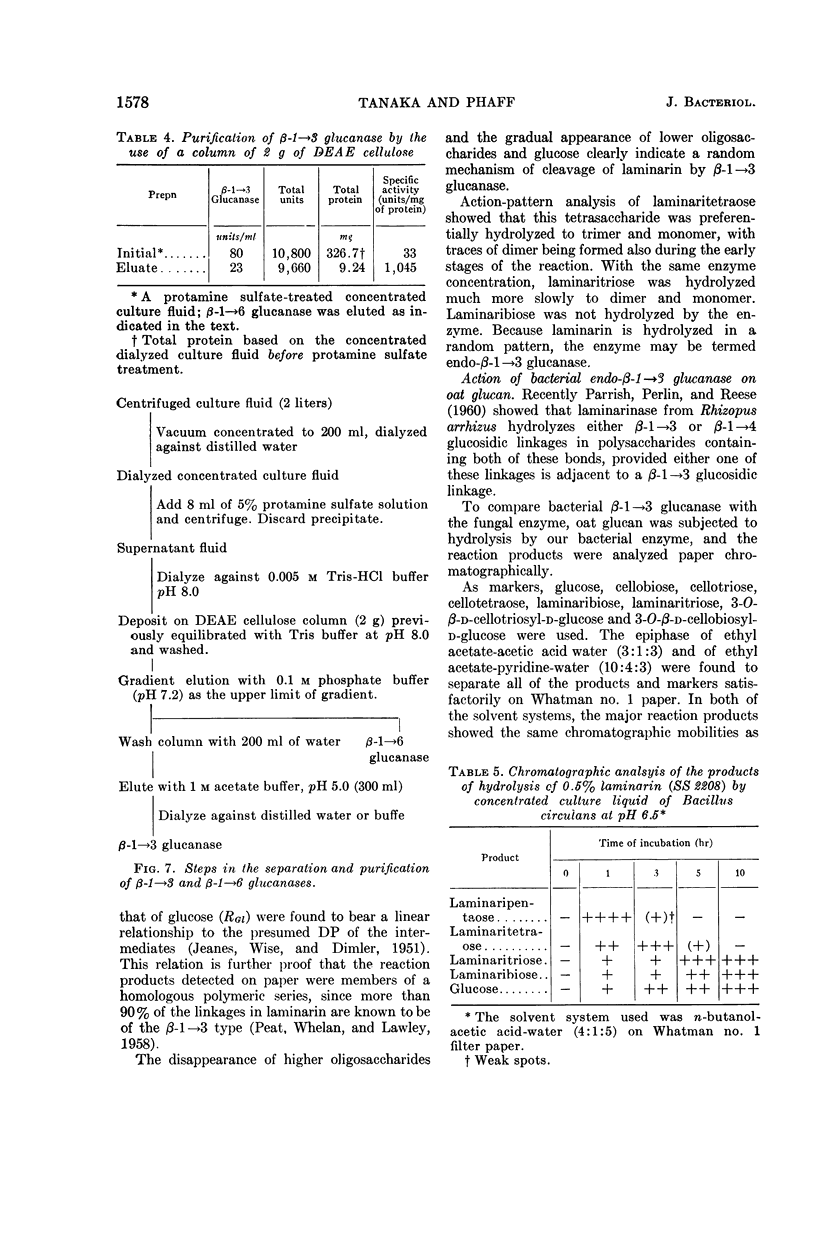
Abstract
Tanaka, Hirosato (University of California, Davis), and Herman J. Phaff. Enzymatic hydrolysis of yeast cell walls. I. Isolation of wall-decomposing organisms and separation and purification of lytic enzymes. J. Bacteriol. 89:1570–1580. 1965.—A number of microorganisms, able to decompose and grow on yeast cell walls, were isolated from soil. These isolates demonstrated various types of attack on yeast walls. A bacterium, identified as Bacillus circulans, and a species of Streptomyces produced clear, lysed zones when grown on an agar medium containing baker's yeast cell walls. The streptomycete formed glucanase, mannanase, and protease, but B. circulans produced only glucanases. Purified mannan could be prepared from the culture fluid of B. circulans grown on baker's yeast cell walls. In a liquid, mineral medium, extracellular lytic enzyme production by B. circulans was optimal after 3 days of aerobic growth at 30 C with 0.5% baker's yeast cell walls as the carbon source. Twelve other carbon sources were ineffective as inducers. Among a number of polysaccharides tested, the crude enzymes of B. circulans hydrolyzed only β-1→3 glucan (laminarin) and β-1→6 glucan (pustulan), both by a random mechanism, to a mixture of dimer and glucose. The β-1→3 and β-1→6 glucanases were separated from each other by diethylaminoethyl cellulose column chromatography. Water-soluble oat glucan, which contains in the linear chain both β-1→3 and β-1→4 bonds, was also hydrolyzed by the bacterial β-1→3 glucanase. The products of this reaction indicated that this enzyme hydrolyzes β-1→3 or β-1→4 glucosidic linkages, provided the β-glucopyranosyl units composing these bonds are substituted in the 3 position by another glucose unit.
Full text
PDF
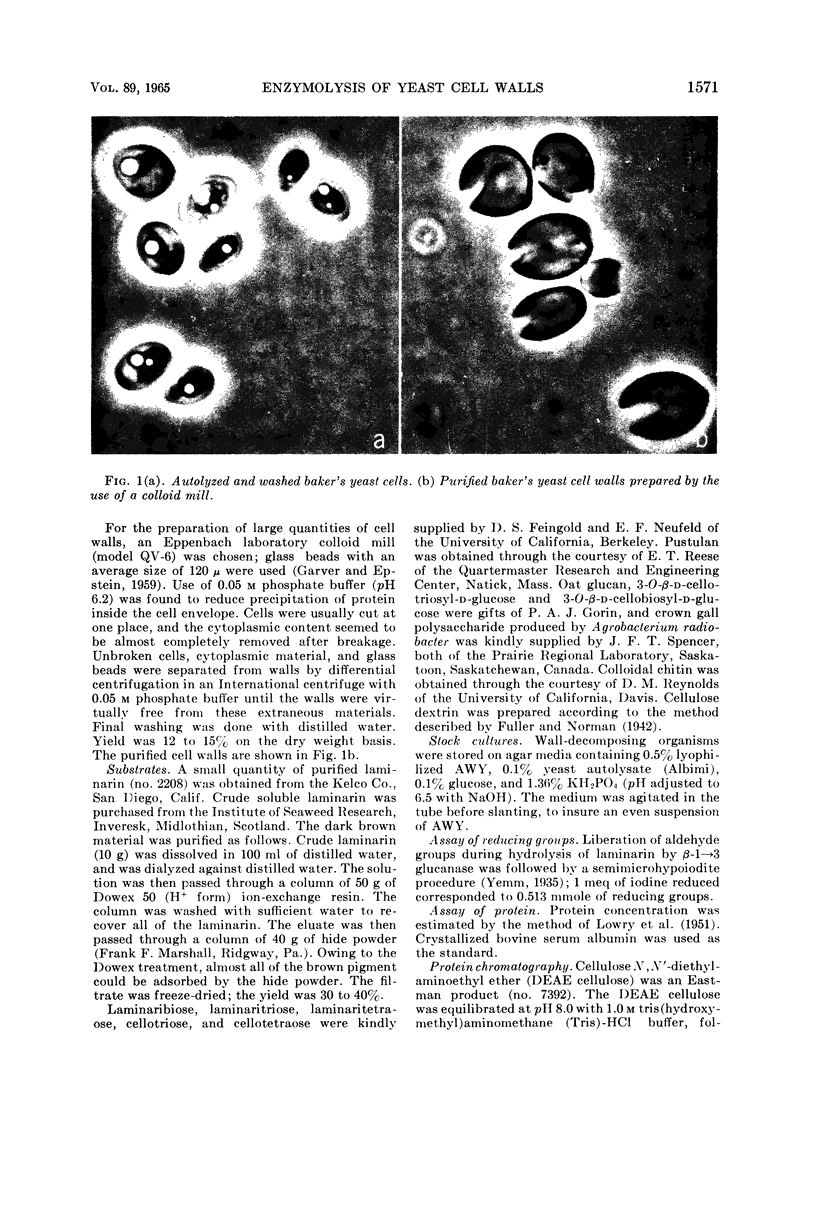


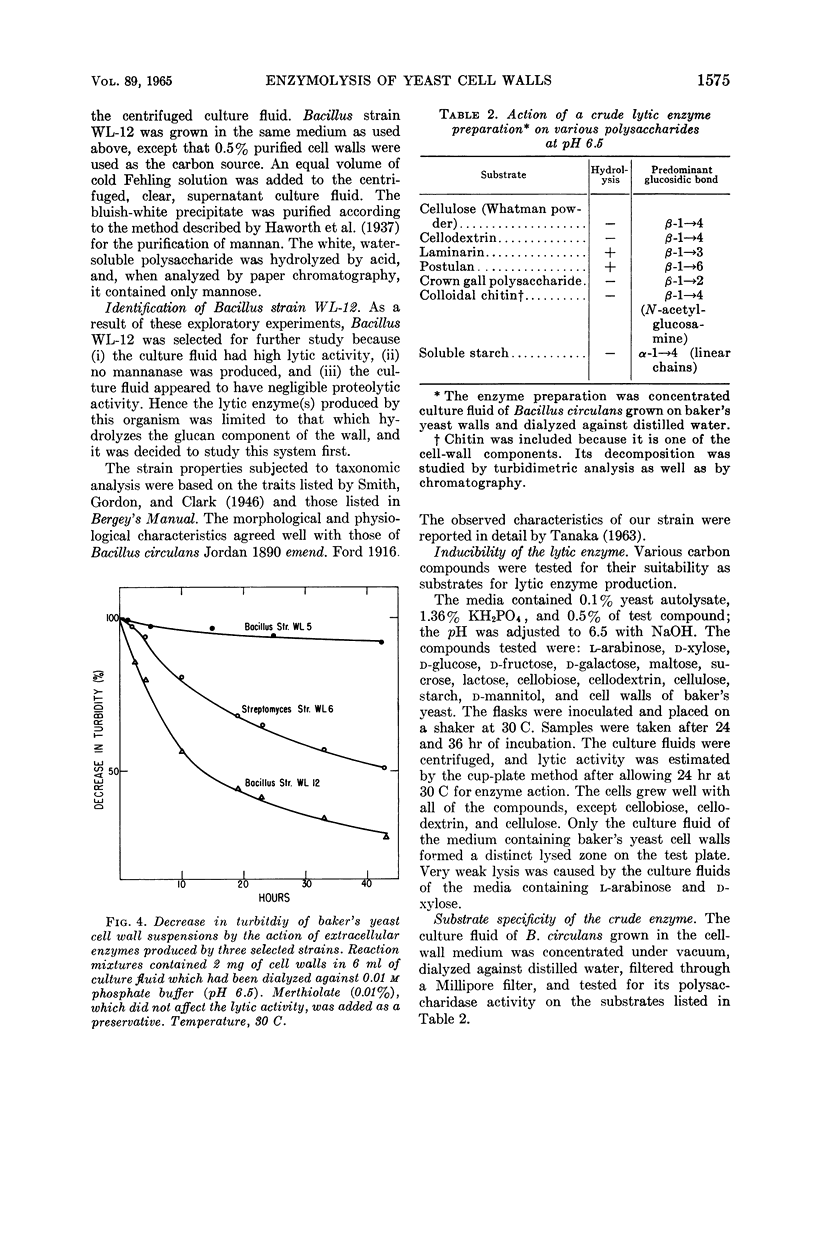

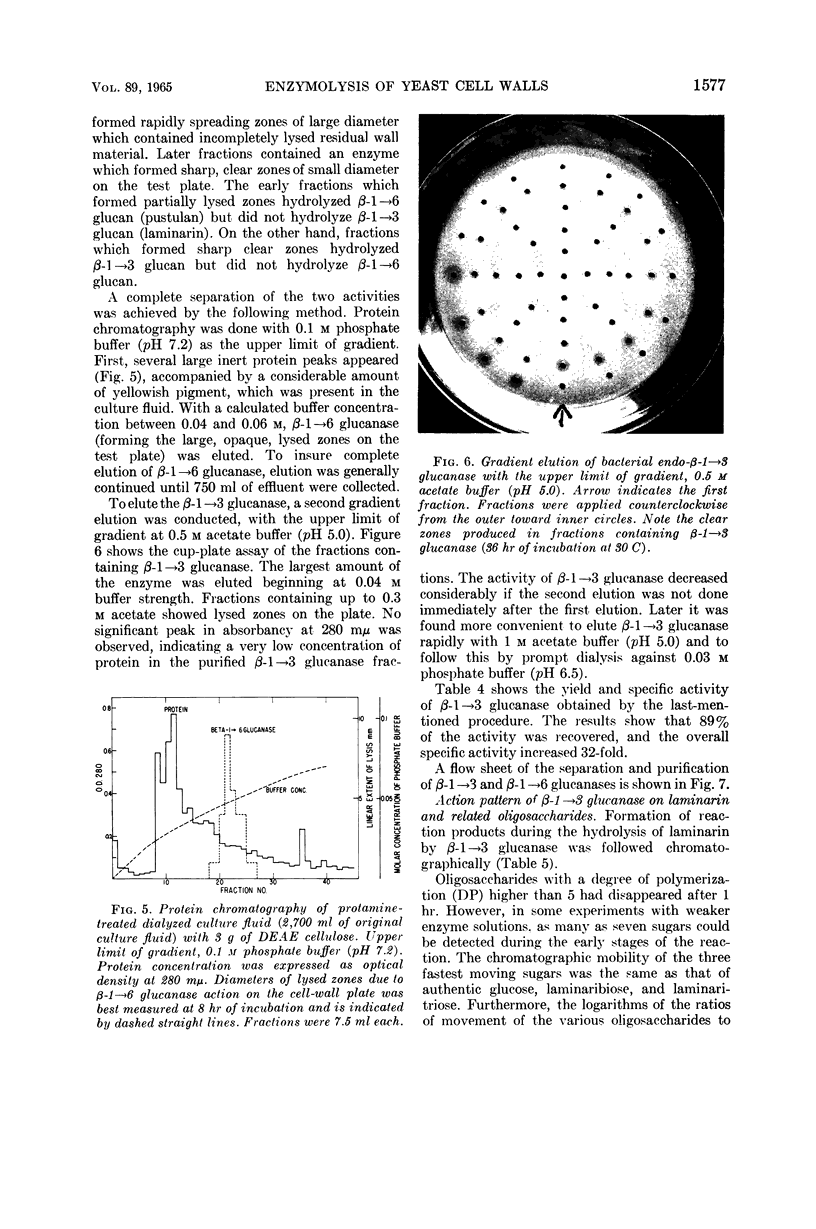


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- GARVER J. C., EPSTEIN R. L. Method for rupturing large quantities of microorganisms. Appl Microbiol. 1959 Sep;7:318–319. doi: 10.1128/am.7.5.318-319.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOLDEN M., TRACEY M. V. A study of enzymes that can break down tobacco-leaf components; digestive juice of Helix on defined substrates. Biochem J. 1950 Oct;47(4):407–414. doi: 10.1042/bj0470407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HORIKOSHI K., KOFFLER H., ARIMA K. Purification and properties of beta-1,3-glucanase from the "lytic enzyme" of Bacillus circulans. Biochim Biophys Acta. 1963 Jun 11;73:267–275. doi: 10.1016/0006-3002(63)90311-1. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- NICKERSON W. J. SYMPOSIUM ON BIOCHEMICAL BASES OF MORPHOGENESIS IN FUNGI. IV. MOLECULAR BASES OF FORM IN YEASTS. Bacteriol Rev. 1963 Sep;27:305–324. doi: 10.1128/br.27.3.305-324.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PARRISH F. W., PERLIN A. S. Steric factors affecting the specificity of polyglycosidases. Nature. 1960 Sep 24;187:1110–1111. doi: 10.1038/1871110b0. [DOI] [PubMed] [Google Scholar]
- PHAFF H. J. CELL WALL OF YEASTS. Annu Rev Microbiol. 1963;17:15–30. doi: 10.1146/annurev.mi.17.100163.000311. [DOI] [PubMed] [Google Scholar]
- REESE E. T., MANDELS M. Use of enzymes in isolation and analysis of polysaccharides. Appl Microbiol. 1959 Nov;7:378–387. doi: 10.1128/am.7.6.378-387.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]







