 215.Shan YX, Yang TL, Mestril R, Wang PH. Hsp10 and Hsp60 suppress ubiquitination of insulin-like growth factor-1 receptor and augment insulin-like growth factor-1 receptor signaling in cardiac muscle: implications on decreased myocardial protection in diabetic cardiomyopathy. J. Biol. Chem. 2003;278:45492–45498. doi: 10.1074/jbc.M304498200. [DOI] [PubMed] [Google Scholar]
- 216.Lai HC, Liu TJ, Ting CT, Yang JY, Huang L, et al. Regulation of IGF-I receptor signaling in diabetic cardiac muscle: dysregulation of cytosolic and mitochondria HSP60. Am. J. Physiol. Endocrinol. Metab. 2007;292:E292–E297. doi: 10.1152/ajpendo.00189.2006. [DOI] [PubMed] [Google Scholar]
- 217.Itoh S, ten Dijke P. Negative regulation of TGF-beta receptor/Smad signal transduction. Curr. Opin. Cell Biol. 2007;19:176–184. doi: 10.1016/j.ceb.2007.02.015. [DOI] [PubMed] [Google Scholar]
- 218.Glasgow E, Mishra L. Transforming growth factor-β signaling and ubiquitinators in cancer. Endocrinol. Relat. Cancer. 2008;15:59–72. doi: 10.1677/ERC-07-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Inman GJ, Nicolas FJ, Hill CS. Nucleocytoplasmic shuttling of Smads 2, 3, and 4 permits sensing of TGF-β receptor activity. Mol. Cell. 2002;10:283–294. doi: 10.1016/s1097-2765(02)00585-3. [DOI] [PubMed] [Google Scholar]
- 220.ten Dijke P, Hill CS. New insights into TGF-β-Smad signalling. Trends Biochem. Sci. 2004;29:265–273. doi: 10.1016/j.tibs.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 221.Kang JS, Saunier EF, Akhurst RJ, Derynck R. The type I TGF-β receptor is covalently modified and regulated by sumoylation. Nat. Cell Biol. 2008;10:654–664. doi: 10.1038/ncb1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Anders RA, Dore JJ, Jr, Arline SL, Garamszegi N, Leof EB. Differential requirement for type I and type II transforming growth factor β receptor kinase activity in ligand-mediated receptor endocytosis. J. Biol. Chem. 1998;273:23118–23125. doi: 10.1074/jbc.273.36.23118. [DOI] [PubMed] [Google Scholar]
- 223.Ohashi N, Yamamoto T, Uchida C, Togawa A, Fukasawa H, et al. Transcriptional induction of Smurf2 ubiquitin ligase by TGF-β. FEBS Lett. 2005;579:2557–2563. doi: 10.1016/j.febslet.2005.03.069. [DOI] [PubMed] [Google Scholar]
- 224.Tajima Y, Goto K, Yoshida M, Shinomiya K, Sekimoto T, et al. Chromosomal region maintenance 1 (CRM1)-dependent nuclear export of Smad ubiquitin regulatory factor 1 (Smurf1) is essential for negative regulation of transforming growth factor-β signaling by Smad7. J. Biol. Chem. 2003;278:10716–10721. doi: 10.1074/jbc.M212663200. [DOI] [PubMed] [Google Scholar]
- 225.Levy L, Howell M, Das D, Harkin S, Episkopou V, Hill CS. Arkadia activates Smad3/Smad4-dependent transcription by triggering signal-induced SnoN degradation. Mol. Cell Biol. 2007;27:6068–6083. doi: 10.1128/MCB.00664-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Nagano Y, Mavrakis KJ, Lee KL, Fujii T, Koinuma D, et al. Arkadia induces degradation of SnoN and c-Ski to enhance transforming growth factor-β signaling. J. Biol. Chem. 2007;282:20492–20501. doi: 10.1074/jbc.M701294200. [DOI] [PubMed] [Google Scholar]
- 227.Mavrakis KJ, Andrew RL, Lee KL, Petropoulou C, Dixon JE, et al. Arkadia enhances Nodal/TGF-β signaling by coupling Phospho-Smad2/3 activity and turnover. PLoS Biol. 2007;5:e67. doi: 10.1371/journal.pbio.0050067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Moren A, Hellman U, Inada Y, Imamura T, Heldin CH, Moustakas A. Differential ubiquitination defines the functional status of the tumor suppressor Smad4. J. Biol. Chem. 2003;278:33571–33582. doi: 10.1074/jbc.M300159200. [DOI] [PubMed] [Google Scholar]
- 229.Lin X, Liang M, Liang YY, Brunicardi FC, Feng XH. SUMO-1/Ubc9 promotes nuclear accumulation and metabolic stability of tumor suppressor Smad4. J. Biol. Chem. 2003;278:31043–31048. doi: 10.1074/jbc.C300112200. [DOI] [PubMed] [Google Scholar]
- 230.Lee PS, Chang C, Liu D, Derynck R. Sumoylation of Smad4, the common Smad mediator of transforming growth factor-β family signaling. J. Biol. Chem. 2003;278:27853–27863. doi: 10.1074/jbc.M301755200. [DOI] [PubMed] [Google Scholar]
- 231.Imoto S, Sugiyama K, Muromoto R, Sato N, Yamamoto T, Matsuda T. Regulation of transforming growth factor-β signaling by protein inhibitor of activated STAT, PIASy through Smad3. J. Biol. Chem. 2003;278:34253–34258. doi: 10.1074/jbc.M304961200. [DOI] [PubMed] [Google Scholar]
- 232.Poncelet AC, Schnaper HW, Tan R, Liu Y, Runyan CE. Cell phenotype-specific down-regulation of Smad3 involves decreased gene activation as well as protein degradation. J. Biol. Chem. 2007;282:15534–15540. doi: 10.1074/jbc.M701991200. [DOI] [PubMed] [Google Scholar]
- 233.Penela P, Ribas C, Mayor F., Jr Mechanisms of regulation of the expression and function of G protein-coupled receptor kinases. Cell Signal. 2003;15:973–981. doi: 10.1016/s0898-6568(03)00099-8. [DOI] [PubMed] [Google Scholar]
- 234.Elorza A, Penela P, Sarnago S, Mayor F., Jr MAPK-dependent degradation of G protein-coupled receptor kinase 2. J. Biol. Chem. 2003;278:29164–29173. doi: 10.1074/jbc.M304314200. [DOI] [PubMed] [Google Scholar]
- 235.Salcedo A, Mayor F, Jr, Penela P. Mdm2 is involved in the ubiquitination and degradation of G-protein-coupled receptor kinase 2. EMBO J. 2006;25:4752–4762. doi: 10.1038/sj.emboj.7601351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Citri A, Gan J, Mosesson Y, Vereb G, Szollosi J, Yarden Y. Hsp90 restrains ErbB-2/HER2 signalling by limiting heterodimer formation. EMBO Rep. 2004;5:1165–1170. doi: 10.1038/sj.embor.7400300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Citri A, Harari D, Shohat G, Ramakrishnan P, Gan J, et al. Hsp90 recognizes a common surface on client kinases. J. Biol. Chem. 2006;281:14361–14369. doi: 10.1074/jbc.M512613200. [DOI] [PubMed] [Google Scholar]
- 238.Xu W, Yuan X, Xiang Z, Mimnaugh E, Marcu M, Neckers L. Surface charge and hydrophobicity determine ErbB2 binding to the Hsp90 chaperone complex. Nat. Struct. Mol. Biol. 2005;12:120–126. doi: 10.1038/nsmb885. [DOI] [PubMed] [Google Scholar]
- 239.Esser C, Alberti S, Hohfeld J. Cooperation of molecular chaperones with the ubiquitin/proteasome system. Biochim. Biophys. Acta. 2004;1695:171–188. doi: 10.1016/j.bbamcr.2004.09.020. [DOI] [PubMed] [Google Scholar]
- 240.Blagosklonny MV. Hsp-90-associated oncoproteins: multiple targets of geldanamycin and its analogs. Leukemia. 2002;16:455–462. doi: 10.1038/sj.leu.2402415. [DOI] [PubMed] [Google Scholar]
- 241.Lange BM, Rebollo E, Herold A, Gonzalez C. Cdc37 is essential for chromosome segregation and cytokinesis in higher eukaryotes. EMBO J. 2002;21:5364–5374. doi: 10.1093/emboj/cdf531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Wang H, Goode T, Iakova P, Albrecht JH, Timchenko NA. C/EBPalpha triggers proteasome-dependent degradation of cdk4 during growth arrest. EMBO J. 2002;21:930–941. doi: 10.1093/emboj/21.5.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Alberti S, Demand J, Esser C, Emmerich N, Schild H, Hohfeld J. Ubiquitylation of BAG-1 suggests a novel regulatory mechanism during the sorting of chaperone substrates to the proteasome. J. Biol. Chem. 2002;277:45920–45927. doi: 10.1074/jbc.M204196200. [DOI] [PubMed] [Google Scholar]
- 244.Demand J, Alberti S, Patterson C, Hohfeld J. Cooperation of a ubiquitin domain protein and an E3 ubiquitin ligase during chaperone/proteasome coupling. Curr. Biol. 2001;11:1569–1577. doi: 10.1016/s0960-9822(01)00487-0. [DOI] [PubMed] [Google Scholar]
- 245.Song J, Takeda M, Morimoto RI. Bag1-Hsp70 mediates a physiological stress signalling pathway that regulates Raf-1/ERK and cell growth. Nat. Cell Biol. 2001;3:276–282. doi: 10.1038/35060068. [DOI] [PubMed] [Google Scholar]
- 246.Wang HG, Takayama S, Rapp UR, Reed JC. Bcl-2 interacting protein, BAG-1, binds to and activates the kinase Raf-1. Proc. Natl. Acad. Sci. USA. 1996;93:7063–7068. doi: 10.1073/pnas.93.14.7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Hwang JR, Zhang C, Patterson C. C-terminus of heat shock protein 70-interacting protein facilitates degradation of apoptosis signal-regulating kinase 1 and inhibits apoptosis signal-regulating kinase 1-dependent apoptosis. Cell Stress Chaperones. 2005;10:147–156. doi: 10.1379/CSC-90R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Daviau A, Proulx R, Robitaille K, Di Fruscio M, Tanguay RM, et al. Down-regulation of the mixed-lineage dual leucine zipper-bearing kinase by heat shock protein 70 and its cochaperone CHIP. J. Biol. Chem. 2006;281:31467–31477. doi: 10.1074/jbc.M607612200. [DOI] [PubMed] [Google Scholar]
- 249.Hunter T. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol. Cell. 2007;28:730–738. doi: 10.1016/j.molcel.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 250.Dai Z, Quackenbush RC, Courtney KD, Grove M, Cortez D, et al. Oncogenic Abl and Src tyrosine kinases elicit the ubiquitin-dependent degradation of target proteins trough a Ras-independent pathway. Genes Dev. 1998;12:1415–1424. doi: 10.1101/gad.12.10.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.