Abstract
OBJECTIVES
The goal of this study was to identify genes upregulated in the heart in human patients with hypertrophic cardiomyopathy (HCM).
BACKGROUND
Hypertrophic cardiomyopathy is a genetic disease caused by mutations in contractile sarcomeric proteins. The molecular basis of diverse clinical and pathologic phenotypes in HCM remains unknown.
METHODS
We performed polymerase chain reaction-select complementary DNA subtraction between normal hearts and hearts with HCM and screened subtracted libraries by Southern blotting. We sequenced the differentially expressed clones and performed Northern blotting to detect increased expression levels.
RESULTS
We screened 288 independent clones, and 76 clones had less than twofold increase in the signal intensity and were considered upregulated. Sequence analysis identified 36 genes including those encoding the markers of pressure overload-induced (“secondary”) cardiac hypertrophy, cytoskeletal proteins, protein synthesis, redox system, ion channels and those with unknown function. Northern blotting confirmed increased expression of skeletal muscle alpha-actin (ACTA1), myosin light chain 2a (MLC2a), GTP-binding protein Gs-alpha subunit (GNAS1), NADH ubiquinone oxidoreductase (NDUFB10), voltage-dependent anion channel 1 (VDAC1), four-and-a-half LIM domain protein 1 (FHL1) (also known as SLIM1), sarcosin (SARCOSIN) and heat shock 70kD protein 8 (HSPA8) by less than twofold. Expression levels of ACTA1, MLC2a and GNAS1 were increased in six additional and FHL1 in four additional hearts with HCM.
CONCLUSIONS
A diverse array of genes is upregulated in the heart in human patients with HCM, which could account for the diversity of clinical and pathologic phenotypes. Markers of secondary hypertrophy are also upregulated, suggesting commonality of pathways involved in HCM and the acquired forms of cardiac hypertrophy. Elucidation of the role of differentially expressed genes in HCM could provide for new therapeutic targets.
Hypertrophic cardiomyopathy (HCM) is a genetic disease caused by mutations in the contractile sarcomeric proteins (1). Hypertrophic cardiomyopathy is diagnosed clinically by the presence of unexplained left ventricular hypertrophy and pathologically by myocyte hypertrophy, disarray and interstitial fibrosis (2). While the causality of mutant contractile proteins in the pathogenesis of HCM is well established, the molecular bases of diverse cardiac phenotypes in HCM remain unknown.
We and others have proposed cardiac hypertrophy in HCM is a “compensatory” phenotype due to increased cardiac myocyte stress or altered Ca+2 sensitivity of the contractile apparatus imparted by the mutant contractile proteins (3). Accordingly, increased myocyte stress leads to expression of a variety of cardiac genes that activate the transcription machinery leading to hypertrophy and other phenotypes of HCM. Thus, the pathogenesis of hypertrophy, a common programmed response of the myocardium to any form of stress, whether caused by a genetic defect or by an acquired condition, involves common pathways. Similarly, pathogenesis of diverse cardiac phenotypes also results from upregulation of expression of a variety of genes in response to the primary impetus provided by the mutant contractile protein (3). To identify genes that are upregulated in the heart in HCM, as the initial step for delineating their role in induction of cardiac phenotypes, we performed subtraction hybridization between normal hearts and hearts with HCM followed by Northern blotting to confirm upregulation of expression of the differentially expressed genes in HCM.
METHODS
Subtraction hybridization and differential screening
Each patient signed an informed consent, and the institutional review board approved the protocols. Subtraction hybridization was performed between two messenger ribo-nucleic acid (mRNA) pools extracted from a heart with HCM and a normal heart, using a polymerase chain reaction (PCR)-select complementary deoxyribonucleic acid (cDNA) subtraction kit (CLONETECH, Palo Alto, California). In brief, left ventricular septal tissue was obtained by myomectomy from a patient who had a long-standing history of symptomatic HCM with left ventricular outflow tract obstruction. A corresponding segment of the interventricular septum was excised from an age- and gender-matched normal donor heart that was not used for cardiac transplantation. Tissues were frozen in liquid nitrogen and stored at −135°C until mRNA extraction. Total RNA and poly A+ RNA were extracted from hearts with HCM and control hearts side-by-side using the same reagents and protocols and quantified by measuring absorption at 260 nm. To select genes that were differentially expressed in the heart in HCM, 2 µg of polyA+ mRNAs extracted were used to generate subtraction cDNA libraries. The cDNA library generated from the normal heart was subtracted from that of the heart with HCM (forward subtraction) through two sets of hybridization, which resulted in a library enriched for genes that were upregulated in the heart with HCM. A reverse subtraction cDNA library was generated by subtracting the cDNA library generated from the heart with HCM from that of the normal heart. To reduce the background, two rounds of suppression PCR were performed to amplify the differentially expressed genes. The enriched cDNA library was subcloned into a T/A cloning plasmid vector, and the resulting products were transformed into Escherichia coli and plated on agar plates.
To confirm that the subtracted clones represented differentially expressed genes and to reduce the number of false positives, colonies were initially screened by PCR-select differential screening (CLONETECH). In brief, the cloned inserts were amplified by PCR using flanking primers, and 288 randomly selected clones in the forward-subtracted cDNA library were arrayed on three sets of 96-spot dot blot membranes in duplicates for subsequent hybridization. Probes were prepared by PCR amplification of the forward and reverse subtraction cDNA libraries, restriction enzyme digestion to remove the adapter sequences in order to reduce the background and radiolabeling with [32P] dCTP a specific activity of more than 109 cpm/µg. Each set of membranes was hybridized either with forward or reverse subtracted probe in ExpressHyb solution (CLONETECH). After washing in high stringency conditions, the membranes were exposed to X-ray films in the presence of intensifying screens for 24 h.
Sequencing
Differentially expressed clones were sequenced using Big Dye Terminator Cycle Sequencing Ready Reaction Kit on an ABI Genetic Analyzer 310 (PE Biosystem, Foster City, California). Sequences were analyzed using BLAST search of gene bank database.
Northern blotting
To confirm differential expression of the putative candidate genes in HCM, Northern blotting was performed on mRNA extracts from hearts with HCM and control hearts. In brief, 10 µg aliquots of total RNA extracts were loaded onto a formaldehyde-agarose gel, subjected to electrophoresis and were transferred to a nylon membrane (Bio-Rad Laboratories, Cambridge, Massachusetts). The forward-subtracted cDNA probes were radiola-beled with [32P] dCTP and hybridized to mRNAs on the membranes in the presence of Denhardt’s reagent (0.1% ficol, 0.1% polyvinylpyrrolidone, 0.1% bovine serum albumin, 100 µg/ml of denatured fragmented salmon sperm DNA) in hybridization solution (6 × sodium chloride-sodium citrate, 0.5% sodium dodecylsulfate). After washing, the membrane was exposed to an X-ray film for 24 to 48 h.
To determine whether the differentially expressed clones were also upregulated in additional hearts with HCM, Northern blotting was performed on mRNA extracts from six additional hearts with HCM and two control hearts.
RESULTS
Genes upregulated in HCM
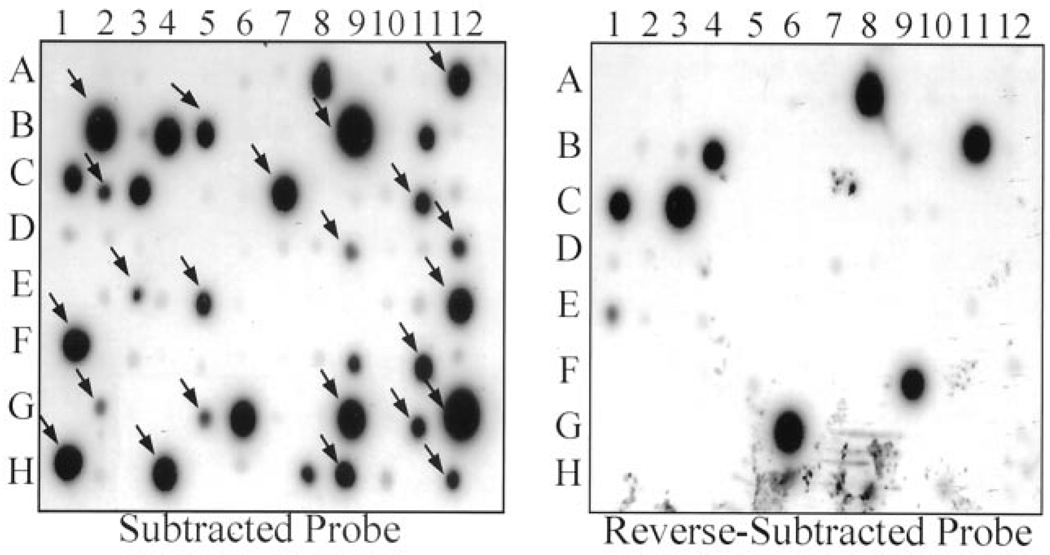
A total of 288 clones from the subtracted library were screened by duplicate dot-blot hybridization. A twofold or greater difference in the intensity of signals detected from colonies hybridized with the forward and reverse-subtraction probes (forward less than reverse) was considered evidence of differential expression. We identified 76 differentially expressed clones (Fig. 1) that were sequenced. Blast analyses of the sequences identified 36 differentially expressed genes (Table 1) that code for a diverse array of proteins with a variety of functions.
Figure 1.
Subtracted clones dot blotted at 96-spot membranes. Membranes hybridized with radiolabeled forward and reverse subtracted complementary deoxyribonucleic acid libraries. Differentially expressed genes, identified by a twofold increase in signal intensity in the membrane hybridized with the forward subtraction probes, are shown by arrows. Clones hybridized with the reverse subtraction probes are considered background.
Table 1.
Differentially Expressed Genes in the Heart in HCM
| Gene | Symbol | Accession # | Locus | # of Clone |
|---|---|---|---|---|
| Sarcomeric | ||||
| Skeletal muscle α-actin | ACTA1 | J00068 | 1q42.13–q42.2 | 12 |
| Myosin light chain 2a | LOC58498 | M94547 | 7p11.21–p11.2 | 7 |
| Myosin light chain 2v | MYL2v | AF020768 | 12q23–q24.3 | 7 |
| β-myosin heavy chain | MYL7 | M58018 | 14q2 | 3 |
| α-myosin heavy chain | MYH6 | D00943 | 14q2 | 2 |
| Skeletal fast myosin light chain 2 | MLC2B | M21812 | ? | 1 |
| Myosin, light chain 3, alkali | MYL3 | M24122 | 3p21.3–p21.2 | 1 |
| Sarcosin | SARCOSIN | AF056929 | ? | 1 |
| Cytoskeletal | ||||
| Four-and-a-half LIM protein 1 | FHL1 | U29538 | Xq26 | 2 |
| SLIMMER | SLIMMER | AF063002 | Xq26 | 2 |
| Skeletal muscle α 2 actinin | ACTN2 | M86406 | 1q42–q43 | 2 |
| Nebulin-related protein | NRAP | U96486 | 10q24–q26 | 1 |
| Desmin | DES | M63391 | 2q35 | 1 |
| Protein synthesis | ||||
| Transcription elongation fac 1α1 | EEF1A1 | X03558 | 6q14 | 4 |
| A+U-rich element RNA binding factor | HNRPDL | D89678 | 4q13–21 | 2 |
| Mitochondrial ribosomal RNA | MTRNR1 | S64650 | mitochondrion | 2 |
| Mitochondrial ribosomal RNA | 16S rRNA | X93334 | mitochondrion | 1 |
| Ribosomal protein S4X | RPS4X | M58458 | Xq13.1 | 1 |
| Ribosomal protein L31 | RPL31 | X15940 | 2 | 1 |
| Ribosomal protein L24 | RPL24 | M94314 | 3q | 1 |
| Redox system | ||||
| NADH ubiquinone oxidoreductase | NDUFB10 | AF088991 | 16 | 3 |
| Ubiquinol-cytochrome c reductase core I | UQCRC1 | D26485 | 3p21.3 | 3 |
| NADH dehydrogenase Fe-S protein 1 | NDUFS1 | X61100 | 2q33–q34 | 1 |
| Ion channels | ||||
| Voltage-dependent anion channel 1 | VDAC1 | L06132 | 5q31 | 1 |
| Na+/H+ exchanger isoform 1 | SLC9A1 | S68616 | 1p36.1–p35 | 1 |
| Solute carrier family 25, Member 3-1a | SLC25A3 | X77337 | 12q23 | 1 |
| Extracellular matrix | ||||
| α-1 (I) chain of procollagen type I | COL1A1 | AF017178 | 17q21.3–q22 | 1 |
| Intracellular signal transduction | ||||
| GTP-binding protein Gs α subunit | GNAS1 | X07036 | 20q13.2–q13.3 | 1 |
| Others and unknowns | ||||
| Heat shock 70kD protein 8 | HSPA8 | Y00371 | 11q23.3–q25 | 2 |
| Smooth muscle myosin light chain 2 | MYRL2 | J02854 | 20 | 2 |
| Nonsarcomeric MLC-regulatory | MLCB | X54304 | 18 | 2 |
| B-type natriuretic peptide precursor | BNNP | M25296 | 1p36.2 | 1 |
| Myoglobin gene | MB | X00372 | 22q13.1 | 1 |
| HSPC040 (CGI-129) | LOC51660 | AF151887 | 6 | 1 |
| Chromosome 1 open reading frame 8 | C1ORF8 | AF047439 | 1p36–p31 | 1 |
HCM = hypertrophic cardiomyopathy.
Northern blot analyses
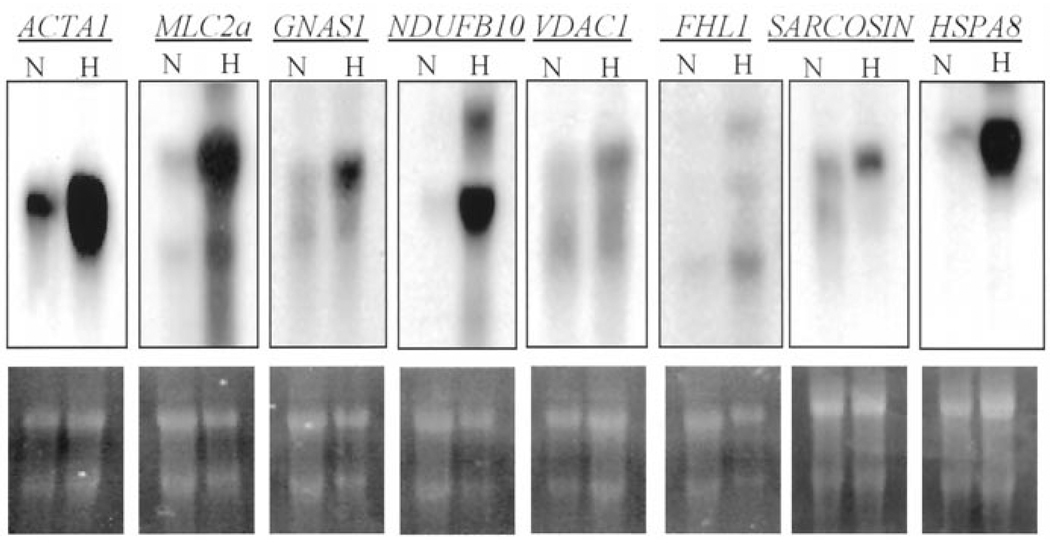
Northern blot analyses were performed to compare expression of skeletal muscle alpha-actin (ACTA1), myosin light chain 2a (MLC2a), guanosine 5′-triphosphate (GTP)-binding protein Gs-alpha subunit (GNAS1), reduced nicotinamide adenine dinucleotide ubiquinone oxidoreductase (NDUFB10), voltage-dependent anion channel 1 (VDAC1), four-and-a-half LIM domain protein 1 (FHL1) (also known as SLIM1), sarcosin (SARCOSIN) and heat shock 70kD protein 8 (HSPA8) between hearts with HCM and control hearts. The results, shown in Figure 2, confirmed upregulation of expression of ACTA1 (fourfold), MLC2a (fivefold), GNAS1 (threefold), NDUFB10 (eightfold), VDAC1 (twofold), FHL1 (twofold), SARCOSIN (twofold) and HSPA8 (ninefold).
Figure 2.
Northern blots showing upregulation of expression of a selected differentially expressed gene identified by subtraction hybridization: the upper panels shows blots representing expression of ACTA1, MLC2a, GNAS1, NDUFB10, HSPA8, VDAC1, FHL1 and SARCOSIN in hearts with hypertrophic cardiomyopathy (H) and normal control (N) hearts. The lower panels show the corresponding agarose gels photographs showing 28 S and 18 S ribonucleic acids. ACTA1 = skeletal muscle alpha-actin; FHL1 = four-and-a-half LIM domain protein 1; GNAS1 = GTP-binding protein Gs-alpha subunit; HSPA8 = heat shock 70kD protein 8; MLC2a = myosin light chain 2a; NDUFB10 = reduced nicotinamide adenine dinucleotide ubiquinone oxidoreductase; VDAC1 = voltage-dependent anion channel 1.
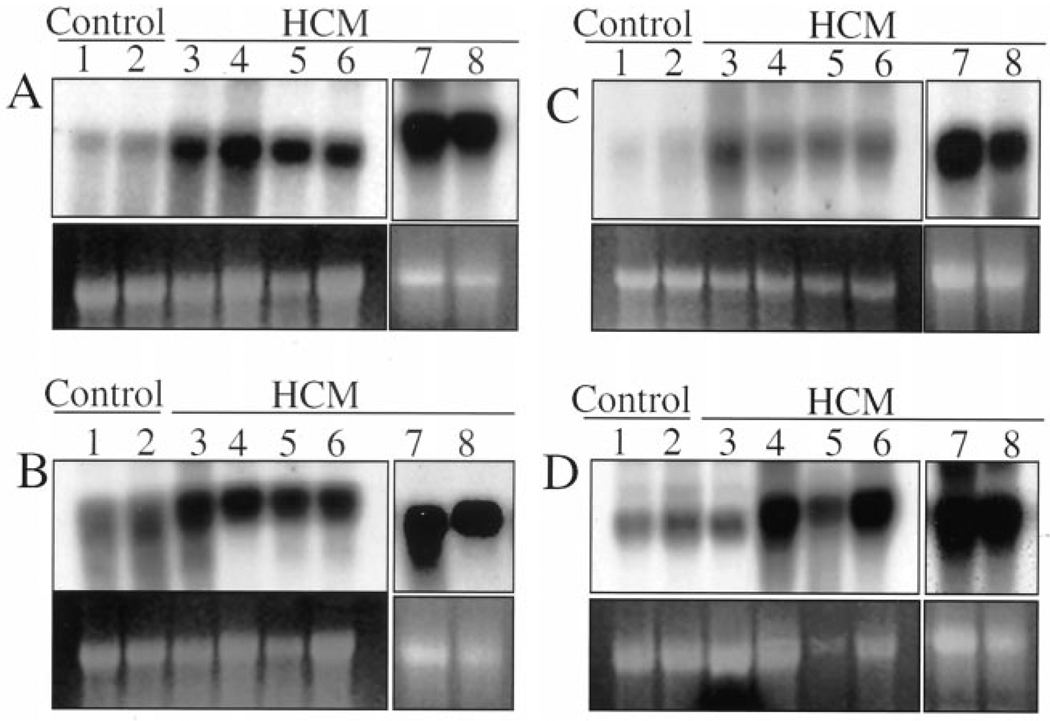
Expression levels of ACTA1, GNAS1, MLC2a and FHL1 genes were determined by Northern blotting in six additional left ventricular tissues from patients with HCM and two normal hearts. The results are shown in Figure 3. As shown, expression levels of ACTA1, GNAS1 and MLC2a were increased in all six hearts by less than twofold, while expression levels of FHL1 were increased by more than twofold in four hearts.
Figure 3.
Northern blots showing the expression of four differentially expressed genes in the left ventricular tissues of six additional patients with hypertrophic cardiomyopathy (HCM). Lanes 1 and 2 represent the left ventricular samples obtained from normal hearts (donor hearts not used for transplantation). Lanes 3 through 8 represent the left ventricular tissues obtained from patients with HCM. Expression levels of skeletal muscle alpha-actin (A), myosin light chain 2A (B) and GTP-binding protein Gs-alpha subunit (C) were increased in all hearts with HCM, as compared with the control hearts. Expression levels of four-and-a-half LIM domain protein 1 (D) were increased by less than twofold in 4 out of 6 hearts with HCM.
DISCUSSION
A diverse array of genes encoding for proteins with variety of functions, including markers of cardiac hypertrophy due to pressure overload (secondary hypertrophy), is upregulated in the human heart in HCM. The diversity of molecular phenotype in HCM is in accord with the diversity of pathologic and clinical phenotypes that encompass not only myocyte hypertrophy and disarray but also interstitial fibrosis, thickening of the media of intramural coronary arteries and arrhythmias (2). Increased expression of the markers of “secondary” cardiac hypertrophy supports the hypothesis that hypertrophy in HCM is also a “secondary” phenotype (3), and common pathways are involved in the induction of cardiac hypertrophy in genetic and nongenetic forms.
We have confirmed increased expression of selected differentially expressed genes (eight genes) by Northern blotting in the myomectomy sample that was used for subtraction subsequently and subsequently for four genes in multiple hearts with HCM, further validating the observed results. We used PCR-select cDNA subtraction hybridization, which is considered a robust technique for identification of differentially expressed genes (4,5). Unlike microar-ray DNA chips, which are restricted to known sequences and are subject to bias for the selection of putative genes, subtraction hybridization affords the opportunity to screen all differentially expressed genes without a selection bias or a priori knowledge of their sequences. However, it is a tedious method, and, in each set of subtraction experiments, a fraction of total differentially expressed genes are identified, and the yield decreases with each additional subtraction. We performed three sets of colony screening from a single cDNA subtraction experiment and selected genes that showed at least a twofold increase in the expression levels. Therefore, the total number of differentially expressed genes in the heart in HCM could be greater. In addition, expression of many genes could be decreased in HCM, and, since we did not screen the reverse subtracted library, they were not detected. Furthermore, cDNA subtraction hybridization detects gene expression at the mRNA stage, which may not correspond to protein expression. Moreover, expression profiling detects only expression of the genes and not the activity of their proteins, which, for many proteins, such as the intracellular signaling kinases, is the primary determinant of protein function. Therefore, it not surprising that upregulation of expression of intracellular signaling kinases, which play fundamental roles in induction of acquired forms of cardiac hypertrophy (6) and, likewise, are expected to modulate cardiac hypertrophy in HCM, were not detected. Furthermore, expression profiling is an initial step that provides a basis for future studies to delineate the role of differentially expressed genes in the pathogenesis of cardiac phenotype.
A variety of genes including those encoding for contractile sarcomeric proteins, cytoskeletal proteins, ion channels, intracellular signal tranducers, protein maintaining the redox state of the myocardium, along with transcriptional and translation machinery were upregulated in HCM. The most common upregulated genes were the markers of “secondary” cardiac hypertrophy, such as skeletal alpha-actin, isoforms of myosin light chain and, less commonly, brain natriuretic factor, also known to be activated in pressure-overload induced (secondary) cardiac hypertrophy. Many of the upregulated genes have not been previously implicated in cardiac hypertrophy, and function of a few, such as sarcosin (7) and SLIM1 (8), is unknown. In addition, structure and function of proteins encoded by HSPC040 (9) and C1ORF8 (10), which have been identified by expressed sequence tags analysis and comparative proteomics, respectively, are unknown. Among the genes with the highest upregulation were HSPA8, also known as HSP73, which is a member of HSP70 multigene family with chaperoning roles for nascent polypeptide facilitating their correct folding, translocation and degradation (11). Expression of NDUFB10 encoding for the first enzyme complex in the electron transport chain of mitochondria (12) was also upregulated significantly. Upregulation of the expression of HSPA8 and NDUFB10 (12), along with increased expression of many ribosomal proteins is in accord with the increased protein synthesis and mitochondria function in hypertrophic states and signifies their role as potential modulators of cardiac phenotype in HCM. Other notable genes included GNAS1, encodes for the GTP-binding protein Gs and couples the hormone-receptor binding to adenylyl cyclase activation (13); VDAC1, encodes for VDAC1 that forms the major pathway for movement of adenine nucleotides through the outer membrane of mitochondria (14) and sodium hydrogen exchanger, which is a ubiquitous membrane-bound enzyme involved in pH regulation and activated by a variety of signals including growth factors (15).
In summary, we have identified a variety of genes that are upregulated in human patients with HCM that could contribute to the diversity of clinical and pathologic phenotypes of HCM. Increased expression of genes encoding for the markers of “secondary” cardiac hypertrophy supports the hypothesis that hypertrophy in HCM, a genetic disease, is also a “secondary” phenotype and involves pathways that are similar to those implicated in the acquired forms. Characterization of the role of upregulated gene in HCM could provide clues to the diversity of clinical and pathologic phenotypes and for new therapeutic and preventive targets.
Acknowledgments
Supported by grants from the National Heart, Lung, and Blood Institute, Specialized Centers of Research (P50-HL42267-01) and an Established Investigator Award (9640133N) from the AHA, National Center, Dallas, Texas.
Abbreviations and Acronyms
- ACTA1
skeletal muscle alpha-actin
- cDNA
complementary deoxyribonucleic acid
- FHL1
four-and-a-half LIM domain protein 1
- GNAS1
guanosine 5′-triphosphate (GTP)-binding protein Gs-alpha subunit
- HCM
hypertrophic cardiomyopathy
- HSPA8
heat shock 70kD protein 8
- MLC2a
myosin light chain 2A
- mRNA
messenger ribonucleic acid
- NDUFB10
reduced nicotinamide adenine dinucleotide ubiquinone oxidoreductase
- PCR
polymerase chain reaction
- SARCOSIN
sarcosin
- VDAC1
voltage-dependent anion channel 1
REFERENCES
- 1.Seidman CE. Hypertrophic cardiomyopathy: from man to mouse. J Clin Invest. 2000;106:S9–S13. [Google Scholar]
- 2.Wigle ED, Rakowski H, Kimball BP, Williams WG. Hypertrophic cardiomyopathy: clinical spectrum and treatment. Circulation. 1995;92:1680–1692. doi: 10.1161/01.cir.92.7.1680. [DOI] [PubMed] [Google Scholar]
- 3.Marian AJ. Pathogenesis of diverse clinical and pathological phenotypes in hypertrophic cardiomyopathy. Lancet. 2000;355:58–60. doi: 10.1016/s0140-6736(99)06187-5. [DOI] [PubMed] [Google Scholar]
- 4.Diatchenko L, Lau YF, Campbell AP, et al. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci U S A. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zuber J, Tchernitsa OI, Hinzmann B, et al. A genome-wide survey of RAS transformation targets. Nat Genet. 2000;24:144–152. doi: 10.1038/72799. [DOI] [PubMed] [Google Scholar]
- 6.Molkentin JD. Calcineurin and beyond: cardiac hypertrophic signaling. Circ Res. 2000;87:731–738. doi: 10.1161/01.res.87.9.731. [DOI] [PubMed] [Google Scholar]
- 7.Taylor A, Obholz K, Linden G, Sadiev S, Klaus S, Carlson KD. DNA sequence and muscle-specific expression of human sarcosin transcripts. Mol Cell Biochem. 1998;183:105–112. doi: 10.1023/a:1006824331819. [DOI] [PubMed] [Google Scholar]
- 8.Brown S, McGrath MJ, Ooms LM, Gurung R, Maimone MM, Mitchell CA. Characterization of two isoforms of the skeletal muscle LIM protein 1, SLIM1. Localization of SLIM1 at focal adhesions and the isoform slimmer in the nucleus of myoblasts and cytoplasm of myotubes suggests distinct roles in the cytoskeleton and in nuclear-cytoplasmic communication. J Biol Chem. 1999;274:27083–27091. doi: 10.1074/jbc.274.38.27083. [DOI] [PubMed] [Google Scholar]
- 9.Mao M, Fu G, Wu JS, et al. Identification of genes expressed in human CD34(+) hematopoietic stem/progenitor cells by expressed sequence tags and efficient full-length cDNA cloning. Proc Natl Acad Sci U S A. 1998;95:8175–8180. doi: 10.1073/pnas.95.14.8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lai CH, Chou CY, Ch’ang LY, Liu CS, Lin W. Identification of novel human genes evolutionarily conserved in Caenorhabditis elegans by comparative proteomics. Genome Res. 2000;10:703–713. doi: 10.1101/gr.10.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tavaria M, Gabriele T, Anderson RL, et al. Localization of the gene encoding the human heat shock cognate protein, HSP73, to chromosome 11. Genomics. 1995;29:266–268. doi: 10.1006/geno.1995.1242. [DOI] [PubMed] [Google Scholar]
- 12.Loeffen JL, Triepels RH, van den Heuvel LP, et al. cDNA of eight nuclear encoded subunits of NADH: ubiquinone oxidoreductase: human complex I cDNA characterization completed. Biochem Biophys Res Commun. 1998;253:415–422. doi: 10.1006/bbrc.1998.9786. [DOI] [PubMed] [Google Scholar]
- 13.Kozasa T, Itoh H, Tsukamoto T, Kaziro Y. Isolation and characterization of the human Gs alpha gene. Proc Natl Acad Sci U S A. 1988;85:2081–2085. doi: 10.1073/pnas.85.7.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blachly-Dyson E, Baldini A, Litt M, McCabe ER, Forte M. Human genes encoding the voltage-dependent anion channel (VDAC) of the outer mitochondrial membrane: mapping and identification of two new isoforms. Genomics. 1994;20:62–67. doi: 10.1006/geno.1994.1127. [DOI] [PubMed] [Google Scholar]
- 15.Sardet C, Franchi A, Pouyssegur J. Molecular cloning, primary structure, and expression of the human growth factor-activatable Na+/H+ antiporter. Cell. 1989;56:271–280. doi: 10.1016/0092-8674(89)90901-x. [DOI] [PubMed] [Google Scholar]