Abstract
The newly recognized bacterial second messenger 3′,5′-cyclic diguanylic acid (cyclic diguanylate (c-di-GMP)) has been shown to regulate a wide variety of bacterial behaviors and traits. Biosynthesis and degradation of c-di-GMP have been attributed to the GGDEF and EAL protein domains, respectively, based primarily on genetic evidence. Whereas the GGDEF domain was demonstrated to possess diguanylate cyclase activity in vitro, the EAL domain has not been tested directly for c-di-GMP phosphodiesterase activity. This study describes the analysis of c-di-GMP hydrolysis by an EAL domain protein in a purified system. The Vibrio cholerae EAL domain protein VieA has been shown to inversely regulate biofilm-specific genes (vps) and virulence genes (ctxA), presumably by decreasing the cellular pool of c-di-GMP. VieA was maximally active at neutral pH, physiological ionic strength, and ambient temperatures and demonstrated c-di-GMP hydrolytic activity with a Km of 0.06 μm. VieA was unable to hydrolyze cGMP. The putative metal coordination site of the EAL domain, Glu170, was demonstrated to be necessary for VieA activity. Furthermore, the divalent cations Mg2+ and Mn2+ were necessary for VieA activity; conversely, Ca2+ and Zn2+ were potent inhibitors of the VieA phosphodiesterase. Calcium inhibition of the VieA EAL domain provides a potential mechanism for regulation of c-di-GMP degradation.
The cyclic dinucleotide 3′,5′-cyclic diguanylic acid (c-di-GMP)2 has been characterized as an important second messenger that affects a range of physiological and behavioral traits in an assortment of bacterial species. C-di-GMP was initially identified as an allosteric activator of cellulose synthase in Gluconacetobacter xylinum (1). Similarly, c-di-GMP has been implicated in regulation of cellulose biosynthesis and biofilm formation in the Gram-negative enteric pathogen Salmonella enterica serovar Typhimurium (2), as well as in survival of S. typhimurium in mice (3). Regulation of multicellular behaviors such as biofilms is not limited to Gram-negative bacteria, since there is evidence that c-di-GMP inhibits biofilm formation in Staphylococcus aureus as well (4). In addition, c-di-GMP was shown to regulate the transition between sessile and motile phenotypes in S. typhimurium, the opportunistic pathogen Pseudomonas aeruginosa, and commensal Escherichia coli (5).
In the classical biotype of the facultative intestinal pathogen Vibrio cholerae, not only does the c-di-GMP control biofilm formation (6), but it also affects expression of virulence factors such as cholera toxin (CT) (7). Specifically, increased intracellular concentrations of c-di-GMP favor biofilm formation by activating expression of Vibrio exopolysaccharide (vps) genes (6), which encode enzymes that produce the extracellular matrix that provides the infrastructure for the characteristic three-dimensional shape of biofilms. Decreased levels of c-di-GMP, on the other hand, are associated with reduced vps expression and increased production of CT (6, 7). The mechanisms involved in c-di-GMP-mediated regulation of these phenotypes are entirely unknown.
The inverse regulation of environment-specific (biofilm) and infection-specific (virulence) genes is of particular interest because of the shift in gene expression that is central to the pathogenic V. cholerae life cycle. In endemic regions, V. cholerae is typically found in aquatic reservoirs, and there is evidence that it forms biofilms on abiotic surfaces, including the chitinous exoskeletons of zooplankton and phytoplankton (8–10). V. cholerae undergoes a shift in gene expression upon entry into the host; vps expression ceases (11), and expression of virulence genes is induced (12–16). Whereas much research has been dedicated to studying environmental forms, such as biofilms, and to elucidating mechanisms of virulence gene regulation, little is understood about the molecular mechanisms that control the shift from the environmental to virulent state of V. cholerae. The intracellular concentration of c-di-GMP and proteins that modulate c-di-GMP levels may be key determinants in the transition from the environmental biofilm state to the virulent state of V. cholerae.
Biosynthesis and degradation of c-di-GMP have been predicted to occur through the activities of diguanylate cyclases and phosphodiesterases, respectively. These activities have been attributed to proteins containing GGDEF and/or EAL domains (17, 18). The putative diguanylate cyclases contain a conserved domain with a Gly-Gly-Asp-Glu-Phe motif (GGDEF, also known as domain of unknown function 1, or DUF1) (18). The GGDEF domain has limited but significant homology to eukaryotic adenylate cyclases (19). Synthesis of c-di-GMP by GGDEF-containing enzymes involves the 3′ to 5′ linkage of two GTP molecules into a cyclic molecule with the release of two pyrophosphate groups (20). A number of GGDEF domain proteins have been demonstrated to have diguanylate cyclase activity in vitro (21).
The putative c-di-GMP phosphodiesterases contain a domain enriched in glutamic acid, alanine, and leucine (EAL; also the domain of unknown function 2, DUF2). The functions were initially assigned to the GGDEF and EAL domains based on the occurrence of these sequences in enzymes with the described activities. For example, in G. xylinum, in which c-di-GMP metabolism was first characterized, six genes were found that control the intracellular concentration of c-di-GMP, each containing a GGDEF and an EAL domain (18, 22). The phosphodiesterase activity attributed to the EAL domain is based on limited homology to eukaryotic nucleotide phosphodiesterases (reviewed in Ref. 23) and is predicted to result in the linearization of c-di-GMP to pGpG, a physiologically inactive dinucleotide; a second, presumably ubiquitous phosphodiesterase, PDE-B, would hydrolyze pGpG to two GMP molecules.
Genes encoding proteins with GGDEF and/or EAL domains are abundant in bacterial genomes, usually in combination with other regulatory or sensory domains. In V. cholerae, 22 open reading frames encode proteins with homology to the EAL module, and 40 encode GGDEF domain proteins. There is considerable overlap of these proteins, as 10 possess both domains. One particular EAL domain protein of interest, VieA, is a putative response regulator with a conserved N-terminal phosphoreceiver domain, an internal EAL domain, and a C-terminal helix-turn-helix DNA binding domain. Encoded in the vieSAB three-component signal transduction system operon, the vieA locus has been demonstrated to be required for virulence of V. cholerae in the infant mouse (24). A vieA mutant demonstrates decreased CT production and 10-fold attenuated virulence (7, 24). Moreover, deletion of vieA results in increased intracellular c-di-GMP, vps expression, and biofilm formation (6). Conversely, overexpression of VieA decreases c-di-GMP levels and vps expression (6). Mutation of the phosphoreceiver or DNA-binding domains of VieA does not affect these phenotypes, whereas mutation of the central EAL domain does (6, 24). Expression of VieA and subsequent c-di-GMP activity during infection may thus be a critical signal, mediated by the modulation of c-di-GMP concentrations, for V. cholerae to down-regulate environment-specific genes and increase production of virulence factors.
There is a large body of indirect evidence that proteins containing the EAL domain possess c-di-GMP phosphodiesterase activity. Recently, the EAL domain was demonstrated to possess phosphodiesterase activity on an unnatural substrate, but the specificity of the EAL domain for c-di-GMP was not addressed (25). In the present work, we show that the EAL domain protein VieA acts as a c-di-GMP phosphodiesterase in a purified system. In addition, we have characterized the enzymatic properties of VieA in terms of optimal reaction conditions, ion requirement, ion inhibition, and kinetics.
EXPERIMENTAL PROCEDURES
Strains and Growth Conditions
Strains used in this study are listed in TABLE ONE. V. cholerae were grown in LB broth containing 100 μg/ml streptomycin at 37 °C with aeration. E. coli were incubated in LB containing 50 μg/ml ampicillin or 10 μg/ml chloramphenicol, as appropriate, at 37 °C with aeration, except during induction of protein expression (see details below).
TABLE ONE.
Strains used in this study
| Strain | Description | Reference/Source |
|---|---|---|
| AC61 | Classical V. cholerae O395 lacZ::res-tet-res. | Ref. 33 |
| AC575 | DH5α pMMB67EH | Ref. 38 |
| AC1596 | V. cholerae O395 vieAE170A | Ref. 6 |
| AC1758 | E. coli DH5α pBAD33::His6-VCA0956 | This work |
| AC1817 | E. coli DH5α pMMB67EH::vieA-His6 | This work |
| AC1835 | E. coli DH5α pMMB67EH::vieAE170A-His6 | This work |
Cloning of EAL Domain Proteins
Full-length vieA and vieAE170A were cloned into expression vector pMMB67EH. Briefly, wild type vieA and the E170A point mutant were amplified from AC61 genomic DNA by PCR using primers vieAF2 (5′-CGAGCTCTTTAGGATACATTTTTATGAAAATAATGATAGTAGAAG-3′) and CHVAR1 (5′-AACTGCAGCTAATGATGATGATGATGATGTTTTAATGTTACAAAACGCAC-3′). Primer vieAF2 introduced a SacI restriction site (underlined), a ribosomal binding site (italicized), and a start codon (boldface type). Each reverse primer introduced a PstI site (underlined), a His6 tag (italicized), and a stop codon (boldface). The PCR fragments were digested with SacI and PstI, ligated into pMMB67EH, and transformed into E. coli DH5α by electroporation. The correct clones were confirmed by digestion and sequencing.
Purification of EAL Domain Proteins
Expression of His6-tagged EAL proteins was induced in exponential growth phase bacteria with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside at 30 °C with aeration. Bacteria were collected by centrifugation and resuspended in His6 buffer consisting of 10 mm Tris, pH 8, 300 mm NaCl, 50 mm NaH2PO4, 10% glycerol, 1 mm phenylmethylsulfonyl fluoride, and 2 mm imidazole. All subsequent steps were carried out at 4 °C. The suspensions were lysed by French press at 16,000 p.s.i. and then briefly sonicated to break up genomic DNA. The lysates were fractionated by centrifugation at 10,000 × g for 30 min. The soluble fraction (supernatant) was incubated with Ni2+-nitrilotriacetic acid-agarose resin (Qiagen) for 1 h with gentle rocking. Subsequently, the samples were applied to a 5-ml polypropylene column (Qiagen), and the resin was allowed to settle. After elution of the His6 buffer, the column was washed twice with wash buffer (His6 buffer, 40 mm imidazole). The His6-tagged proteins were eluted from the column with elution buffer (His6 buffer, 250 mm imidazole) and then dialyzed overnight in Slide-A-Lyzer 10,000 molecular weight cut-off dialysis cassettes (Pierce) against 75 mm Tris, pH 8, 250 mm NaCl, 25 mm KCl, 10 mm MgCl2, 30% glycerol. The lysates, column washes, and elutions were analyzed by SDS-PAGE and Coomassie Brilliant Blue staining to assess protein purity. The concentrations of the purified protein samples were determined using the modified Lowry protein assay kit (Pierce). Dialyzed proteins were stored at –20 °C.
Synthesis of c-di-GMP Substrate
C-di-GMP substrate for the phosphodiesterase assays was synthesized enzymatically using the VCA0956 protein, which contains a GGDEF diguanylate cyclase domain. VCA0956 was amplified from AC61 genomic DNA with primers A0956F2 (5′-GCTCTAGATTTAGGATACATTTTTATGCATCATCATCATCATCATATGACAACTGAAGATTTCAAAA-3′) and A0956R (5′-ACATGCATGCTTAGAGCGGCATGACTCGAT-3′). Primer A0956F2 introduced an XbaI restriction site (underlined), a ribosomal binding site (boldface type), and an N-terminal His6 tag (italicized). Primer A0956R introduced an SphI site (underlined) after the stop codon. The PCR fragment was digested and ligated into the arabinose-inducible expression vector pBAD33 and transformed into E. coli DH5α. Clones were confirmed by digestion and sequencing. Protein expression was induced in exponential phase cells using 0.2% l-arabinose. His6-VCA0956 was purified by Ni2+-nitrilotriacetic acid affinity chromatography using methods similar to those described for purification of the EAL domain proteins, except the His6 binding buffer contained 35 mm imidazole, and the His6 wash buffer contained 50 mm imidazole.
Purified His6-VCA0956 (60 ng/μl) was incubated 4 h to overnight at 23 °C with 3 μl of [α-32P]GTP (3000 Ci/mmol, 3.3 μm; PerkinElmer Life Sciences) in buffer containing 75 mm Tris, pH 8, 250 mm NaCl, 25 mm KCl, and 10 mm MgCl2. When higher concentrations of c-di-GMP were desired, unlabeled GTP was included at 0.01 or 0.1 mm final concentrations. The reactions were treated with calf intestinal phosphatase (New England Biolabs) for 30 min to hydrolyze noncyclized phosphate groups. The c-di-GMP product was separated from proteins by centrifugation through an Ultrafree-MC 5000 molecular weight cut-off column (Centricon) for 45 min at 5000 × g. Reaction products were separated by thin layer chromatography using polyethyleneimine (PEI)-cellulose plates and 1.5 m KH2PO4, pH 3.65, buffer (1) and then analyzed by phosphorimagery and volume analysis using ImageQuant 1.2. The final concentration and yield of c-di-GMP were calculated based on the percentage incorporation of radiolabeled GTP into radiolabeled c-di-GMP. Products were stored at –20 °C.
Phosphodiesterase Assays
The standard phosphodiesterase assay involved incubating purified protein with 11 nm [32P]c-di-GMP (no unlabeled c-di-GMP) for 30 min to 1 h at room temperature in 75 mm Tris, pH 8, 250 mm NaCl, 25 mm KCl, and 10 mm MgCl2. A no enzyme negative control and a snake venom phosphodiesterase (SVPD; Amersham Biosciences, product number E20240Y) positive control were included in all assays. The reactions were stopped by spotting 0.5 μl of the reaction on PEI-cellulose and allowing it to air-dry. Nucleotides were separated by TLC in 1.5 m KH2PO4, pH 3.65, and visualized by phosphorimagery.
The following experiments were done to determine the optimum parameters for phosphodiesterase activity (1). To determine the optimal reaction temperature, a master mix of the above reaction was aliquoted (30 μl) and incubated in heat blocks at 4, 16, 23, 30, 37, 45, 52, 65, or 80 °C (2). For optimization of reaction pH, the pH of the standard buffer described above was adjusted with HCl or NaOH to pH 3.5–12 (3). The optimum Mg2+ concentration for VieA activity was determined using 75 mm Tris, pH 8, and 150 mm KCl; MgCl2 was added to the reaction at 0–100 mm final concentrations (4). The optimal ionic strength for the reaction was determined by measuring activity in 75 mm Tris, pH 8, 25 mm MgCl2, and 0–500 mm KCl. For each experiment, 0.5 μl were analyzed by TLC after 45 min. Experiments addressing the inhibitory effects of divalent cations were performed at 23 °C using 75 mm Tris, pH 8, 25 mm KCl, 25 mm MgCl2, and 0.33 μm c-di-GMP and analyzed as described. For all experiments, the percentage hydrolysis of c-di-GMP was determined. All assays were performed in triplicate.
The requirement of VieA for divalent cations was evaluated by first chelating divalent cations from the protein and substrate preparations. Briefly, Chelex-100 was added to 100-μl aliquots of VieA or c-di-GMP at ~10 mg of Chelex-100 per ml and incubated for 10 min at room temperature. The protein or substrate was then transferred to fresh Chelex-100; this was repeated for a total of five treatments. The final incubation was allowed to proceed for 1.5 h at room temperature. Chelated samples were stored at –20 °C. The protein samples were confirmed to lack phosphodiesterase activity by incubating with [32P]c-di-GMP in reaction buffer lacking MgCl2. Divalent cations from MgCl2, CaCl2, MnCl2, and ZnCl2 were reconstituted into 75 mm Tris, pH 8, reaction buffer at 50 mm final concentrations. VieA was incubated in the buffer for 1 min prior to the addition of substrate. Phosphodiesterase activity was assayed in reactions containing each divalent cation alone or in combination (equimolar concentrations) with Mg2+ for 1 h at room temperature, analyzed by TLC, and quantified as percentage hydrolysis of c-di-GMP.
VieA kinetics experiments were done using the determined optimized conditions of 75 mm Tris, pH 8, 25 mm KCl, 25 mm MgCl2 at 23 °C. Measurements were done with independently purified protein preparations. Because substrate concentrations were limiting, VieA was diluted to yield linear initial reaction rates. For the data shown, 0.013 μg of protein was used. C-di-GMP was included in the reactions at a range of concentrations between 11 nm and 0.91 μm. At 30-s intervals, aliquots of the phosphodiesterase reactions were spotted on PEI-cellulose plates for TLC and densitometry analysis. Mol of c-di-GMP hydrolyzed at each time point were calculated based on the percentage of total c-di-GMP hydrolyzed. Initial velocities of the reactions were determined by calculating the slope of the linear part of the reaction. Km and Vmax values were calculated using the Michaelis-Menten equation and Lineweaver-Burk methods.
To address the substrate specificity of VieA, phosphodiesterase assays were performed as described for c-di-GMP, except 1 μg of VieA and 13 nm [3H]cGMP (MP Biomedicals) were used. The reactions were incubated at 23 °C for 30 min to 16 h, and then the products were analyzed by TLC using PEI-cellulose plates and development in a solvent consisting of 5:2 methanol, 1 m ammonium acetate (26). The dried plates were cut to separate the lanes of the individual samples and then divided into 40 sections 0f 0.5 cm each. Each section was placed in Ecoscint-A scintillation fluid (National Diagnostics), and counts were measured using a Beckman LS 5000TD counter. The presence of [3H]cGMP and [3H]GMP was determined by comparing the location of sections with radioactivity relative to the published RF values of these molecules (26). A no enzyme negative control and SVPD-positive control were included in these experiments.
Purification of pGpG
c-di-GMP, synthesized and purified as described above, was used as substrate in a reaction with VieA, and nucleotides were separated by TLC. Spots corresponding to pGpG in the PEI-cellulose matrix were removed from the flexible backing of the TLC plate. pGpG was extracted from the material twice by resuspending in 0.5 m (NH4)2CO3 (1) and vortexing vigorously to solubilize the nucleotides. After a 15-min incubation at 23 °C, the supernatant containing pGpG was collected and centrifuged to remove any remaining PEI-cellulose debris.
VieA-His6, VieAE170A-His6, and mock-purified protein from the vector control were tested for the ability to degrade pGpG to GMP in phosphodiesterase assays set up as described above, except pGpG was substituted for c-di-GMP. Reaction products were analyzed by TLC.
RESULTS
Detection of c-di-GMP Phosphodiesterase Activity by VieA
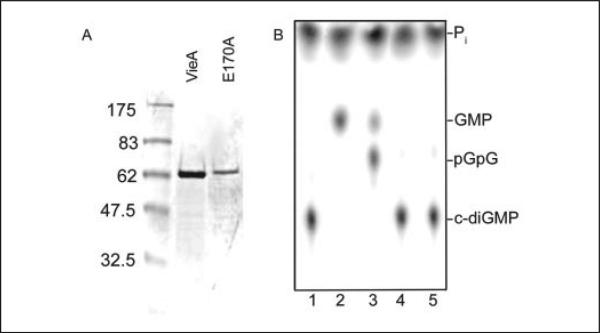
His6-tagged VieA and VieAE170A, which contains a point mutation in the putative magnesium coordination site, were expressed and purified in E. coli DH5α to greater than 95% purity as determined by SDS-PAGE and Coomassie staining (Fig. 1A). These preparations, as well as that from the vector control processed in parallel, were tested for the ability to hydrolyze 32P-labeled c-di-GMP. Substrate and products were detected using TLC and phosphorimagery (1). All RF values for nucleotide species detected in these assays agreed with those identified previously (1). The SVPD control rapidly hydrolyzed c-di-GMP (RF = 0.27) to GMP (RF = 0.61). It was apparent that the VieA preparation was capable of degrading c-di-GMP due to the disappearance of the spot at RF = 0.27 (Fig. 1B). Interestingly, the hydrolytic products present in the VieA reaction included pGpG (RF = 0.55) as well as GMP (RF = 0.61). The appearance of GMP as a product of VieA activity is addressed below. Furthermore, whereas SVPD was able to hydrolyze cGMP, VieA was not, indicating that VieA phosphodiesterase activity was specific to c-diGMP (data not shown).
FIGURE 1. Protein expression and detection of enzyme activity.
His6-tagged VieA and VieAE170A were purified using Ni2+-nitrilotriacetic acid column chromatography and tested for the ability to hydrolyze enzymatically synthesized, radiolabeled c-di-GMP substrate. A, purity of VieA and VieAE170A protein preparations as determined by SDS-PAGE analysis and Coomassie staining. His6-tagged VieA and VieAE170A are ~66 kDa. B, VieA (1 μg), VieAE170A (5 μg), and controls were incubated with [32P]c-di-GMP, and then reaction products were analyzed by TLC. VieA, but not VieAE170A or the vector control, demonstrated c-di-GMP phosphodiesterase activity. Lane 1, no enzyme control; lane 2, SVPD; lane 3, VieA; lane 4, VieAE170A; lane 5, vector control. Products of the reactions are labeled on the right. Cyclic diguanylate (c-di-GMP) migrated at RF = 0.27; pGpG migrated at RF = 0.55; and GMP migrated at RF = 0.61.
The highly conserved Glu residue of the EAL motif, Glu170, was predicted to be part of the coordination site for a divalent cation, specifically Mg2+ (23). Previous studies by our laboratory showed that mutation of conserved Glu170 to Ala abolished the ability of VieA to repress vps expression; the same effect was seen with deletion of vieA and had the downstream effect of increasing biofilm formation (6). This phenotype was attributed to an increase in intracellular c-di-GMP due to mutation of the putative EAL PDE. Our studies confirmed that the VieAE170A protein is unable to hydrolyze c-di-GMP (Fig. 1B).
Activation and Inhibition of VieA Activity by Divalent Cations
Ross et al. characterized c-di-GMP phosphodiesterase activity in cellulose synthase-containing membranes of G. xylinum (1). This activity was stimulated by the addition of 10 mm Mg2+ to the membrane extracts and strongly inhibited by the presence of 2 mm Ca2+. Based on these data, the effect of these divalent cations, as well as Mn2+ and Zn2+, on VieA phosphodiesterase activity was investigated.
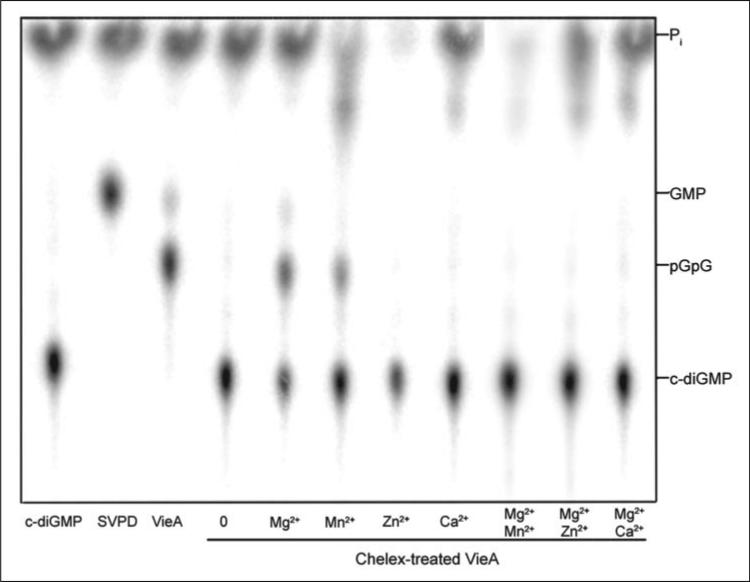
Aliquots of VieA and [32P]c-di-GMP substrate were treated with Chelex-100 to remove divalent cations present in the storage buffers until no phosphodiesterase activity was evident (Fig. 2, lane 4). The reactions were reconstituted with Mg2+, Mn2+, Zn2+, or Ca2+ individually or in combination. Mg2+ was able to restore phosphodiesterase activity to VieA, although not to the level present in non-Chelex-treated protein (Fig. 2, lane 3 versus lane 5). This is possibly due to degradation of the protein during Chelex-100 treatment at room temperature. Alternatively, chelation of Mg2+ may have yielded a fraction of VieA in which proper folding could not be restored upon reintroduction of Mg2+. Mn2+ could restore phosphodiesterase activity to ~55% of that seen with Mg2+. Neither Zn2+ nor Ca2+ permitted detectable hydrolysis of c-di-GMP in these assays. Interestingly, the addition of these cations in equimolar amounts as Mg2+ abolished VieA activity, suggesting an inhibitory role for Zn2+ and Ca2+. Furthermore, providing equimolar amounts of Mn2+ and Mg2+ to the reaction gave activity similar to providing Mn2+ alone (57% activity). These results suggest that Mn2+, Ca2+, and Zn2+ may have higher affinity than Mg2+ for the VieA metal coordination site.
FIGURE 2. Divalent cation requirement of VieA.
Divalent cations were removed from the VieA preparation by treatment with Chelex-100 until no activity was detected (lane A4). To determine which cations could support VieA phosphodiesterase activity, the protein was reconstituted with Mg2+, Mn2+, Zn2+, and Ca2+ individually or in combination with Mg2+, each at a concentration of 50 mm, and then tested for the ability to hydrolyze c-di-GMP. Mg2+ and, to a lesser extent, Mn2+ could restore VieA phosphodiesterase activity (lanes 5 and 6). Ca2+ and Zn2+ not only failed to reinstate activity (lanes 7 and 8) but also abolished c-di-GMP hydrolysis by VieA in the presence of equimolar concentrations of Mg2+ (lanes 10 and 11).
To further investigate VieA inhibition by Ca2+ and Mn2+, the concentration of these cations that reduced VieA phosphodiesterase activity to 50% was determined. VieA activity was extremely sensitive to Ca2+; in reactions containing 0.43 mm VieA and 25 mm MgCl2, 100 μm CaCl2 was able to reduce activity to one-half Vmax. In contrast, 50–75 mm MnCl2 was required to reduce VieA activity to one-half Vmax, and even 500 mm MnCl2 was unable to inhibit activity completely, decreasing the reaction rate to ~37% of Vmax. The Mg2+ concentration necessary for optimal VieA activity was determined by performing the phosphodiesterase assays using a range of MgCl2 from 0 to 100 mm. VieA phosphodiesterase activity increased with increasing Mg2+ up to 25 mm MgCl2 (data not shown). Further increases in Mg2+ had no positive or negative effects on activity.
Optimization of VieA Reaction Conditions
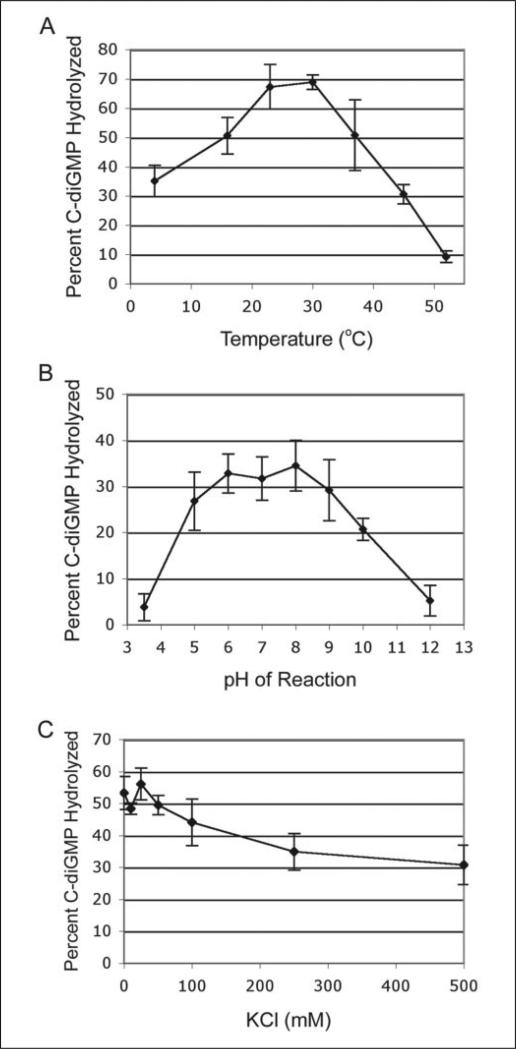
The parameters for the VieA phosphodiesterase reaction were optimized for temperature, pH, and ionic strength (Fig. 3, A–C). For determination of optimal temperature, VieA activity was analyzed at temperatures ranging from 4 to 80 °C. VieA activity was significantly impaired at 52 °C and above, and no phosphodiesterase activity was apparent at 65 °C or above (not shown). Reaction rates were best and comparable at 23 and 30 °C after a 45-min incubation, suggesting that VieA phosphodiesterase is most active at ambient temperatures; subsequently, all reactions were incubated at 23 °C.
FIGURE 3. Optimization of VieA reaction conditions.
A, the VieA phosphodiesterase reaction was performed at a range of temperatures from 4 to 80 °C. The highest rate of c-di-GMP hydrolysis was achieved at 23 and 30 °C. No activity was detectable at temperatures above 52 °C, but some c-di-GMP hydrolysis occurred at low temperatures. B, the pH of the phosphodiesterase reaction was adjusted to 3.5–12 prior to the addition of enzyme and substrate. VieA activity was comparable at pH between 5 and 9 and was inhibited at extreme basic or acidic pH. C, the ionic strength was adjusted upwards from the standard reaction buffer by the addition of 0–500 mm KCl. VieA activity was essentially the same with up to 50 mm added KCl; the total ionic strength of the reaction buffer is physiologically relevant. Further increases in ionic strength inhibited VieA phosphodiesterase activity, but only slightly. A–C, the data shown represent the mean percentage of substrate that was hydrolyzed in three independent experiments.
Rates of c-di-GMP hydrolysis were measured at a wide range of buffer pH (3.5–12). Activity peaked at neutral pH; there were no significant differences in reaction rates at pH between 5 and 9, indicating that VieA is not extremely sensitive to pH. Because pH 8 consistently gave somewhat higher reaction rates than the others tested, pH 8 was used in subsequent assays.
The ionic strength of the VieA phosphodiesterase reaction buffer was also optimized. To the VieA buffer containing 75 mm Tris, pH 8, and 25 mm MgCl2, 0–500 mm KCl was added. C-di-GMP hydrolysis was comparable for 0–50 mm KCl and decreased with further increases in KCl, suggesting inhibition of activity at high ionic strength, possibly due to masking of charges and reduced electrostatic interaction between Mg2+ and the metal coordination site of VieA. Similar results were apparent when NaCl was used to adjust ionic strength (data not shown). The use of 25 mm KCl in the reaction consistently gave the highest VieA activity and was used in subsequent experiments. The ionic strength of this reaction is ~190 mm, which falls in the physiologically relevant range of 150–200 mm. Examination of activity at very low ionic strength was limited by the minimum amount of MgCl2 required for maximal c-di-GMP hydrolytic activity.
Kinetics of VieA c-di-GMP Phosphodiesterase Activity
The optimal reaction conditions determined above were used to analyze the kinetics of VieA c-di-GMP hydrolytic activity. VieA phosphodiesterase activity demonstrated standard Michaelis-Menten kinetics. Km and Vmax values were determined for VieA using two independently expressed and purified preparations of enzyme. The Km and Vmax calculated for each protein preparation were very similar. VieA demonstrated a Km of 0.06 μm, suggesting high affinity of VieA for c-di-GMP. Vmax was determined to be 20.5 μmol min–1 mg–1 protein.
Phosphodiesterase B Activity of VieA
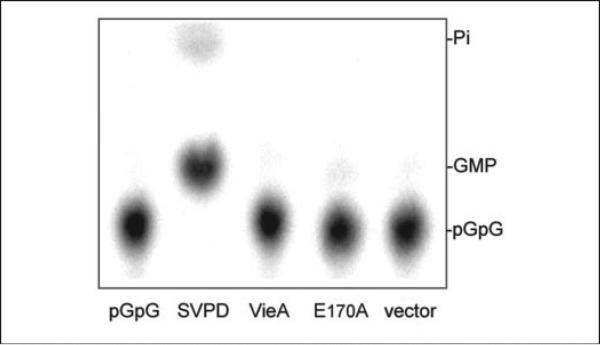
It has been suggested that a phosphodiesterase different from the EAL domain phosphodiesterase, PDE-B, hydrolyzes pGpG to GMP (27). Our experiments with VieA showed that GMP appeared only when higher concentrations of protein were used (data not shown). This introduced the possibility that a second phosphodiesterase was present in the VieA preparations. To address whether the appearance of GMP was due to a PDE-B contaminant, VieA, VieAE170A, and vector controls were used in phosphodiesterase assays using purified pGpG as substrate. Whereas SVPD was able to hydrolyze pGpG to GMP, neither VieA, VieAE170A, nor the vector control reactions showed GMP product after a 2-h incubation (Fig. 4). Even after 24 h, only a trace amount of GMP (<3%) was apparent in these reactions, compared with <1% for the c-di-GMP only control. Thus, if small quantities of a PDE-B are present, they are not responsible for the substantial amount of GMP that appears after a 45-min incubation of VieA with c-di-GMP. Because VieA was unable to hydrolyze pGpG to GMP when given pGpG as a substrate, it is possible that VieA can hydrolyze pGpG to GMP at a low rate when bound as an intermediate during c-di-GMP hydrolysis but cannot bind pGpG when introduced exogenously.
FIGURE 4. Hydrolysis of pGpG.
The potential presence of contaminants with PDE-B activity in the protein preparations was addressed by incubating VieA, VieAE170, and vector control samples with radiolabeled pGpG as the substrate. Whereas SVPD could hydrolyze pGpG (RF = 0.55) to GMP (RF = 0.61), no other reaction showed GMP product after 24 h, verifying that no PDE-B contaminant was present.
DISCUSSION
There has been increasing interest in the novel second messenger c-di-GMP and in its effect on bacterial gene regulation. A role for c-di-GMP has been demonstrated in regulation of exopolysaccharide biosynthesis, motility, and virulence in a variety of bacteria, both Gram-negative and Gram-positive (1–5, 25). With the exception of cellulose biosynthesis in G. xylinum and A. tumefaciens, in which c-di-GMP functions as an allosteric activator of cellulose synthase (1, 17), the mechanism by which c-di-GMP modulates these phenotypes is unknown. However, there has been significant progress in understanding the metabolism of c-di-GMP. Genetic studies of genes encoding the GGDEF domain, which possesses weak homology to eukaryotic adenylate cyclases, provided evidence that the GGDEF domain functions as a c-di-GMP cyclase (17). Recently, GGDEF proteins from bacteria representing the major phylogenetic branches were purified and demonstrated to have diguanylate cyclase activity in vitro (21).
Similar to the GGDEF domain, the assignment of c-di-GMP hydrolytic activity was initially ascribed to the EAL domain based on genetic evidence (5, 6, 18). For example, mutation of the EAL-encoding gene vieA resulted in increased levels of c-di-GMP in nucleotide extracts (6). This work demonstrates c-di-GMP hydrolytic activity by the EAL-containing protein VieA. VieA was selected as the EAL protein of interest because of its role in regulation of vps expression and biofilm formation, and in CT expression and virulence of V. cholerae, a facultative intestinal pathogen. Furthermore, previous work investigating EAL domain function could not specifically attribute c-diGMP phosphodiesterase activity to the EAL domain, because the GGDEF domain was present in these proteins as well or because the protein contained both domains but showed primarily diguanylate cyclase activity (18). VieA lacks a GGDEF domain, which allows unambiguous assignment of c-diGMP phosphodiesterase activity to the EAL domain.
Using enzymatically synthesized c-di-GMP substrate, VieA was demonstrated to possess c-di-GMP hydrolytic activity. Protein preparations of vector alone, processed in parallel, demonstrated no activity. The conserved Glu residue (Glu170) in the EAL domain, which was predicted to be a divalent ion coordination site and probably required for EAL activity (23), was confirmed to be necessary for VieA activity, since an E170A mutation abolished c-di-GMP phosphodiesterase activity of the protein. The measured Km of VieA for c-di-GMP was calculated to be 0.06 μm. This Km is considerably lower than that of other bacterial cyclic nucleotide phosphodiesterases. For example, CpdA, a cAMP phosphodiesterase of E. coli, has a Km of 0.5 mm cAMP (28). A Vibrio fischeri zinc-containing cyclic nucleotide phosphodiesterase showed a lower Km of 73 μm cAMP (29). The measured Km for VieA is also 4-fold lower than the Km measured for phosphodiesterase-containing membranes of G. xylinum, 0.25 μm (27), suggesting high affinity of VieA for this substrate.
The VieA EAL domain protein appears to be specific for the c-di-GMP substrate. Whereas SVPD is a promiscuous phosphodiesterase that can hydrolyze among others c-di-GMP, pGpG, cGMP, and cAMP, c-di-GMP was the only substrate tested that could be hydrolyzed by VieA. Previous studies by Ross et al. (27) addressed the ability of phosphodiesterase-containing membranes to hydrolyze cGMP and c-di-GMP analogues, including c-di-IMP, c-di-AMP, c-di-CMP, and diastereoisomers of c-di-GMP, so the latter were not considered in this work.
Ross et al. (27) predicted that EAL phosphodiesterases function solely in the first step of c-di-GMP hydrolysis to yield pGpG and that a second enzyme, PDE-B, degrades pGpG to GMP. Analysis of the VieA reaction products by TLC showed that not only was the linear pGpG product formed, but GMP was produced as well. Because GMP was detectable only in reactions containing 10–100-fold higher concentration of protein than was used in the kinetics studies, the possibility arose that a PDE-B was present as a contaminant in the protein preparation. However, direct testing of VieA, VieAE170A, and the vector control for the ability to hydrolyze pGpG as a substrate failed to detect a PDE-B. This was not due to a defect in the purification of pGpG, since the SVPD-positive control was able to hydrolyze pGpG to GMP, implying that PDE-B activity detected in these assays can be ascribed to VieA. Because the rate of GMP formation was orders of magnitude slower than that of pGpG, the PDE-B activity of VieA is probably not physiologically significant.
An intriguing result of this assay was that GMP product was not formed in the VieA reaction when pGpG was provided exogenously as the substrate. It is possible that VieA does not bind exogenous pGpG efficiently and that GMP is only produced when pGpG is already bound, immediately following the initial hydrolysis of c-di-GMP. Preliminary binding studies investigating the ability of VieA to bind c-di-GMP and pGpG support the hypothesis that VieA has very low affinity for pGpG relative to c-di-GMP (data not shown). Alternatively, because VieA also is predicted to be a response regulator, which often bind DNA as dimers to control transcription (30–32), it is possible that dimerization of VieA also affects its enzyme activity, specifically in mediating hydrolysis of the pGpG intermediate product. It is unlikely that VieA dimerization is required for the initial hydrolytic cleavage of c-di-GMP, since the conserved response regulator domains can be deleted from VieA without affecting its regulatory activity on vps expression (6). Determination of the crystal structure of VieA may elucidate the potential overlap between the presumed regulatory function of VieA and its enzymatic activity.
As a potential dual function protein, VieA phosphodiesterase and response regulator activities may be affected by phosphorylation. Phosphorylation of response regulators of two- and three-component regulatory systems typically increases affinity of the protein for its target promoter DNA and thus increases expression of that gene. Phosphorylation of VieA may positively or negatively affect its enzyme activity as well. Preliminary studies with VieA phosphorylated in vitro with acetyl phosphate suggest that there is no significant change in the rate of VieA hydrolysis of c-di-GMP (data not shown). In addition, deletion of the VieA phosphoreceiver domain, as stated above, or mutation of the conserved aspartic acid residue that is predicted to be the site of phosphorylation of VieA had no effect on the ability of the VieA EAL domain to repress expression of vps genes (6). Thus, we do not expect phosphorylation to affect the phosphodiesterase activity of VieA.
VieA was determined to have optimal activity at neutral pH, physiological ionic strength, and ambient temperatures. The in vitro optimum conditions with respect to pH and ionic strength coincided with those expected to be experienced by VieA in the bacterial cytoplasm. The temperature optimum is consistent with the facultative pathogen nature of V. cholerae, which includes growth in the human small intestine at 37 °C and growth in aqueous reservoirs in temperate zones around the world. V. cholerae encodes 21 additional EAL domain proteins and 41 GGDEF proteins (10 of these have both domains) (23), each of which could potentially affect the intracellular pools of c-diGMP. Interestingly, like VieA, other GGDEF and EAL proteins often contain sensory/regulatory modules that could participate in signal transduction. This introduces the likelihood that subsets of GGDEF and/or EAL domain proteins are expressed under specific conditions. The vieSAB locus, for example, is expressed at low levels during growth in LB broth in vitro but is induced during infection of the mouse as well as during growth in minimal medium supplemented with the amino acids Asn, Arg, Glu, and Ser (6, 33, 34). Other GGDEF and EAL proteins may dominate under other, as yet undefined conditions.
Strong inhibition of VieA c-diGMP phosphodiesterase activity by Ca2+ was observed in these studies, supporting earlier findings using phosphodiesterase-containing membrane preparations (1). In fact, Ca2+ and Zn2+ inhibited activity even when VieA was preincubated with MgCl2, so VieA presumably was saturated with Mg2+, suggesting that Ca2+ is capable of displacing Mg2+ from the metal coordination site. These findings may be important for understanding the function of proteins in which both GGDEF diguanylate cyclases and EAL phosphodiesterase domains are present. In such proteins with domains of opposing functions, regulation of the individual domains is likely to be critical, and Ca2+ may provide a means by which c-di-GMP hydrolysis can be inhibited. In this model, the presence of Ca2+ would repress c-di-GMP hydrolysis, allowing c-di-GMP to accumulate through the activity of GGDEF activity. Regulation of GGDEF activity by divalent cations has not been described.
Ca2+ inhibition of c-di-GMP degradation by EAL domain proteins has particularly interesting implications with regard to V. cholerae biofilm formation. Formation of most V. cholerae biofilms is dependent on the production of exopolysaccharide (VPS) (35). However, VPS-independent biofilms that form in seawater have been described (36, 37). These biofilms have an altered architecture and cell-cell interaction that appear to be mediated by contacts between the lipopolysaccharide O-antigen of adjacent bacteria and are dependent on the presence of Ca2+ in the growth medium. Previous studies by our laboratory have demonstrated that mutation of an EAL can cause increased intracellular c-di-GMP and increased VPS-dependent biofilm formation. Similarly, inhibition of EAL activity by Ca2+ may positively regulate VPS-independent biofilm development as well. Because biofilm formation is thought to promote survival of V. cholerae in the environment, and because biofilm-associated V. cholerae are predicted to be more infectious due to protection from a variety of antimicrobial agents, regulation of c-di-GMP metabolism by Ca2+ may be important to V. cholerae fitness in certain environments.
The bacterial second messenger c-di-GMP is being implicated in a wide variety of bacterial phenotypes, especially multicellular behaviors such as biofilms. In addition, there is evidence in V. cholerae and S. typhimurium that c-di-GMP can regulate virulence phenotypes as well. C-di-GMP may be a more global regulator of bacterial gene and protein regulation than is realized, so it is critical that we understand the metabolism of this compound. These experiments provide the first direct evidence that an EAL domain protein, VieA, possesses c-di-GMP hydrolytic activity in a purified system. Moreover, Ca2+ inhibition may provide a means of regulating EAL-mediated degradation of c-di-GMP.
Footnotes
This work was supported by National Institutes of Health Grant AI45746 (to A. C.) and Center for Gastroenterology Research on Absorptive and Secretory Processes, New England Medical Center, Grant P30DK34928. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The abbreviations used are: c-di-GMP, 3′,5′-cyclic diguanylic acid; CT, cholera toxin; PEI, polyethyleneimine; SVPD, snake venom phosphodiesterase; PDE, phosphodiesterase.
REFERENCES
- 1.Ross P, Aloni Y, Weinhouse H, Michaeli D, Weinberger-Ohana P, Mayer R, Benziman M. Carbohydr. Res. 1986;149:101–117. [Google Scholar]
- 2.Garcia B, Latasa C, Solano C, Garcia-del Portillo F, Gamazo C, Lasa I. Mol. Microbiol. 2004;54:264–277. doi: 10.1111/j.1365-2958.2004.04269.x. [DOI] [PubMed] [Google Scholar]
- 3.Hisert KB, MacCoss M, Shiloh MU, Darwin KH, Singh S, Jones RA, Ehrt S, Zhang Z, Gaffney BL, Gandotra S, Holden DW, Murray D, Nathan C. Mol. Microbiol. 2005;56:1234–1245. doi: 10.1111/j.1365-2958.2005.04632.x. [DOI] [PubMed] [Google Scholar]
- 4.Karaolis DK, Rashid MH, Chythanya R, Luo W, Hyodo M, Hayakawa Y. Antimicrob. Agents Chemother. 2005;49:1029–1038. doi: 10.1128/AAC.49.3.1029-1038.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simm R, Morr M, Kader A, Nimtz M, Romling U. Mol. Microbiol. 2004;53:1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- 6.Tischler AD, Camilli A. Mol. Microbiol. 2004;53:857–869. doi: 10.1111/j.1365-2958.2004.04155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tischler AD, Lee SH, Camilli A. J. Bacteriol. 2002;184:4104–4113. doi: 10.1128/JB.184.15.4104-4113.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huq A, Small EB, West PA, Huq MI, Rahman R, Colwell RR. Appl. Environ. Microbiol. 1983;45:275–283. doi: 10.1128/aem.45.1.275-283.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tamplin ML, Gauzens AL, Huq A, Sack DA, Colwell RR. Appl. Environ. Microbiol. 1990;56:1977–1980. doi: 10.1128/aem.56.6.1977-1980.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watnick PI, Kolter R. Mol. Microbiol. 1999;34:586–595. doi: 10.1046/j.1365-2958.1999.01624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schoolnik GK, Voskuil MI, Schnappinger D, Yildiz FH, Meibom K, Dolganov NA, Wilson MA, Chong KH. Methods. Enzymol. 2001;336:3–18. doi: 10.1016/s0076-6879(01)36573-4. [DOI] [PubMed] [Google Scholar]
- 12.Herrington DA, Hall RH, Losonsky G, Mekalanos JJ, Taylor RK, Levine MM. J. Exp. Med. 1988;168:1487–1492. doi: 10.1084/jem.168.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee SH, Hava DL, Waldor MK, Camilli A. Cell. 1999;99:625–634. doi: 10.1016/s0092-8674(00)81551-2. [DOI] [PubMed] [Google Scholar]
- 14.Lee SH, Butler SM, Camilli A. Proc. Natl. Acad. Sci. U. S. A. 2001;98:6889–6894. doi: 10.1073/pnas.111581598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller VL, Mekalanos JJ. J. Bacteriol. 1985;163:580–585. doi: 10.1128/jb.163.2.580-585.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taylor RK, Miller VL, Furlong DB, Mekalanos JJ. Proc. Natl. Acad. Sci. U. S. A. 1987;84:2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ausmees N, Mayer R, Weinhouse H, Volman G, Amikam D, Benziman M, Lindberg M. FEMS Microbiol. Lett. 2001;204:163–167. doi: 10.1111/j.1574-6968.2001.tb10880.x. [DOI] [PubMed] [Google Scholar]
- 18.Tal R, Wong HC, Calhoon R, Gelfand D, Fear AL, Volman G, Mayer R, Ross P, Amikam D, Weinhouse H, Cohen A, Sapir S, Ohana P, Benziman M. J. Bacteriol. 1998;180:4416–4425. doi: 10.1128/jb.180.17.4416-4425.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pei J, Grishin NV. Proteins. 2001;42:210–216. doi: 10.1002/1097-0134(20010201)42:2<210::aid-prot80>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 20.Chan C, Paul R, Samoray D, Amiot NC, Giese B, Jenal U, Schirmer T. Proc. Natl. Acad. Sci. U. S. A. 2004;101:17084–17089. doi: 10.1073/pnas.0406134101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryjenkov DA, Tarutina M, Moskvin OV, Gomelsky M. J. Bacteriol. 2005;187:1792–1798. doi: 10.1128/JB.187.5.1792-1798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong HC, Fear AL, Calhoon RD, Eichinger GH, Mayer R, Amikam D, Benziman M, Gelfand DH, Meade JH, Emerick AW. Proc. Natl. Acad. Sci. U. S. A. 1990;87:8130–8134. doi: 10.1073/pnas.87.20.8130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galperin MY, Nikolskaya AN, Koonin EV. FEMS Microbiol. Lett. 2001;203:11–21. doi: 10.1111/j.1574-6968.2001.tb10814.x. [DOI] [PubMed] [Google Scholar]
- 24.Tischler A. D. a. A. C. Infect. Immun. 2005 doi: 10.1128/IAI.73.9.5873-5882.2005. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bobrov AG, Kirillina O, Perry RD. FEMS Microbiol. Lett. 2005 doi: 10.1016/j.femsle.2005.04.036. [DOI] [PubMed] [Google Scholar]
- 26.Keirns JJ, Wheeler MA, Bitensky MW. Anal. Biochem. 1974;61:336–348. doi: 10.1016/0003-2697(74)90400-x. [DOI] [PubMed] [Google Scholar]
- 27.Ross P, Mayer R, Weinhouse H, Amikam D, Huggirat Y, Benziman M, de Vroom E, Fidder A, de Paus P, Sliedregt LA. J. Biol. Chem. 1990;265:18933–18943. [PubMed] [Google Scholar]
- 28.Imamura R, Yamanaka K, Ogura T, Hiraga S, Fujita N, Ishihama A, Niki H. J. Biol. Chem. 1996;271:25423–25429. doi: 10.1074/jbc.271.41.25423. [DOI] [PubMed] [Google Scholar]
- 29.Callahan SM, Cornell NW, Dunlap PV. J. Biol. Chem. 1995;270:17627–17632. doi: 10.1074/jbc.270.29.17627. [DOI] [PubMed] [Google Scholar]
- 30.Dziejman M, Mekalanos JJ. Mol. Microbiol. 1994;13:485–494. doi: 10.1111/j.1365-2958.1994.tb00443.x. [DOI] [PubMed] [Google Scholar]
- 31.Hidaka Y, Park H, Inouye M. FEBS Lett. 1997;400:238–242. doi: 10.1016/s0014-5793(96)01396-8. [DOI] [PubMed] [Google Scholar]
- 32.Scarlato V, Rappuoli R. J. Bacteriol. 1991;173:7401–7404. doi: 10.1128/jb.173.22.7401-7404.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Camilli A, Mekalanos JJ. Mol. Microbiol. 1995;18:671–683. doi: 10.1111/j.1365-2958.1995.mmi_18040671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee SH, Angelichio MJ, Mekalanos JJ, Camilli A. J. Bacteriol. 1998;180:2298–2305. doi: 10.1128/jb.180.9.2298-2305.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yildiz FH, Schoolnik GK. Proc. Natl. Acad. Sci. U. S. A. 1999;96:4028–4033. doi: 10.1073/pnas.96.7.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kierek K, Watnick PI. Proc. Natl. Acad. Sci. U. S. A. 2003;100:14357–14362. doi: 10.1073/pnas.2334614100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kierek K, Watnick PI. Appl. Environ. Microbiol. 2003;69:5079–5088. doi: 10.1128/AEM.69.9.5079-5088.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yildiz FH, Schoolnik GK. J. Bacteriol. 1998;180:773–784. doi: 10.1128/jb.180.4.773-784.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]