Abstract
Objective
Heterozygous missense mutations in the coding region of angiogenin (ANG), an angiogenic ribonuclease, have been reported in amyotrophic lateral sclerosis (ALS) patients. However, the role of ANG in motor neuron physiology and the functional consequences of these mutations are unknown. We searched for new mutations and sought to define the functional consequences of these mutations.
Methods
We sequenced the coding region of ANG in an independent cohort of North American ALS patients. Identified ANG mutations were then characterized using functional assays of angiogenesis, ribonucleolysis, and nuclear translocation. We also examined expression of ANG in normal human fetal and adult spinal cords.
Results
We identified four mutations in the coding region of ANG from 298 ALS patients. Three of these mutations are present in the mature protein. Among the four mutations, P(-4)S, S28N, and P112L are novel, and K17I has been reported previously. Functional assays show that these ANG mutations result in complete loss of function. The mutant ANG proteins are unable to induce angiogenesis because of a deficiency in ribonuclease activity, nuclear translocation, or both. As a correlate, we demonstrate strong ANG expression in both endothelial cells and motor neurons of normal human spinal cords from the developing fetus and adult.
Interpretation
We provide the first evidence that ANG mutations, identified in ALS patients, are associated with functional loss of ANG activity. Moreover, strong ANG expression, in normal human fetal and adult spinal cord neurons and endothelial cells, confirms the plausibility of ANG dysfunction being relevant to the pathogenesis of ALS.
Amyotrophic lateral sclerosis (ALS) is a progressive neurodegenerative disease that typically presents in the fifth to sixth decades of life with upper and lower motor neuron signs. Initially, there are symptoms that include distal muscle weakness and wasting, increased muscle tone with hyperreflexia, and at times diaphragmatic and/or bulbar weakness. Atypical forms can include symptoms of dementia, parkinsonism, or both. All forms of ALS inexorably progress to generalized amyotrophy, culminating in respiratory failure and death after an average duration of 3 to 5 years.1
The incidence of ALS is estimated at 0.4 to 1.8 per 100,000 people.2,3 Approximately 80 to 90% of ALS cases occur in individuals with no known family history, whereas the remaining cases are attributable to familial inheritance in either an autosomal dominant or recessive manner.3,4 Mutations in the Cu/Zn superoxide dismutase gene 1 (ALS1; SOD1; OMIM 147450), have been identified in 12 to 23% of familial5–7 and in 0 to 7% of sporadic8–10 ALS patients. Currently, SOD1 is the only known autosomal dominant gene in which mutations have been functionally associated with ALS, although three other loci have been identified for typical autosomal dominant ALS by linkage (ALS3, 18q21, OMIM 606640; ALS6, 16q12, OMIM 608030; and ALS7, 20ptel-p13, OMIM 608031).2,3 Other dominantly inherited genetic loci, associated with an atypical ALS phenotype, have also been identified (ALS with dementia/parkinsonism, MAPT, OMIM 157140; progressive lower motor neuron disease, DCTN1, OMIM 601143; and ALS8, VAPB, OMIM 608627). In autosomal dominant ALS with frontotemporal dementia (OMIM 105550), genetic linkage has been reported to 9q21-q22.11 Mutations in the SETX gene (OMIM 608465) have been identified in juvenile-onset autosomal dominant ALS. Lastly, genetic loci identified for juvenile-onset autosomal recessive disease include ALS2 (ALS2, 2q33, OMIM 606352) and linkage for ALS5 to 15q15.1-q21.2,3
Recent linkage analysis in Irish and Scottish ALS populations identified chromosome 14q11.2 as a candidate region and then angiogenin (ANG), a 14.1kDa angiogenic ribonuclease (RNase), as an ALS candidate gene.12,13 Seven heterozygous missense mutations in ANG were identified by sequence screening of 1,629 patients with ALS.13 Analysis of the ANG crystal structure suggested that these mutations may disrupt the structure and result in functional loss. However, the functional consequences of these mutations are unknown.13 We now report herein the identification of four mutations in the coding region of the ANG gene on screening an independent cohort of 298 SOD1-negative ALS patients. Three of these mutations occur in the mature protein and one in the signal peptide sequence. Using angiogenesis, ribonucleolysis, and nuclear translocation assays, we demonstrate that these mutations result in complete loss of function. Moreover, we show ANG expression in both endothelial cells and motor neurons of normal human fetal and adult spinal cord. Our data suggest that ANG plays a role in motor neuron health and provide evidence that ANG mutations, identified in ALS patients, are associated with functional loss of angiogenic activity.
Patients and Methods
Patients
Clinical specimens used in this study were those submitted to the Massachusetts General Hospital Clinical Neurogenetics DNA Diagnostics Laboratory for mutational analysis of the SOD1 gene. These specimens were identified as anonymous under a discarded tissue protocol approved by the authors’ institutional review board. In general, the clinical diagnosis of ALS is believed to be accurate in greater than 95% of cases.1,14 Neurologists were the primary referring physicians for these cases. This cohort consists of both apparently sporadic and familial ALS patients. Because specific data on ethnicity of each case are not available, we have referenced this as a North American cohort. Heterozygous missense mutations in the coding region of ANG gene were found in four patients.
PATIENT 1
Patient 1 is a man of English extraction, and apparently a sporadic case, who had onset of disease at 60 years of age. He has typical ALS clinical features including onset of weakness in distal extremities, upper and lower motor neuron signs, and stiffness with cramping. The patient’s reported history and examination documented a progressive amyotrophy, spasticity with brisk reflexes, and normal sensory function. No bulbar signs or diaphragmatic weakness are present now 3 years into the disease course.
PATIENT 2
Patient 2 is a 62-year-old woman of German and Eastern European mixed extraction, who evidenced onset of disease at 61 years of age with typical clinical features of distal extremity weakness and mild lower extremity spasticity. Fasciculations were noted clinically but were unconfirmed by electrophysiology studies. No bulbar or respiratory symptoms are in evidence now 1 year into the course of her disease. There is no known family history of ALS.
PATIENT 3
Patient 3 is an 83-year-old man of German and English mixed extraction, with a positive family history. He had a 4- to 5-year history of progressive weakness that was more prominent in the lower than upper extremities. At the time of the submission of this manuscript, the patient was wheelchair bound and had difficulty with speech and swallowing, as well as respiratory function. During the interval of manuscript review, the patient died. DNA from the proband’s affected son is not yet available for DNA analysis. Results when available will be published separately.
PATIENT 4
Patient 4 is a Hispanic woman who presented at the relatively early age of 28 years. Two years into her disease, she has primarily distal weakness of the finger and pretibial lower extremity muscles but shows no evidence of long tract signs or upper motor neuron abnormalities. She has a history of early-onset painful neuropathy, as has her father. However, the submitting physician indicated that the painful neuropathy is not a typical clinical feature of ALS but is within the spectrum of what was clinically perceived to be an “atypical case.”
Extended genealogical data for the four patients with ANG mutations are not available. In these ANG-positive ALS patients, no atypical distinguishing clinical features were recognized by the referring physicians apart from the neuropathy described in Patient 4. Moreover, no suggestion of angiogenesis dysfunction was noticed.
Mutation Screening
Genomic DNA was extracted from patients’ peripheral leukocytes using PureGene DNA Purification System by Gentra (Minneapolis, MN). Exon 2 (ANG coding sequence) and portions of the adjacent intronic sequences of the ANG gene were amplified with the following polymerase chain reaction (PCR) primer pair: forward, 5′-TTTGG TGATG CTGTT CTTGG-3′; reverse, 5′TGGGG GAAAG ATCAA TATGC-3′. The PCR amplification was performed using Taq DNA polymerase, and a protocol comprising an initial hold of 5 minutes at 94°C, then 30 cycles (30 seconds at 95°C, 30 seconds at 60°C, and 30 seconds at 72°C), followed by an extension of 7 minutes at 72°C. PCR products were purified by shrimp alkaline phosphatase and Exonuclease I with incubation at 37°C for 15 minutes followed by at 85°C for 15 minutes. Amplicons were sequenced bidirectionally on an ABI 3130×1 (Applied Biosystems, Foster City, CA) by capillary gel electrophoresis. The sequencing reaction was conducted using BigDye Terminator v1.1 (Applied Biosystems). Raw sequence data were analyzed using SeqScape Software for Mutation Profiling, version 5.0 (Applied Biosystems). Positive mutations were identified by comparison of sequence information in both directions against reference sequence (ANG, RefSeq NM_001145) and further confirmed by independent reamplification and bidirectional sequencing from the patients’ original DNA aliquots. Structural viewing of mutant location in wild-type (WT) ANG was performed using Protein Workshop (from http://www.rcsb.org/pdb/home/home.do using the Protein Data Bank accession ID = 1ANG).
Preparation of Recombinant Proteins
A DNA fragment encoding ANG was amplified from a previous clone by PCR with the following primers: forward, 5′-AGCGG ATCCC AGGAT AACTC CAGGT AC-3′; reverse, 5′-AGCGA ATTCT TACTA TAGAC TGAAA AATGA-3′, which contain an EcoRI and a BamHI cleavage site, respectively. This fragment was inserted into the expression vector pGEX-4T-2 between the BamHI and EcoRI sites. Mutations were generated by two-step PCR15 using the following primers: S28N: forward, 5′-TACTG TGAAA ACATC ATGAG G-3′; reverse, 5′-CCTCA TGATG TTTTC ACAGT A-3′; K17I: forward, 5′-CTATG ATGCC ATACC ACAGG GC-3′; reverse, 5′-GCCCT GTGGT AT-GGC ATCAT AG-3′; P112L: forward, 5′-AATGG CTTAC TTGTC CACTT G-3′; reverse, 5′-CAAGT GGACA AG-TAA GCCA TT-3′. The inserted ANG gene and mutations were confirmed by sequencing. All plasmids were transformed into BL21, and protein expression was induced by IPTG. Cells were disrupted by treatment with lysozyme in phosphate-buffered saline (1.5mg per gram of wet BL21) containing 5mM phenylmethyl sulfonyl fluoride. The cleared supernatant of the lysate was applied to a glutathione–Sepharose 4B affinity column. Bound glutathione-S-transferase fusion proteins were cleaved with 250 units thrombin directly from the column. The flow-through was then applied to a diethylaminoethanol-Sephadex column equilibrated with 10mM tris(hydroxymethyl)aminomethane (Tris)-HCl, pH 8.0, at 4°C to remove thrombin. Postdiethylaminoethanol fractions were applied to an SP-Sephadex column equilibrated with 25mM Tris-HCl, pH 8.0, containing 0.2M NaCl. Recombinant proteins were eluted with 25mM Tris-HCl, pH 8.0, containing 0.8M NaCl, dialyzed, and lyophilized.
Angiogenesis Assay
Angiogenic activities of the WT and mutant ANG were determined by assessing endothelial cell tube formation assay on fibrin gels.16 The Fibrin In Vitro Angiogenesis Assay Kit (Chemicon International, Temecula, CA) was used with the following modifications to the manufacturer’s instructions: Fibrin gels were formed by mixing 120μl fibrinogen solution with 80μl thrombin solution in each well of a 48-well tissue culture plate. Human umbilical vein endothelial cells (HUVECs) were seeded at 3 × 104 cells/well in human endothelial serum-free basal growth medium (HEM) containing 1% bovine serum albumin and incubated with 0.2μg/ml of WT or mutant ANG at 37°C under humidified 5% CO2 and for 18 hours. The cells were overlaid with another layer of fibrin gel and incubated for another 48 hours.
Ribonucleolysis Assay
Ribonucleolytic activities of the WT and mutant ANG were examined using yeast transfer RNA as the substrate. Proteins to be tested were added to an assay mixture containing 0.6mg yeast transfer RNA, 30μg RNase-free bovine serum albumin, 30mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 6.8, and 30mM NaCl in a final volume of 300μl. After incubation for 2 hours at 37°C, 700μl of 3.4% ice-cold perchloric acid was added, and the mixture was vortexed and kept on ice for 10 minutes. Specimens were then centrifuged at 15,000g for 10 minutes at 4°C. The absorbance of the supernatants was measured at 260nm.
Nuclear Translocation of Angiogenin
HUVECs were seeded at a density of 5 × 103 cells/mm2 on a coverslip and cultured in HEM + 5% fetal bovine serum and 5ng/ml basic fibroblast growth factor for 24 hours, washed with HEM, and incubated with 1μg/ml WT or mutant ANG proteins at 37°C for 30 minutes. Cells were washed with phosphate-buffered saline, fixed with methanol at −20°C for 10 minutes, and washed again with phosphate-buffered saline containing 30mg/ml bovine serum albumin. The fixed cells were then incubated with 10μg/ml of anti-ANG monoclonal antibody 26-2F for 1 hour, washed, and incubated with Alexa 488 –labeled goat F(ab′)2 anti–mouse IgG at 1:100 dilution for 1 hour. Cell nuclei were stained with 46′-diamidino-2-phenylindole-2 HCl.
Immunohistochemistry and Immunofluorescence
Normal spinal cord tissues were collected from anonymous autopsy materials with approval of authors’ institutional review board. Specimens were selected carefully with the expertise of a neuropathologist and considered the full autopsy report details to exclude clinical cases in which spinal cord pathology might be expected. Specimens were fixed in formalin and embedded in paraffin. Tissue sections of 4μm were cut, deparaffinized with xylene, rehydrated in ethanol, and microwaved for 15 minutes in 10mM citrate buffer, pH 6.0. For immunohistochemistry, endogenous peroxidase was blocked by treatment with 0.3% hydrogen peroxide in methanol for 30 minutes. Sections were blocked in 5% dry milk for 10 minutes and incubated with 10μg/ml of 26-2F at 4°C for 16 hours. Bound antibody was detected with Dako’s Envision system (Dako, Carpinteria, CA). Sections were counterstained with hematoxylin. For immunofluorescence, sections were incubated with 26-2F (30μg/ml) at 4°C overnight and with anti–von Willebrand factor (1:500 dilution) at 37°C for 1 hour. After washing, the sections were incubated with a mixture of Alexa 488 –labeled goat F(ab′)2 anti–mouse IgG and Alexa 555–labeled goat F(ab′)2 anti–rabbit IgG, both at 1:100 dilution, for 1 hour.
Results
Novel Mutations in the Coding Region of Angiogenin
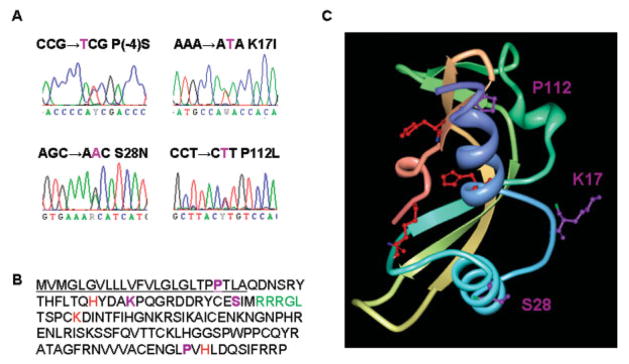
To examine whether ANG mutations occur in a different ALS population than the original cohort Green-way and colleagues13 reported and to understand the functional consequences of ANG mutations, we sequenced the coding region of ANG in an independent cohort of 298 North American patients who have the clinical phenotype of ALS but did not have SOD1 mutations. We identified four mutations in the coding region of ANG, three of which have not been reported previously (Fig 1A). Three mutations are in the mature protein region, and one is in the signal peptide sequence (see Fig 1B). One of these mutations occurs at P112, located in close proximity to H114, which together with H13 and K40 form the catalytic triad17 for RNase activity (see Fig 1C). The P112L mutation may alter the orientation of H114, thereby affecting the enzymatic activity of ANG. Because the RNase activity of ANG is essential for angiogenesis,18 this mutation may thus abolish its biological activity. The other two mutations occur at S28 and K17, relatively distant from the catalytic triad, and their effect on RNase activity of ANG is not intuitive from the crystal structure (see Fig 1C). The K17I mutation was reported previously and was suggested to alter the activity of ANG based on comparative analysis with ANG-RNaseA hybrids.13 The S28N mutation is located adjacent to the nuclear localization sequence (NLS)19 and may thus interfere with the nuclear translocation process of ANG. The fourth mutation, P(-4)S, occurs in the signal peptide of the ANG precursor and may thus affect processing of the precursor and secretion of the mature protein. However, because the mature protein does not contain this mutation, it cannot be further studied using the experimental methods herein.
Fig 1.
Angiogenin (ANG) mutations identified in Northern American amyotrophic lateral sclerosis (ALS) patients. (A) DNA sequence traces of mutations identified by bidirectional sequencing. Mutations are indicated using single-letter amino acid code. (B) Amino acid sequence of ANG with the signal peptide underlined. Mutations identified in this study, P(-4)S, K17I, S28N, and P112L, are shown in purple. The RNase catalytic residues (H13, K40, and H114) are shown in red. The nuclear localization signal (31 RRRGL35) is shown in green. (C) Crystal structure of ANG (from www.rcsb.org; 1ANG) showing the positions of mutated residues (purple). The catalytic triad is shown in red.
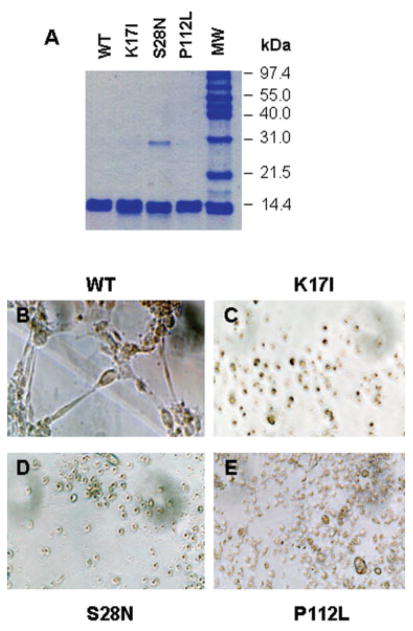
Mutant Angiogenin Proteins Are Not Angiogenic
To directly assess the functional consequences of these coding mutations, we prepared recombinant proteins using site-directed mutagenesis. As shown in Figure 2A, all three mutant proteins and the WT ANG were purified to homogeneity, with a trace dimeric form of S28N. We have shown previously that ANG dimmer formation does not alter its biological activity.20 A standard in vitro angiogenesis assay using HUVEC tube formation on fibrin gel was next performed to assess the angiogenic activity of the mutant ANG. HUVECs cultured on fibrin gel will form tubular structures on treatment with an active angiogenic factor.16 WT ANG (see Fig 2B) induces the formation of HUVEC tubes, whereas K17I (see Fig 2C), S28N (see Fig 2D), and P112L (see Fig 2E) do not, indicating that all three mutant proteins have completely lost their angiogenic activities. Thus, ANG mutations, identified in ALS patients, are loss-of-function mutations.
Fig 2.
Angiogenic activity of wild-type (WT) and mutant angiogenin (ANG) proteins. Recombinant WT and mutant ANG proteins were expressed and purified. (A) Sodium dode-cyl sulfate polyacrylamide gel electrophoresis and Coomassie blue staining. Five micrograms of the proteins were loaded in each lane. (B–E) Endothelial cell tube formation assay. Human umbilical vein endothelial cells (HUVECs) were cultured on fibrin gels in the presence of (B) WT, (C) K17I, (D) S28N, and (E) P112L. Formation of tubular structures was evaluated using a phase-contrast microscope. Pictures shown are from a representative experiment of five independent repeats. Original magnification ×40.
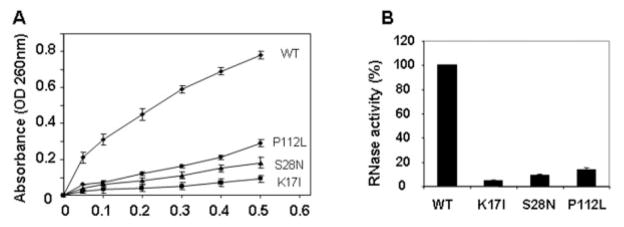
Ribonucleolytic Activity of Mutant Angiogenin Proteins
To explore the underlying mechanisms that result in the loss of function for each of these mutants, we performed ribonuclease assays using yeast transfer RNA as a substrate21 because this activity is essential for angiogenesis.18 As shown in Figure 3A, all three mutant proteins have substantially decreased activity compared with the WT ANG. Quantitative analysis (see Fig 3B) indicates that P112L, S28N, and K17I have 14, 9, and 5%, respectively, of the ribonucleolytic activity of WT ANG (n = 4; p < 0.001). Thus, all three mutant proteins have impaired RNase activity, which may account for the loss of angiogenic activity.
Fig 3.
Ribonucleolytic activity of wild-type (WT) and mutant angiogenin (ANG) proteins. RNase activity was measured with yeast transfer RNA (tRNA) as the substrate. Increasing concentration of WT and mutant ANG proteins were incubated with yeast tRNA (2mg/ml) at 37°C for 2 hours. Undigested tRNA was precipitated by perchloric acid. (A) Absorbance of the supernatants at 260nm. (B) Relative RNase activity of mutant ANG as compared with that of WT ANG (100%). The amount of enzyme required to generate 0.1 optical density (OD) is compared with that of WT ANG to generate the same OD unit. Student’s t test of four independent experiments shows that the difference between WT and each of the three mutant proteins is significant (n = 4; p < 0.001).
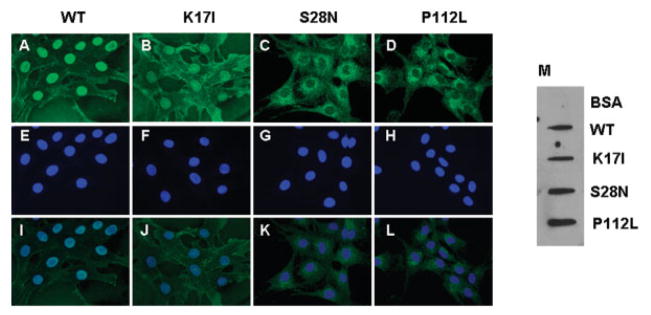
Nuclear Translocation of Mutant Angiogenin Protein in Endothelial Cells
We next examined whether these mutant ANG undergo nuclear translocation, another essential requirement for mediating angiogenesis.19 We have shown previously that nuclear translocation of ANG requires receptor-mediated endocytosis but is independent of lysosomes and microtubules.22 WT and mutant ANG were added to cultured HUVECs and detected by immunofluorescence with anti-ANG monoclonal antibody 26-2F.20 Figures 4A to D show immunofluorescent staining of ANG proteins. Figures 4E to H document 46′-diamidino-2-phenylindole-2 HCl staining of the cell nuclei. A merge of the two panels show that WT (see Fig 4I) and K17I (see Fig 4J) ANG are able to translocate to the nucleus, but that S28N (see Fig 4K) and P112L (see Fig 4L) ANG cannot. The primary antibody (26-2F) used in this experiment is specific for human ANG.23 X-ray structural analysis of ANG-antibody complex has shown that 26-2F interacts with two segments consisting of residues 34 to 41 and 85 to 91, respectively.24 These two regions are apart in the primary but close in the three-dimensional structures. A slot blotting experiment was performed to confirm that 26-2F is still able to recognize the mutant ANG protein (see Fig 4M) so that the decrease in nuclear ANG is not an artifact of staining. Because nuclear translocation is essential for ANG to induce angiogenesis, S28N and P112L are unlikely to be angiogenic even though they retain 14 and 9%, respectively, of the ribonucleolytic activity (see Fig 3B).
Fig 4.
Nuclear translocation of wild-type (WT) and mutant angiogenin (ANG) proteins. Human umbilical vein endothelial cells (HUVECs) were incubated with 1μg/ml of (A, E, I) WT, (B, F, J) K17I, (C, G, K) S28N, and (D, H, L) P112L ANG at 37°C for 30 minutes and fixed with −20°C methanol. ANG was visualized by immunofluorescence with the anti-ANG monoclonal antibody 26-2F and Alexa 488 –labeled goat F(ab′)2 anti–mouse IgG (A–D). (E–H) 46′-diamidino-2-phenylindole-2 HCl staining of the cell nuclei. (I–L) Merge of the green and blue fluorescence. Images are from a representative experiment of five independent repeats. Original magnification ×600. BSA = bovine serum albumin.
Expression of Angiogenin in Human Spinal Cord
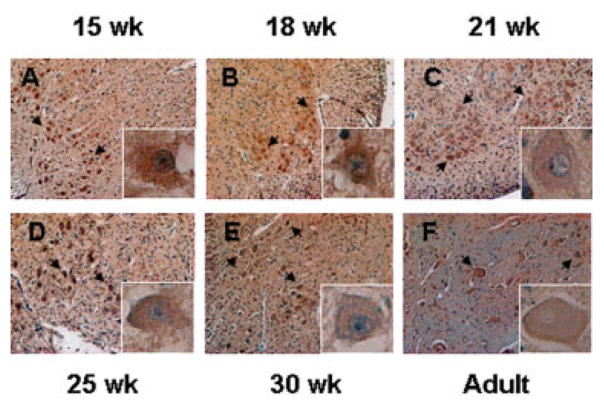
To explore the plausibility of ANG mutations being relevant to motor neuron disease in humans, we used immunohistochemistry to detect the ANG protein in normal spinal cords obtained from fetal (ranging from 15–30 weeks’ gestation) and adult human autopsies. Strong ANG staining was observed in spinal cord ventral horn motor neurons of both fetal and adult cases (Figs 5A–F). Of note, strong ANG staining was also detected in the extracellular matrix and interstitial tissues of all cases, consistent with ANG being a secreted protein.25 Currently, it is unclear whether the ANG protein detected in motor neurons is expressed by the neurons or is a consequence of cellular uptake of secreted ANG from other types of cells. Nevertheless, strong cytoplasmic and nuclear accumulation of ANG in motor neurons of both prenatal and adult spinal cords (see Fig 5, insets) suggest a physiological role of ANG, both early in development and later in adulthood, and supports the hypothesis that ANG mutations are likely relevant to ALS pathology. To the best of our knowledge, this is the first demonstration of ANG expression in human spinal cords.
Fig 5.
Immunohistochemical staining of angiogenin (ANG) in fetal and adult human spinal cords. Spinal cords of (A) 15-, (B) 18-, (C) 21-, (D) 25-, and (E) 30-week-old fetuses and of an (F) adult were collected, fixed in formalin, and embedded in paraffin. Sections of 4μM were cut and stained immunohistochemically for ANG with 26-2F. Images are from the ventral horn area of the spinal cords where motor neurons are located. Arrows denote ANG staining in motor neurons. Original magnification ×100. Insets are the high-magnification (A–E, ×400; F, ×200) images of the motor neuron and its surroundings.
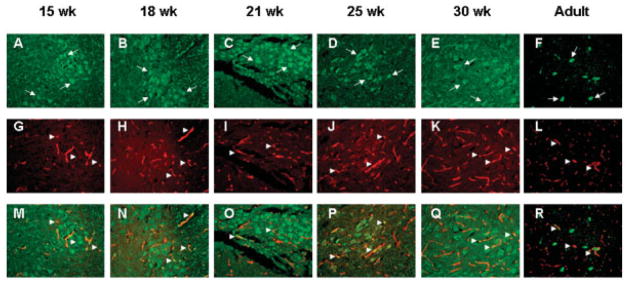
Double immunofluorescence with anti-ANG (green, Figs 6A–F) monoclonal and anti–von Willebrand factor (red, Figs 6G–L) polyclonal antibodies was conducted to determine whether ANG is also localized in endothelial cells of spinal cord tissues. As shown in Figure 6, besides strong expression in motor neurons (see Figs 6A–F, arrows), ANG also colocalizes with von Willebrand factor in the blood vessels (see Figs 6M–R, arrowheads) of both prenatal and adult spinal cords. These results suggest that ANG also may mediate angiogenesis in the spinal cord and may play a role in maintaining the physiological health of motor neurons. Thus, ANG abnormalities may have a dual role in ALS: directly through motor neuron function and/or indirectly through endothelial cells and aberrant angiogenesis.
Fig 6.
Angiogenin (ANG) expression in motor neurons and blood vessels of fetal and adult human spinal cords. (A–F) Green immunofluorescence for ANG with 26-2F and Alexa 488 –labeled goat anti–mouse IgG. Arrows indicate representative ANG staining in motor neurons. (G–L) Red immunofluorescence for blood vessels with anti–von Willebrand factor polyclonal antibody and Alexa 555–labeled goat anti–rabbit IgG. Arrowheads indicate representative blood vessels. (M–R) Merge of green and red fluorescence. Arrowheads indicate colocalization of ANG and von Willebrand factor. Images are from a representative area of the ventral horns of spinal cords of (A, G, M) 15-, (B, H, N) 18-, (C, I, O) 21-, (D, J, P) 25-, and (E, K, Q) 30-week-old fetuses and an (F, L, R) adult. Original magnification ×100.
Discussion
ANG is an angiogenic protein originally isolated from human tumor conditioned medium based on its angiogenic activity.26 It is upregulated in numerous human cancers and plays a role in tumor angiogenesis.27 Mechanistic studies indicate that ANG undergoes nuclear translocation in endothelial cells where it binds to the promoter region of rDNA, stimulates rRNA transcription, and is essential for cell proliferation.28 ANG-mediated rRNA transcription has been shown to be required for angiogenesis induced by vascular endothelial cell growth factor (VEGF),29 an essential angiogenic protein that has also been implicated in ALS.30 In a mouse model of ALS, disruption of the promoter element of VEGF results in selective motor neuron degeneration.31 In SOD1G93A rats, treatment with intraventricular VEGF results in substantially improved motor function, delayed disease onset, and extended survival.32 In humans, VEGF was shown to be a modifier of ALS by protecting motor neurons from ischemic injury and death.30 A potential role of ANG in ALS is thus envisioned from its involvement in VEGF-mediated angiogenesis.
Independently, linkage analysis identified ANG as a potential ALS susceptibility gene.12 Allelic association studies of Irish and Scottish ALS populations identified chromosome 14q11.2 where ANG gene is located as a candidate region. A synonymous single nucleotide polymorphism (rs 11701) was found to be associated with Irish and Scottish ALS populations from sequencing 1,629 ALS patients.12 Thereafter, seven heterozygous missense mutations in ANG were identified in 15 patients with either familial or sporadic ALS by sequencing the same 1,629 ALS patients.13 From this work, ANG was proposed as the second angiogenic molecule to be involved in ALS.33 However, direct functional testing was not performed, and it is unknown whether these mutations affect the biological activity of ANG.13
Our study has identified, in an independent North American ALS cohort, three novel mutations (P(-4)S, S28N, P112L) in ANG and confirms one previously documented mutation (K17I). In addition, we demonstrate abolished angiogenic activity in all three mutations that occur in the mature ANG protein. Mature ANG consists of only 123 amino acid residues, but 3 distinct functional sites; that is, the receptor binding site, the catalytic triad, and the NLS, have been identified. All three functional sites need to be intact for ANG to be angiogenic. K17I is capable of nuclear translocation (see Fig 4B), suggesting that the loss of its angiogenic activity is likely due to its diminished (by 95%) ribonucleolytic activity (see Fig 3B). S28N has lost its nuclear translocation ability (see Fig 4C), perhaps consistent with it being located near the NLS (see Fig 1B). Moreover, this variant has only 9% of the ribonucleolytic activity of the WT ANG. The loss of angiogenic activity of S28N could be due to the loss of the enzymatic activity, incapacitated nuclear translocation, or both. The findings that P112L retains partial ribonucleolytic activity but loses completely nuclear translocation ability are rather unexpected. P112 is positioned only two amino acid residues away from the essential catalytic residue H114, but it is distant from the known NLS. Substitution of Pro by Leu would likely change the local structure and could thereby significantly alter the catalytic center, but its effect on the structure of the NLS would be expected to be minimal. Further structural work is needed to specifically elucidate how P112 loses its nuclear translocation property and yet retains limited enzymatic activity.
Our data suggest that ANG is the first gene in which loss-of-function mutations are documented in ALS patients. It is currently unknown whether the mutant ANG proteins have a dominant negative function in angiogenesis or in motor neuron function, or in both. Alternatively, haploinsufficiency might account for the ANG-related pathobiology in ALS because all the mutations so far identified are heterozygous.13 It is possible that homozygous loss of ANG is lethal. A role for ANG in ALS pathobiology is not unexpected because dysfunctional angiogenesis has been implicated in ALS pathogenesis.30,33–36 We have previously shown that ANG is a permissive factor for angiogenesis induced by VEGF,29 the first angiogenic molecule shown to play a role in ALS.30,31,37,38 Furthermore, we demonstrated that ANG-mediated ribosomal biogenesis is a general requirement for endothelial cell proliferation.29 Our finding here, that ANG is expressed in endothelial cells of spinal cords of both normal fetal and adult humans, and that mutant ANG completely lacks angiogenic activity, further supports the importance of angiogenesis in motor neuron physiology, in general, and its dysfunction in ALS, in particular.
The role of ANG in neuroprotection probably extends beyond its effect on endothelial cells, as significant immunostaining of ANG was observed in ventral horn motor neurons of both normal human fetal and adult spinal cords (see Fig 5). We have recently shown that ANG may have functions independent of angiogenesis, as it is involved more broadly in ribosomal biogenesis.28,29,39 ANG binds to the promoter region of rDNA and stimulates rRNA transcription.28 As such, ANG function in motor neurons may be related to ribosomal biogenesis and protein translation. A defect in this pathway, as a consequence of ANG mutation in ALS, may lead to insufficient synthesis of ribosomes, thereby affecting motor neuron viability. Efforts to create and characterize ANG transgenic and knockout mice and to determine the expression and function of ANG in motor neurons and glia cells are under way.
Acknowledgments
This work was supported by the NCI (R01 CA105241, G.H.) and the Stanley L. Robbins Memorial Research Fund, Department of Pathology, Brigham and Women’s Hospital (D.W.).
We thank the patients, their families, and physicians for their participation in this project. We also thank Dr M.B. Feany for helpful discussions.
References
- 1.Rowland LP, Shneider NA. Amyotrophic lateral sclerosis. N Engl J Med. 2001;344:1688–1700. doi: 10.1056/NEJM200105313442207. [DOI] [PubMed] [Google Scholar]
- 2.Gros-Louis F, Gaspar C, Rouleau GA. Genetics of familial and sporadic amyotrophic lateral sclerosis. Biochim Biophys Acta. 2006;1762:956–972. doi: 10.1016/j.bbadis.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 3.Pasinelli P, Brown RH. Molecular biology of amyotrophic lateral sclerosis: insights from genetics. Nat Rev Neurosci. 2006;7:710–723. doi: 10.1038/nrn1971. [DOI] [PubMed] [Google Scholar]
- 4.Boillee S, Vande Velde C, Cleveland DW. ALS: a disease of motor neurons and their nonneuronal neighbors. Neuron. 2006;52:39–59. doi: 10.1016/j.neuron.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 5.Andersen PM, Nilsson P, Keranen ML, et al. Phenotypic heterogeneity in motor neuron disease patients with CuZn-superoxide dismutase mutations in Scandinavia. Brain. 1997;120(pt 10):1723–1737. doi: 10.1093/brain/120.10.1723. [DOI] [PubMed] [Google Scholar]
- 6.Cudkowicz ME, McKenna-Yasek D, Sapp PE, et al. Epidemiology of mutations in superoxide dismutase in amyotrophic lateral sclerosis. Ann Neurol. 1997;41:210–221. doi: 10.1002/ana.410410212. [DOI] [PubMed] [Google Scholar]
- 7.Niemann S, Joos H, Meyer T, et al. Familial ALS in Germany: origin of the R115G SOD1 mutation by a founder effect. J Neurol Neurosurg Psychiatry. 2004;75:1186–1188. doi: 10.1136/jnnp.2003.028324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Battistini S, Giannini F, Greco G, et al. SOD1 mutations in amyotrophic lateral sclerosis. Results from a multicenter Italian study. J Neurol. 2005;252:782–788. doi: 10.1007/s00415-005-0742-y. [DOI] [PubMed] [Google Scholar]
- 9.Corrado L, D’Alfonso S, Bergamaschi L, et al. SOD1 gene mutations in Italian patients with sporadic amyotrophic lateral sclerosis (ALS) Neuromuscul Disord. 2006;16:800–804. doi: 10.1016/j.nmd.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 10.Jones CT, Swingler RJ, Simpson SA, Brock DJ. Superoxide dismutase mutations in an unselected cohort of Scottish amyotrophic lateral sclerosis patients. J Med Genet. 1995;32:290–292. doi: 10.1136/jmg.32.4.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vance C, Al-Chalabi A, Ruddy D, et al. Familial amyotrophic lateral sclerosis with frontotemporal dementia is linked to a locus on chromosome 9p13.2-21.3. Brain. 2006;129:868–876. doi: 10.1093/brain/awl030. [DOI] [PubMed] [Google Scholar]
- 12.Greenway MJ, Alexander MD, Ennis S, et al. A novel candidate region for ALS on chromosome 14q11.2. Neurology. 2004;63:1936–1938. doi: 10.1212/01.wnl.0000144344.39103.f6. [DOI] [PubMed] [Google Scholar]
- 13.Greenway MJ, Andersen PM, Russ C, et al. ANG mutations segregate with familial and ‘sporadic’ amyotrophic lateral sclerosis. Nat Genet. 2006;38:411–413. doi: 10.1038/ng1742. [DOI] [PubMed] [Google Scholar]
- 14.Rowland LP. Diagnosis of amyotrophic lateral sclerosis. J Neurol Sci. 1998;160(suppl 1):S6–S24. doi: 10.1016/s0022-510x(98)00193-2. [DOI] [PubMed] [Google Scholar]
- 15.Higuchi R, Recombinant PCR. In: PCR protocols: a guide to methods and applications. Innis MA, editor. New York: Academic Press; 1990. pp. 178–183. [Google Scholar]
- 16.Chalupowicz DG, Chowdhury ZA, Bach TL, et al. Fibrin II induces endothelial cell capillary tube formation. J Cell Biol. 1995;130:207–215. doi: 10.1083/jcb.130.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shapiro R, Weremowicz S, Riordan JF, Vallee BL. Ribonucleolytic activity of angiogenin: essential histidine, lysine, and arginine residues. Proc Natl Acad Sci U S A. 1987;84:8783–8787. doi: 10.1073/pnas.84.24.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shapiro R, Vallee BL. Site-directed mutagenesis of histidine-13 and histidine-114 of human angiogenin. Alanine derivatives inhibit angiogenin-induced angiogenesis. Biochemistry. 1989;28:7401–7408. doi: 10.1021/bi00444a038. [DOI] [PubMed] [Google Scholar]
- 19.Moroianu J, Riordan JF. Nuclear translocation of angiogenin in proliferating endothelial cells is essential to its angiogenic activity. Proc Natl Acad Sci U S A. 1994;91:1677–1681. doi: 10.1073/pnas.91.5.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu G, Xu C, Riordan JF. Human angiogenin is rapidly translocated to the nucleus of human umbilical vein endothelial cells and binds to DNA. J Cell Biochem. 2000;76:452–462. doi: 10.1002/(sici)1097-4644(20000301)76:3<452::aid-jcb12>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 21.Shapiro R, Riordan JF, Vallee BL. Characteristic ribonucleolytic activity of human angiogenin. Biochemistry. 1986;25:3527–3532. doi: 10.1021/bi00360a008. [DOI] [PubMed] [Google Scholar]
- 22.Li R, Riordan JF, Hu G. Nuclear translocation of human angiogenin in cultured human umbilical artery endothelial cells is microtubule and lysosome independent. Biochem Biophys Res Commun. 1997;238:305–312. doi: 10.1006/bbrc.1997.7290. [DOI] [PubMed] [Google Scholar]
- 23.Fett JW, Olson KA, Rybak SM. A monoclonal antibody to human angiogenin. Inhibition of ribonucleolytic and angiogenic activities and localization of the antigenic epitope. Biochemistry. 1994;33:5421–5427. doi: 10.1021/bi00184a010. [DOI] [PubMed] [Google Scholar]
- 24.Chavali GB, Papageorgiou AC, Olson KA, et al. The crystal structure of human angiogenin in complex with an antitumor neutralizing antibody. Structure. 2003;11:875–885. doi: 10.1016/s0969-2126(03)00131-x. [DOI] [PubMed] [Google Scholar]
- 25.Kurachi K, Davie EW, Strydom DJ, et al. Sequence of the cDNA and gene for angiogenin, a human angiogenesis factor. Biochemistry. 1985;24:5494–5499. doi: 10.1021/bi00341a032. [DOI] [PubMed] [Google Scholar]
- 26.Fett JW, Strydom DJ, Lobb RR, et al. Isolation and characterization of angiogenin, an angiogenic protein from human carcinoma cells. Biochemistry. 1985;24:5480–5486. doi: 10.1021/bi00341a030. [DOI] [PubMed] [Google Scholar]
- 27.Tello-Montoliu A, Patel JV, Lip GY. Angiogenin: a review of the pathophysiology and potential clinical applications. J Thromb Haemost. 2006;4:1864–1874. doi: 10.1111/j.1538-7836.2006.01995.x. [DOI] [PubMed] [Google Scholar]
- 28.Xu ZP, Tsuji T, Riordan JF, Hu GF. Identification and characterization of an angiogenin-binding DNA sequence that stimulates luciferase reporter gene expression. Biochemistry. 2003;42:121–128. doi: 10.1021/bi020465x. [DOI] [PubMed] [Google Scholar]
- 29.Kishimoto K, Liu S, Tsuji T, et al. Endogenous angiogenin in endothelial cells is a general requirement for cell proliferation and angiogenesis. Oncogene. 2005;24:445–456. doi: 10.1038/sj.onc.1208223. [DOI] [PubMed] [Google Scholar]
- 30.Lambrechts D, Storkebaum E, Morimoto M, et al. VEGF is a modifier of amyotrophic lateral sclerosis in mice and humans and protects motoneurons against ischemic death. Nat Genet. 2003;34:383–394. doi: 10.1038/ng1211. [DOI] [PubMed] [Google Scholar]
- 31.Oosthuyse B, Moons L, Storkebaum E, et al. Deletion of the hypoxia-response element in the vascular endothelial growth factor promoter causes motor neuron degeneration. Nat Genet. 2001;28:131–138. doi: 10.1038/88842. [DOI] [PubMed] [Google Scholar]
- 32.Storkebaum E, Lambrechts D, Dewerchin M, et al. Treatment of motoneuron degeneration by intracerebroventricular delivery of VEGF in a rat model of ALS. Nat Neurosci. 2005;8:85–92. doi: 10.1038/nn1360. [DOI] [PubMed] [Google Scholar]
- 33.Lambrechts D, Lafuste P, Carmeliet P, Conway EM. Another angiogenic gene linked to amyotrophic lateral sclerosis. Trends Mol Med. 2006;12:345–347. doi: 10.1016/j.molmed.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 34.Lambrechts D, Storkebaum E, Carmeliet P. VEGF: necessary to prevent motoneuron degeneration, sufficient to treat ALS? Trends Mol Med. 2004;10:275–282. doi: 10.1016/j.molmed.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 35.Storkebaum E, Lambrechts D, Carmeliet P. VEGF: once regarded as a specific angiogenic factor, now implicated in neuroprotection. Bioessays. 2004;26:943–954. doi: 10.1002/bies.20092. [DOI] [PubMed] [Google Scholar]
- 36.Vande Velde C, Cleveland DW. VEGF: multitasking in ALS. Nat Neurosci. 2005;8:5–7. doi: 10.1038/nn0105-5. [DOI] [PubMed] [Google Scholar]
- 37.Azzouz M, Ralph GS, Storkebaum E, et al. VEGF delivery with retrogradely transported lentivector prolongs survival in a mouse ALS model. Nature. 2004;429:413–417. doi: 10.1038/nature02544. [DOI] [PubMed] [Google Scholar]
- 38.Ilzecka J. Does VEGF represent a potential treatment for amyotrophic lateral sclerosis? Curr Opin Investig Drugs. 2006;7:54–59. [PubMed] [Google Scholar]
- 39.Tsuji T, Sun Y, Kishimoto K, et al. Angiogenin is translocated to the nucleus of HeLa cells and is involved in ribosomal RNA transcription and cell proliferation. Cancer Res. 2005;65:1352–1360. doi: 10.1158/0008-5472.CAN-04-2058. [DOI] [PubMed] [Google Scholar]