Abstract
Both selective and random processes can affect the outcome of natural hybridization. A recent analysis in BMC Evolutionary Biology of natural hybridization between an introduced and a native salamander reveals the mosaic nature of introgression, which is probably caused by a combination of selection and demography.
Natural interspecific hybridization has been observed in a diverse array of taxa [1-3], and the production of such hybrids can have a wide range of evolutionary consequences. For instance, natural hybridization can act as a 'destructive' force, resulting in the fusion of taxa that do not reveal strong reproductive isolation [1]. Alternatively, it can result in evolutionary novelty via the production of new reproductively isolated hybrid species [1-3], or by the transfer of potentially adaptive traits through a process known as introgressive hybridization. In a recent article in BMC Evolutionary Biology, Fitzpatrick et al. [4] tested for introgressive hybridization by performing a genome-wide scan of native salamander (Ambystoma californiense) populations, and identified genomic regions that have rapidly fixed alternative alleles derived from a recently introduced, invasive species (Ambystoma mavortium). This rapid assimilation of non-native alleles suggests that selective processes were involved in their fixation.
Introgressive hybridization is a multi-step process by which genetic material from one population infiltrates into another, genetically differentiated population [5]. In order for introgressive hybridization to take place, F1 hybrids must initially be produced between the two species. In nature, such hybrid formation is often a rare event owing to the presence of multiple prezygotic and postzygotic reproductive barriers [1], and indeed in most species-pairs studied to date, the frequencies of F1 hybrid formation are low [1]. However, even if the F1 hybrids possess only partial fitness, they may mate with one or both progenitor species to produce first-generation backcross (BC1) hybrids. These BC1 hybrids may further backcross with pure-species individuals, ultimately resulting in the transfer of novel alleles between the hybridizing lineages/species [5].
A key question with respect to introgressive hybridization is 'To what degree is interspecific gene flow the result of adaptive processes?' Whereas hybridization can certainly result in the transfer of alleles and traits across species boundaries [1], such genetic exchange does not necessarily involve the transfer of adaptations. Stochastic processes (that is, random genetic drift) could potentially explain introgression events, and deciphering the roles of stochastic and deterministic (that is, natural selection) processes has often proved difficult. Only a handful of examples are currently available that provide rigorous tests for adaptive introgression [6-11]. Given the scarcity of such analyses, the recent study of Fitzpatrick et al. [4] reporting on signatures of adaptive introgression in salamanders is a significant contribution. Here, we review four examples (including [4]) that have used four different methodologies to test for adaptive introgression.
Evidence for adaptive introgression in plants
Helianthus
Heiser [6] inferred the annual sunflower subspecies, Helianthus annuus texanus, to be a stabilized hybrid resulting from the introgression of a small number of Helianthus debilis genes into a predominately H. a. annuus genome. Kim and Rieseberg [7] demonstrated that the introgression of only three small H. debilis genomicregions (containing three morphological quantitative trait loci (QTLs)) was sufficient to recapitulate the H. a. texanus phenotype (see [12] for a discussion of the parameters of QTL analyses). This unique phenotype includes a combination of traits gained apparently from either H. a. annuus (for example, the shape of bracts surrounding the flower head and the shape of the fruits) or H. debilis (for example, stem speckling and fruit size) [6,7]. Significantly, Whitney et al. [8] presented evidence that the introgression of H. debilis genomic material into a H. a. annuus genetic background not only accounted for the unique morphology of H. a. texanus, but also resulted in adaptive introgression. Specifically, the increased herbivore resistance of H. debilis was transferred into BC1 hybrids towards H. a. annuus.
Iris
The second example of adaptive trait transfer comes from studies of two species of Louisiana Iris: the flood-tolerant Iris fulva and the dry-adapted Iris brevicaulis. Martin et al. [9] combined an analysis of QTLs (using reciprocal BC1 mapping populations) with a highly selective, natural field experiment. The highly selective nature of the experiment involved an intensive, long-term flooding event in which the various genotypes were submerged for approximately four months. This study determined that I. fulva genotypes survived flooding at significantly higher frequencies than any other genotypic class (Table 1), including I. brevicaulis (none of the latter survived [9]). The ability to withstand these extreme conditions was shown to have a genetic basis, with survivorship of backcrosses towards I. brevicaulis strongly influenced by the presence of a number of introgressed alleles from flood-tolerant I. fulva [9]. Furthermore, in the backcrosses towards I. fulva, two QTLs of major effect were detected that influenced flood tolerance, one of which (perhaps surprisingly) included introgressed alleles from the dry-adapted I. brevicaulis. These findings, and those for Helianthus, demonstrate the potential for adaptive introgression to affect the evolutionary trajectory of hybrid plant populations.
Table 1.
Number and fraction of Iris fulva, Iris brevicaulis and their hybrids that survived an extended flooding event
| Genotypic class | Alive | Dead | Fraction survived |
| I. brevicaulis | 0 | 13 | 0 |
| BCIB | 23 | 393 | 0.055 |
| BCIF | 33 | 325 | 0.092 |
| I. fulva | 3 | 8 | 0.273 |
BCIB and BCIF, BC1 hybrids towards I. brevicaulis and I. fulva, respectively, as judged by genotype. Data adapted from [9].
Evidence for adaptive introgression in animals
Mus
Surveys of numerous loci across natural hybrid zones between the mouse species Mus musculus and Mus domesticus have been made. The detail obtained by applying a genomics approach to natural populations has allowed tests for deviations from neutral introgression - reflecting both positive and negative selection [10,11]. Payseur et al. [10] detected X-chromosome loci that were apparently prevented from introgressing across a zone of hybridization between the two mouse species in southern Germany and western Austria. The lack of penetration was hypothesized to mark genomic regions contributing to the partial reproductive isolation between M. musculus and M. domesticus, as caused by selection against hybrids containing an admixture of heterospecific loci (that is, loci from the other species) [10]. In contrast, some X-chromosome alleles of M. domesticus were found at higher than expected frequencies in populations of mice on the M. musculus side of the hybrid zone. These loci suggested
"...the possibility that positive selection may act to drive the spread of alleles from one species on to the genomic background of the other species..." (that is, reflecting adaptive trait introgression [10]). Selection can thus act in different 'directions' (either positively or negatively) on different portions of the genomes of hybrids. This differential selection results in introgression being favored or disfavored, depending upon the traits generated from the loci contained in the various genomic regions.
Loci distributed across the Mus autosomes also exhibited patterns of differential introgression. For example, Teeter et al. [11] reported restricted introgression for numerous loci, indicative of epistatic interactions contributing to lower hybrid fitness. These 'reproductive isolation' loci included genes that affect reproductive characteristics and responses to environmental cues [11]. Although there was evidence for selection against the introgression of certain genomic regions, patterns of genetic variation also identified regions of high-frequency introgression. Consistent with previous findings for the X-chromosome loci [10], most of the introgression involved the occurrence of alleles from M. domesticus in a largely M. musculus genetic background [11]. These loci included genes that underlie cell signaling, olfaction and response to pheromones, and are thus good candidates for adaptive introgression.
Ambystoma
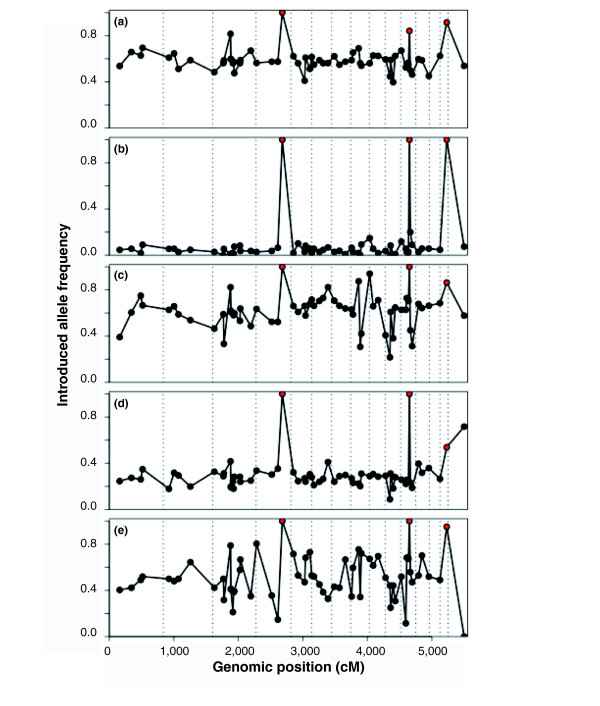
In their work with Ambystoma, Fitzpatrick et al. [4] have provided an excellent test of the hypothesis of mosaicism in the genomes of hybridizing taxa. Their analysis is particularly powerful because they possessed historical records indicating that natural hybridization between the native salamander species, A. californiense, and the introduced A. mavortium (or A. t. mavortium) had begun during the 1940s. The short time period since the formation of the hybrid zone between these species allowed a sensitive assay for the presence or absence of deviations from expected allele frequencies. Such deviations (or lack thereof) would be indicative of the impact of selective and/or stochastic factors on different genomic regions. The analysis of 64 loci, spread across the salamander genomes, from animals collected inside the hybrid zone detected mosaics of conspecific and heterospecific alleles. Both stochastic and deterministic processes accounted for this mosaicism [4]. However, Figure 1 summarizes the "most striking result" [4] from the analysis of hybrid Ambystoma populations. Specifically, Fitzpatrick et al. [4] detected three out of the 64 loci that were fixed, or nearly fixed, for the non-native alleles. The observation that the frequencies of these three non-native alleles were elevated across all five of the ponds sampled substantiates the inference of selection-favored (that is, adaptive) introgression [4]. Only one genomic region revealed evidence for selection against heterozygotes, while the remaining 60 markers did not deviate from neutral expectations.
Figure 1.
Allele frequencies for 64 marker loci across the Ambystoma genome. (a-e) Allele frequency data from five separate ponds [4]. The points marked in red reflect three loci that show selection-driven introgression from the introduced species A. mavortium (or A. t. mavortium) into the native A. californiense.
Mosaic genomes
As discussed above, and by previous authors [1-3], both deterministic and stochastic factors will affect the evolutionary and ecological trajectories of hybrid popula tions. In particular, it has been suggested that selection against hybrids balanced by continual hybridization between the interacting taxa will play the major role in hybrid zone evolution [1]. In contrast, others have argued for a differential selective coefficient depending upon which portions of the genomes of the hybridizing taxa are examined [1-4,7-11]. In that conceptual framework, the genomes of hybridizing organisms would reflect the combined action of negative selection, positive selection and neutral diffusion of alleles from the alternative taxon. The genomes of organisms affected by differential introgression would thus be mosaics, possessing some cassettes of genomic material from the 'foreign' evolutionary lineage while retaining large stretches of non-introgressed genomic material [1-3].
Determining the evolutionary forces, whether stochastic or deterministic, that resulted in the introgression (or lack thereof) of foreign DNA is a primary question for students of natural hybridization. In this regard, the findings for Helianthus, Iris, Mus and Ambystoma indicate that models of evolutionary change remain inadequate when they neglect the role of divergence in the face of gene flow. When the process of introgression is one of the factors tested for in evolutionary analyses, it is often found. This genetic transfer, as reflected by genomes with an admixture of components from various lineages, is almost certainly indicative of the role of both stochastic and deterministic processes. Furthermore, unlike previous conclusions drawn when data were lacking (see [1] for a discussion of this debate), it is now apparent that the deterministic portion of the equation includes selection favoring the introgression of some alleles and traits (see, for example, [7-11]).
Acknowledgments
Acknowledgements
NHM was supported by National Science Foundation grant DEB-0816905.
References
- Arnold ML. Evolution Through Genetic Exchange. Oxford: Oxford University Press; 2006. [Google Scholar]
- Rieseberg LH. Hybrid origins of plant species. Annu Rev Ecol Syst. 1997;28:359–389. doi: 10.1146/annurev.ecolsys.28.1.359. [DOI] [Google Scholar]
- Seehausen O. Hybridization and adaptive radiation. Trends Ecol Evol. 2004;19:198–207. doi: 10.1016/j.tree.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick BM, Johnson JR, Kump DK, Shaffer HB, Smith JJ, Voss SR. Rapid fixation of non-native alleles revealed by genome-wide SNP analysis of hybrid Tiger Salamanders. BMC Evol Biol. 2009;9:176. doi: 10.1186/1471-2148-9-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson E, Hubricht L. Hybridization in Tradescantia. III. The evidence for introgressive hybridization. Amer J Bot. 1938;25:396–402. doi: 10.2307/2436413. [DOI] [Google Scholar]
- Heiser CB., Jr Variation and subspeciation in the common sunflower, Helianthus annuus. Amer Midl Nat. 1954;51:287–305. doi: 10.2307/2422222. [DOI] [Google Scholar]
- Kim S-C, Rieseberg LH. Genetic architecture of species differences in annual sunflowers: Implications for adaptive trait introgression. Genetics. 1999;153:965–977. doi: 10.1093/genetics/153.2.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney KD, Randell RA, Rieseberg LH. Adaptive introgression of herbivore resistance traits in the weedy sunflower Helianthus annuus. Am Nat. 2006;167 doi: 10.1086/504606. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Martin NH, Bouck AC, Arnold ML. Detecting adaptive trait introgression between Iris fulva and I. brevicaulis in highly selective field conditions. Genetics. 2006;172:2481–2489. doi: 10.1534/genetics.105.053538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payseur BA, Krenz JG, Nachman MW. Differential patterns of introgression across the X chromosome in a hybrid zone between two species of house mice. Evolution. 2004;58:2064–2078. doi: 10.1111/j.0014-3820.2004.tb00490.x. [DOI] [PubMed] [Google Scholar]
- Teeter KC, Payseur BA, Harris LW, Bakewell MA, Thibodeau LM, O'Brien JE, Krenz JG, Sans-Fuentes MA, Nachman MW, Tucker PK. Genome-wide patterns of gene flow across a house mouse hybrid zone. Genome Res. 2008;18:67–76. doi: 10.1101/gr.6757907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TF. Genetic analysis of quantitative traits. J Biol. 2009;8:23. doi: 10.1186/jbiol133. [DOI] [PMC free article] [PubMed] [Google Scholar]