Abstract
Particles formed by the bacteriophage MS2 coat protein mutants with insertions in their surface loops induce a strong immune response against the inserted epitopes. The covalent dimers created by fusion of two copies of the coat protein gene are more tolerant to various insertions into the surface loops than the single subunits. We determined a 4.7-Å resolution crystal structure of an icosahedral particle assembled from covalent dimers and compared its stability with wild-type virions. The structure resembled the wild-type virion except for the intersubunit linker regions. The covalent dimer orientation was random with respect to both icosahedral twofold and quasi-twofold symmetry axes. A fraction of the particles was unstable in phosphate buffer because of assembly defects. Our results provide a structural background for design of modified covalent coat protein dimer subunits for use in immunization.
Keywords: virus, structure, assembly, stability
Introduction
The virion of bacteriophage MS2 consists of 180 coat protein subunits, a single molecule of the positive sense RNA genome, and one copy of the maturation protein.1 The capsid has a diameter of 280 Å and follows T = 3 icosahedral quasisymmetry. There are three coat protein subunits in the icosahedral asymmetric unit, and these are named A, B, and C [Fig. 1(a)]. The coat protein subunit consists of an N-terminal hairpin with β-strands A and B, a five-stranded antiparallel β-sheet that forms the inner face of the capsid with strands C to G, and a C-terminal arm composed of two α-helices, A and B. The two helices of one subunit fit into a groove formed by the C-terminal arm and the N-terminal hairpin of the other subunit. In the dimer the five-stranded β-sheets of the two monomers join into one. Because the coat protein subunits make extensive pairwise interactions, the capsid can be considered to be assembled from 90 dimers.2 There are two types of dimers in the capsid: AB dimers located at icosahedral quasi-twofold axes and CC dimers located at icosahedral twofold axes [Fig. 1(a)].
Figure 1.
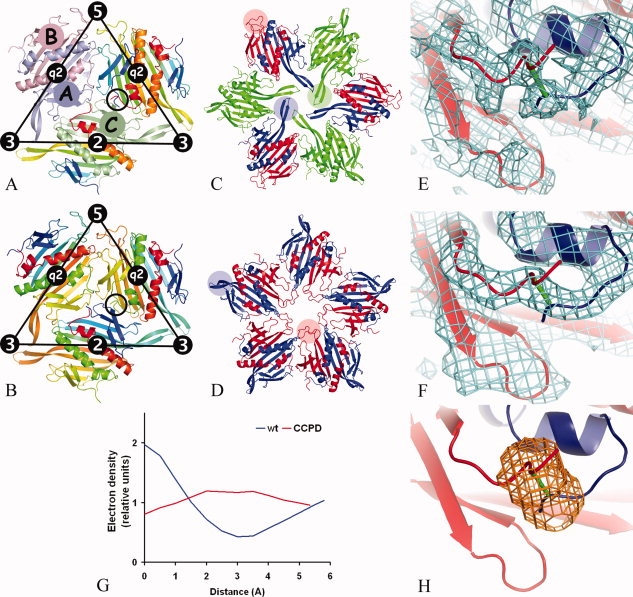
Comparison of the icosahedral subunit arrangement of the wild-type MS2 coat protein (a) and the putative CCPD arrangement (b). The location of icosahedral symmetry elements is shown together with the borders of the icosahedral asymmetric unit. In (a), subunit A1 is painted light-blue, B5 light-red, C1 light-green, and the remaining subunits are rainbow colored from the N-terminus in blue to the C-terminus in red. The black circle indicates the position of the C-terminus of subunit A2 and the N-terminus of subunit B1, which are located in close proximity to each other. Wild-type MS2 subunits were used to represent the CCPD subunits in (b). The subunits were rainbow colored as if the dimer-forming subunits were one chain. For example, subunit A2 was colored from the N-terminus in blue to the C-terminus in yellow-green. The coloring then continues in subunit B1 starting from the N-terminus in yellow-green to the C-terminus in red. The N-terminus of B1 and the C-terminus of A2 in the black circle represent the position of the covalent intersubunit linker. The CCPD subunits represented by the A2B1 and A1B5 dimers have opposite directionality that makes the fivefold symmetry only approximate. Arrangement of MS2 coat protein subunits around threefold (c) and fivefold (d) symmetry axes. The subunits are colored as follows: A in blue, B in red, and C in green. FG loops of selected subunits are highlighted with transparent circles of corresponding color. Comparison of electron density between the N-terminus of the B subunit and the C-terminus of the A subunit from the icosahedral particle of wild-type VLP (e) and CCPD (f). The CCPD subunit is represented by wild-type subunits and the putative link between the subunits is shown in green (f). Subunit B is shown in red and subunit A in blue. Displayed parts of wild-type and CCPD subunits correspond to the circles in (a) and (b), respectively. (g) Electron density distribution along a line connecting atoms CA of residue A128 and N of residue B3 in wild-type and CCPD particles. The lines are shown in Figure 1(e,f), respectively. The interpolated values of electron density at given points were brought on relative scale by dividing by the average electron density for the respective structure. (h) A mask shoving a region that was used in comparison of the electron density distribution between the C- and N-termini of the subunits.
The structures of the three icosahedrally independent subunits of wild-type MS2 are similar to each other. The only structurally important differences between the subunits are located in the FG loop connecting strands F and G. The FG loops participate in the formation of three quasiequivalent contacts. Two conformational variants of the FG loop are part of a sixfold arrangement at a threefold axis, whereas the remaining one is part of a pentameric interface [Fig. 1(c,d)]. It has been shown that the FG loop interactions play a complex role in MS2 particle assembly and stability. Many coat protein variants with mutations in the FG loop have failed to form particles.3–6 Other FG loop mutants have been able to assemble into particles, but the phage with the modified gene was not viable.7,8 A mutant of bacteriophage fr with a four-residue deletion in the FG loop was still capable of forming normal particles even if the loops did not interact, but the particles had lower stability than the wild type.9 The function of the FG loop in the levivirus assembly provides an interesting topic for research as it is a model for studying how one peptide sequence can participate in mediating different protein–protein interactions.
In the coat protein dimmer, the N-terminus of one subunit is located close to the C-terminus of the other subunit [Fig. 1(a)]. It has been shown that it is possible to create a covalent coat protein dimer (CCPD) by placing one copy of the coat protein gene immediately behind the other one while removing the stop codon of the first gene.10 The CCPD folds properly and retains the ability to form virus-like particles (VLPs). We have earlier reported the crystallization of CCPD in space group F432 in which the protein did not crystallize as icosahedral particles. Instead the crystal was built from subunits in an octahedral arrangement.11
We have now determined the 4.7-Å resolution structure of the icosahedral CCPD particle and compared its stability with the wild-type particles to provide additional information on the crystallization of the CCPD in the octahedral arrangement. We have also studied the temperature stability of complexes formed by CCPD subunits with mutations in one of the two FG loops of the covalent dimer to analyze from a new angle the importance of FG loop interactions in MS2 particle assembly and stability.
Results
The CCPD icosahedral particle model
The crystal structure of the CCPD particle has been determined at 4.7 Å resolution. The electron density map resulting from 20-fold noncrystallographic averaging shows features identical to the wild-type MS2 particles except for the region of the intersubunit linker of the CCPD. Because of the low resolution, a complete model of the CCPD dimer was not built. The structure is represented by the wild-type MS2 subunits (pdb entry 2ms2). The data quality statistics are listed in Table I. The Rmerge is high because of weak data especially at higher resolution. Omitting the high-resolution data from the electron density calculation resulted in obliteration of details in the map. All measured reflections were used in the refinement.
Table I.
Scaling, Phasing, and Refinement Statistics
| Space group | P213 |
| Unit cell dimensions (Å) | 368.2 |
| Resolution limits (Å) | 50–4.7 |
| High-resolution bin (Å) | 4.91–4.70 |
| Completeness | 84% |
| Completeness in high-resolution bin | 51% |
| Rmergea | 0.251 |
| Rmergea in high-resolution bin | 1.000 |
| Average redundancy | 3.4 |
| Average redundancy in high-resolution bin | 2.0 |
| 〈I〉/〈σ〉 | 2.9 |
| 〈I〉/〈σ〉 in high-resolution bin | 0.6 |
| R-factor | 0.318 |
| R-factor (high-resolution bin) | 0.365 |
| Correlation coefficient after convergence of map based on model (15 Å) | 0.936 |
| Correlation coefficient after convergence of map after phase extension (4.7 Å) | 0.924 |
| Number of unique reflections | 72,743 |
Rmerge = ΣhΣj |Ihj − 〈Ih〉|/ΣΣ|Ihj|.
The positions of the subunits were refined as rigid bodies. In comparison to the MS2 virion, the CCPD particle has an ∼0.5 Å smaller radius and the subunits are also tilted 1° toward the quasi-threefold axis. Group B factors were refined for the subunits. The resulting B factors of the subunits are similar: 136, 140, and 140 Å2 for the A, B, and C subunits, respectively. No attempts to rebuild the model were made because of the limited resolution of the data. If calculated, the Rfree would be similar to the R-value because of the high noncrystallographic symmetry.12 The coordinates together with the structure factors have been deposited in the protein data bank as entry 2wbh.
The electron density of the CCPD particles showed a connection between the C-terminus of the A subunit and the N-terminus of the B subunit that was not present in the electron density of the wild-type VLP particle calculated at the same resolution [Fig. 1(e,f)]. The distributions of the electron density along a line connecting the C-alpha atoms of residue A128 and the main chain nitrogen of residue B3 of wild type, and CCPD VLPs are shown in Figure 1(g). There is continuous electron density in the CCPD particles, whereas there is a low-density region in the middle in the wild-type particles. Similar connecting density was also observed between the C-terminus of the B subunit and the N-terminus of the A subunit as well as between the C- and N-termini of the C subunits of the CCPD particles. The correlation coefficient between the electron density distribution at the A-B and C-C intersubunit linkers is 94% and that of B-A and C-C is 96%. A mask showing the volume used in the comparison is shown in Figure 1(h). The electron density distribution at the linker region between the C- and N-termini of the C subunits most likely corresponds to 50% occupancy of the linker region because the orientations of the CCPD dimers at the corresponding CC positions are unlikely to be the same in all the particles in the crystal. The correlation coefficient of the electron density distribution at the A-B and B-A linker regions is 94%. The high similarity of the electron density distribution at the A-B and B-A linker regions to each other and to the C-C linker region suggests that the orientation of the CCPD subunits is random with respect to the quasi-twofold axis.
Particle stability measurements
Particles of the bacteriophage MS2 formed a sharp band when run on an agarose gel. The speed of migration of the particles corresponds approximately to a 1.2-kb-long molecule of double-stranded DNA [Figs. 2(a) and 3(a)]. The electrophoresis provided a convenient way of checking for the presence of virions or VLPs in the sample. The particles when heated to a certain temperature (the melting temperature of the particle) are disrupted, which changes the migration behavior of the material on the agarose gel. Apparently, the disrupted particles aggregate and do not enter the gel but appear as a sharp band at the position of the sample application. The agarose gels were prepared from 40 mM Tris, 20 mM acetic acid, and 1 mM EDTA buffer as described in the Materials and Methods section. The buffer used in the temperature stability measurements was 40 mM Tris-HCl, pH 8.0, and 200 mM NaCl. The gels were stained with ethidium bromide and subsequently with Coomassie blue to detect the presence of both RNA and protein components of the particles (Figs. 2 and 3). In the temperature stability experiments, the melting temperature of MS2 phage and wild-type VPLs was 66°C, whereas it was 62°C for CCPD VLPs [Figs. 2(a,d,g) and 3(a,d,g)]. Weak bands corresponding to the migration of 400—200-bp DNA fragments were observed in MS2 and wild-type VLPs preparations [Figs. 2(a,d) and 3(a,d)]. These bands are probably due to the small amounts of contaminating RNA. The preparations of CCPD VLPs contained material that migrated slower than the MS2 virions [Figs. 2(j) and 3(j)]. The slower band may correspond to the particle dimers.13 Most of it precipitated at 55°C and probably corresponded to defective particles.
Figure 2.
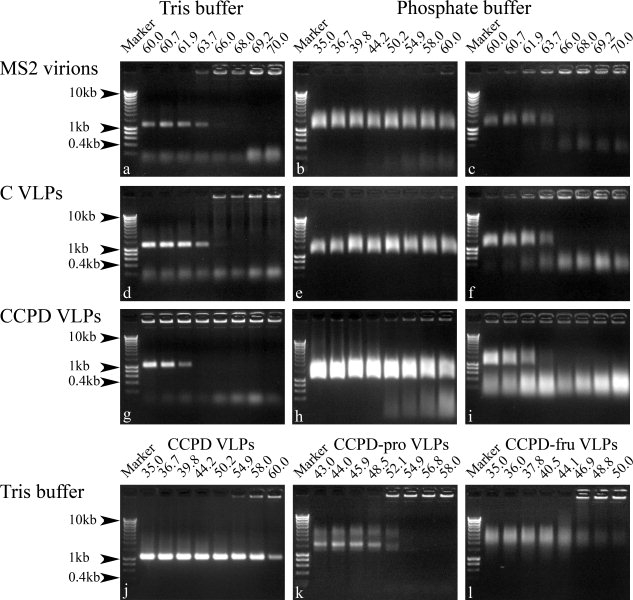
Agarose gel electrophoresis of samples incubated at the indicated temperatures (°C) and buffer compositions. The gels were stained with ethidium bromide and visualized under UV light. The experiments were performed as described in the Materials and Methods section.
Figure 3.
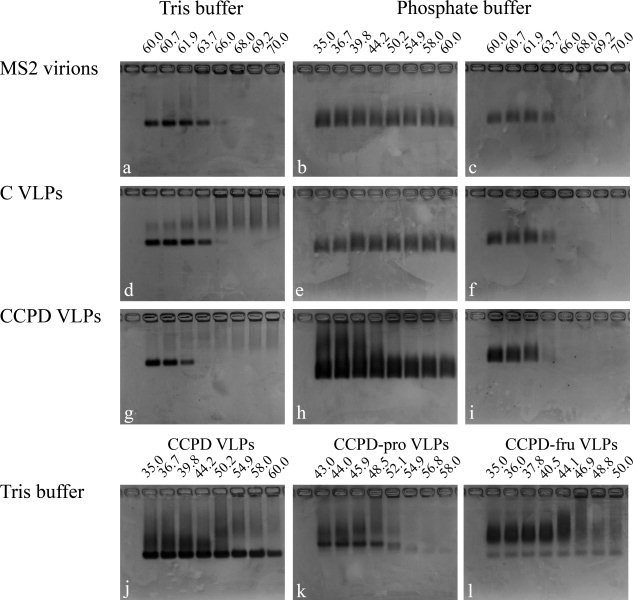
Coomassie blue stain of gels shown in Figure 2. The samples were incubated at the indicated temperatures (°C) and buffer compositions.
We also tested the temperature stability of CCPD variants with mutations in only one of the FG loops. The mutant CCPD-pro had a single residue substitution leu77 → pro (Fig. 4), which was previously shown to impair the ability of MS2 coat protein to form particles.3 The mutant CCPD-FruR had an insertion of 54 residues between the coat protein residues 70–78 (Fig. 4). The protein FruR (pdb code 1uxc) was chosen for the insertion because its C and N-termini are located close to each other.14 The melting temperature of CCPD-pro was 55°C and that of CCPD-FruR was 44°C [Figs. 2(k,l) and 3(k,l)]. The behavior of the complexes assembled from CCPDs with mutations in the FG loop on the agarose gel differed from that of the wild-type particles. The CCPD-pro migrated as two bands. The sharp and faster one corresponded to 1.5-kb DNA molecules, whereas the diffuse and slower corresponded to 3–6-kb DNA fragments [Figs. 2(k) and 3(k)]. The CCPD-FruR complexes migrated as a diffuse band as would 4–10-kb DNA molecules [Figs. 2(l) and 3(l)].
Figure 4.
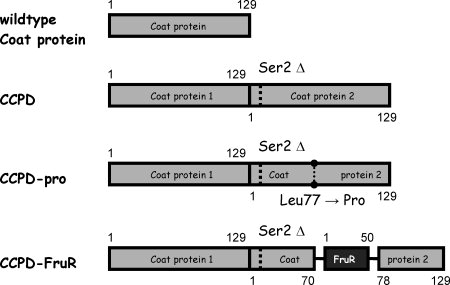
MS2 coat protein variants. The wild-type coat protein has 129 residues. The CCPD contains two copies of the coat protein with a single residue deletion of Ser 2 from the second copy of the coat protein. The CCPD-pro has a single residue substitution Leu77 → Pro in the second copy of the coat protein. The CCPD-FruR has a 54-residue insertion between residues 70–78 of the second copy of the coat protein. The insertion consists of residues Lys and Leu (HindIII restriction site), then 50 residues of the FruR protein and two extra residues Ala and Ser (Nhe1 restriction site). A detailed description of the CCPD sequences is given in the Materials and Methods section.
Measurements with identical temperature ranges were performed in 0.32M Na2HPO4, 0.08M NaH2PO4, and 5% PEG 8000 buffer, because these were the crystallization conditions when CCPD formed the octahedral arrangement. The bands of MS2 virions and VLPs incubated in phosphate buffer were more diffused than the corresponding bands in the Tris buffer (Figs. 2 and 3). The smearing effect was probably caused by the higher ionic strength of the phosphate buffer in the sample. The melting temperatures were not affected, but a fraction of the CCPD particles precipitated at lower than melting temperatures [Fig. 2(h)]. A corresponding precipitation was not observed for MS2 phage and wild-type VLPs [Fig. 2(b,e)]. A diffuse band corresponding to DNA molecules of 200–1000 bp appeared after some higher incubation temperatures in the phosphate buffer samples [Fig. 2(c,f,i)]. This band probably represents partly degraded RNA molecules released from the particles as it was not detected with Coomassie blue staining [Fig. 3(c,f,i)].
Discussion
CCPD icosahedral particle structure and stability
The CCPD subunit consists of 257 residues representing two copies of the MS2 coat protein in which serine 2 of the second subunit is missing (Fig. 4). The molecule itself has an approximate twofold symmetry, but the presence of the intersubunit linker prevents it from having perfect symmetry [Fig. 1(b)]. The approximate symmetry of the CCPD subunit results in two possible orientations of the subunit with respect to the icosahedral twofold and quasi-twofold symmetry axes when incorporated into the particle [Fig. 1(b)]. The 4.7-Å resolution electron density map of the CCPD icosahedral particles indicated that the orientation of the CCPD dimer was probably random with respect to both icosahedral twofold and quasi-twofold axes [Fig. 1(f–h)]. If built, the model of the CCPD icosahedral asymmetric unit would consist of three CCPD chains with 50% occupancy, analogous to the crystallographic asymmetric unit of the CCPD molecules in the F432 crystals.11 The presence of the quasi-twofold symmetrical subunit at the icosahedral twofold symmetry axis disturbs the ideal symmetry of the icosahedral particle [Fig. 1(b)]. The deviation of the CCPD particles from the ideal arrangement are reflected in the lower melting temperature of the CCPD icosahedral particles (62°C), compared with the melting temperature of the MS2 virions and wild-type VPLs (66°C) [Figs. 2(a,d,g) and 3(a,d,g)]. The intersubunit linkers make different interactions at the quasi-threefold interface from the wild-type N- and C-termini, which may also contribute to the lower CCPD particle stability.
The FG loop in MS2 particle assembly and stability
The ability of the CCPD subunits to form particles in which the orientation of the CCPD subunit is random with respect to the icosahedral twofold and quasi-twofold symmetry axes provides an interesting opportunity for studying whether all intersubunit interactions are necessary for MS2 particle assembly and stability. To explore this, we created two CCPD variants with mutations in only one of the FG loops. The substitution leu77 → pro was chosen because the single subunit mutant is incapable of forming VLPs and exists mostly as dimers.3 The protein FruR was selected for insertion to completely eliminate the possibility of the FG loops contacts. The expression of the CCPD subunits with mutation in one of the FG loops could have resulted in the assembly of pentamers or hexamers only. The behavior of the CCPD FG loop mutants on the agarose electrophoresis [Figs. 2(k,l) and 3(k,l)] and size exclusion chromatography (data not shown) indicated that they could form aggregates of the size of the wild-type particles or bigger. The most likely explanation is that they form VLPs. The different speeds of migration of the mutant particles in the agarose gel in comparison to wild-type VLPs could be caused by assembly defects and in the case of CCPD-FruR by different overall charge of the particles. We were not able to obtain representative electron microscopy images of particles assembled from CCPD with FG loop mutations. Most of the particles were probably destroyed during sample preparation (negative staining) because of their low stability. The problem of observing the complexes assembled by CCPD FG loop mutants may be overcome by performing cryoelectron microscopy, which is an interesting topic for future research. The assembly incompetence of the mutated FG loop was compensated for by interactions of the remaining loop and other parts of the subunits in the capsid. The positioning of the mutated FG loops is probably random at the fivefold and sixfold contacts.
The melting temperatures of CCPD-pro and CCPD-FruR complexes were 55°C and 44°C, respectively [Figs. 2(k,l) and 3(k,l)]. The lowered temperature stability of the putative particles formed by CCPD FG loop mutants may be caused by the missing FG loop interactions or assembly defects. One functional FG loop per subunit dimer may be sufficient for particle assembly, but the presence of all FG loops is required for a wild-type level of particle stability. The architecture of the bacteriophage MS2 capsid is robust—the multiplicity of interactions allows it to tolerate some defective subunits.
CCPD icosahedral particles and crystallization of CCPD subunits in F432 space group
We previously showed that the CCPD dimer can be crystallized in space group F432 in which the arrangement of the subunits in the asymmetric unit, together with the crystallographic symmetry, corresponds to T = 3 octahedral particles.11 We did not detect the octahedral particles when the CCPD preparations were analyzed by electron microscopy. A necessary step in the formation of F432 crystals appears to be the initial disassembly of the icosahedral CCPD particles, which were present in the samples of CCPD. Our current results show that the melting temperature of the CCPD particles was only 4°C lower than the melting temperature of the MS2 phage and wild-type VLPs [Figs. 2(a,d,g) and 3(a,d,g)]. This slight decrease in the CCPD particle stability may not explain the formation of the F432 crystals.
When the temperature stability experiments were performed in a phosphate buffer (the crystallization buffer where the F432 crystals formed), a fraction of the CCPD material precipitated already at 35°C (Fig. 2h). In contrast there was no precipitation in the MS2 phage or wild-type VLPs samples at this temperature [Fig. 2(b,e)]. The precipitation of a fraction of CCPD particles at the lower temperature may be an indication that some of the CCPD particles had assembly defects. The crystallization resulting in the formation of the F432 crystals was performed at 37°C and precipitation together with phase separation was observed in the crystallization drops. We, therefore, speculate that the particles with assembly defects could have been a source of the CCPD subunits for the F432 crystal growth.
VLPs assembled from MS2 coat protein mutants with insertions in the N-terminal hairpin are highly immunogenic epitope carriers.15,16 A limitation of the single subunits is their low tolerance to insertions into the N-terminal hairpin loop. To a great extent the folding problem can be overcome by using modified CCPDs.17 We have shown that a fraction of the CCPD particles may have assembly defects that lower their stability in certain buffer compositions. The orientation of the CCPD subunits is most likely random with respect to icosahedral twofold and quasi-twofold axes. Introducing mutations into the FG loop provides an effective mean to regulate the CCPD particle stability. Thus, our findings provide a structural background for design of the CCPD antigen-presenting particles.
Materials and Methods
Cloning and production of CCPD VLPs and MS2 phage
The leu77 → pro mutation and FruR insertion were made in the second subunit of the CCPDs. The leu77 → pro mutation was introduced using primers MS2CCprof GGTGGTGTAGAGCCTCCTGTAGCCGC and MS2CCpror GCGGCTACAGGAGGCTCTACACCACC. The FruR gene was PCR amplified with the primers FruR5 CACTGAAGCTTATGAAACTGGATGAAATCGCTCGG and FruR3 CGACTGCTAGCGTTCGGGTGGTAATTGTGCTCACG introducing the HindIII restriction site at 5′ and Nhe1 restriction site at 3′ end. Corresponding restriction sites were introduced into the MS2 coat protein gene using primers Insert53 (GTGCCTAAAGTGGCAACCCAGAAGCTTGATGTACATCTAGCTAGCCCTGTAGCCGCATGGCGTTCG) and Insert35 (CGAACGCCATGCGGCTACAGGGCTAGCTAGATGTACATCAAGCTTCTGGGTTGCCACTTTAGGCAC). The FruR gene was inserted between residues 70–78 of the MS2 coat protein. The sequence of the HindIII restriction site introduced residues Lys and Leu after residue 70 of the coat protein, before residue 1 of the FruR protein. The sequence of the Nhe1 restriction site introduced residues Ala and Ser after residue 50 of the FruR protein, before residue 78 of the coat protein. The CCPD FG loop mutants were cloned into the expression plasmid pBAD under the control of the arabinose promotor. The sequence was verified (data not shown). The CCPD as in Peabody and Lim10 was cloned, produced, and purified as described in Plevka et al.11 MS2 phage was produced according to Valegård et al.18
Crystallization and data collection
Crystals of CCPD VLPs were obtained using the hanging-drop vapor diffusion technique with a reservoir solution containing 0.1M Bicine, pH 9.0, and 20% PEG 5000-monoethyl. The drops were prepared by mixing 1.5 μL of reservoir solution with an equal volume of CCPD VLP solution (10–20 mg/mL in 20 mM tris-HCl, pH 8.0). Triangular- and tetrahedron-shaped crystals were formed within 1–3 days at 20°C. For data collection, crystals were vitrified in liquid nitrogen. Data were collected from a single crystal at 100 K on the ADSC Quantum3 CCD detector at beamline ID 29 at the ESRF synchrotron radiation source in Grenoble, France. An oscillation range of 0.2° was used during data collection. The crystal diffracted to 4.7 Å resolution. Data were processed and scaled using the HKL2000 package.19 Statistics from data collection and scaling is shown in Table I.
CCPD-VLP structure determination
The CCPD-VLP crystals were of space group P213. One-third of a virus particle occupied a crystallographic asymmetric unit, as one of the icosahedral threefolds was congruent with crystallographic threefold axis. Therefore, one parameter for the particle orientation and one for the position had to be determined. The locked self-rotation function in the program GLRF was used to identify the particle orientation.20 Reflections between 16 and 5.0 Å were used for the calculations. The radius of integration was set to 280 Å. The results suggested that the model particle needs to be rotated with ϕ = 21.3°, φ = 18.3°, and κ = 47.0° from the standard icosahedral orientation according to the XYK polar angle convention. Translational parameters were identified by the phaser translation function21 using a correctly oriented MS2 particle (generated from pdb entry 2ms2). The pdb file representing the entire MS2 capsid was generated using the program Xpand (Gerard Kleywegt's unpublished program). The particle center was identified at x = 0 Å, y = 184 Å, z = 184 Å. The MS2 model of the icosahedral asymmetric unit was used for the molecular replacement. The model was oriented and positioned in the unit cell and was used to calculate the initial phases for reflections up to 15 Å resolution in the program CNS.22 The phases were refined by 15 cycles of averaging by the program AVE,23 using the noncrystallographic symmetry. Phase extension was applied to obtain phases for higher resolution reflections. Addition of a small fraction of higher resolution data (one index at a time) was followed by four cycles of averaging. This procedure was repeated until phases were obtained for all the reflections up to 4.7 Å resolution. The program SigmaA24 was used to use weights in electron density calculations. The positions and orientations of the subunits were subjected to rigid-body refinement in CNS.22 Because of the low resolution of the data, a single B factor was refined for each subunit. A model for the interconnecting loop between the N- and C-termini of the subunits in the dimer was not built. Statistics describing the fit of the model to the experimental data are shown in Table I. Because we did not do any model rebuilding, the model quality indicators remain identical to the original 2ms2 pdb file and are not listed.
Calculation of 4.7-Å resolution density of wild-type MS2 VLP
Because experimental data for the 2ms2 model are not available, we decided to use the 5msf25 model for calculation of 4.7-Å resolution electron density map instead. The model represents MS2 VLP with an RNA aptamer soaked in it. The model was used to calculate the initial phases for reflections up to 4.7 Å resolution. The phases were refined by 10 cycles of averaging, using the noncrystallographic symmetry. The resulting phases were used for electron density calculation.
Temperature stability measurements
The VLP or MS2 phage sample (0.5–1.0 mg/mL) in an appropriate buffer (40 mM Tris-HCl, pH 8.0, 200 mM NaCl or 0.32M Na2HPO4, 0.08M NaH2PO4, 5% PEG 8000) was incubated for 15 min at the selected temperature. The Bio-Rad (CA, USA) PCR machine MyCycler temperature gradient program was used. This resulted in uneven temperature increase between samples as seen by temperature values in Figures 2 and 3. Immediately after heating, the samples were loaded onto a 40 mM Tris, 20 mM acetic acid, 1 mM EDTA, and 0.6 μg/mL ethidium bromide buffer in 1% agarose gel. The particle position was visualized under UV illumination. Subsequently, the gel was stained for 1 h with Coomassie blue (0.05% Coomassie blue G250, 2% TCA, 20% ethanol) and destained overnight in distilled water.
Acknowledgments
The authors thank the staff at the ESRF, France. P.P. acknowledges advice on various calculations from Alwyn Jones and Gerard Kleywegt and help with manuscript preparation from Terese Bergfors.
References
- 1.Witherell GW, Gott JM, Uhlenbeck OC. Specific interaction between RNA phage coat proteins and RNA. Prog Nucl Acid Res Mol Biol. 1991;40:185–220. doi: 10.1016/s0079-6603(08)60842-9. [DOI] [PubMed] [Google Scholar]
- 2.Valegård K, Liljas L, Fridborg K, Unge T. The three-dimensional structure of the bacterial virus MS2. Nature. 1990;345:36–41. doi: 10.1038/345036a0. [DOI] [PubMed] [Google Scholar]
- 3.Peabody DS, Ely KR. Control of translational repression by protein-protein interactions. Nucleic Acids Res. 1992;20:1649–1655. doi: 10.1093/nar/20.7.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peabody DS. The RNA binding site of bacteriophage MS2 coat protein. EMBO J. 1993;12:595–600. doi: 10.1002/j.1460-2075.1993.tb05691.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pushko P, Kozlovskaya T, Sominskaya I, Brede A, Stankevica E, Ose V, Pumpens P, Grens E. Analysis of RNA phage fr coat protein assembly by insertion, deletion and substitution mutagenesis. Protein Eng. 1993;6:883–891. doi: 10.1093/protein/6.8.883. [DOI] [PubMed] [Google Scholar]
- 6.Stockley PG, Stonehouse NJ, Walton C, Walters DA, Medina G, Macedo MB, Hill HR, Goodman STS, Talbot SJ, Tewary HK, Golmohammadi R, Liljas L, Valegård K. Molecular mechanism of RNA-phage morphogenesis. Biochem Soc Trans. 1993;21:627–633. doi: 10.1042/bst0210627. [DOI] [PubMed] [Google Scholar]
- 7.Stonehouse NJ, Valegård K, Golmohammadi R, van den Worm S, Walton C, Stockley PG, Liljas L. Crystal structure of MS2 capsids with mutations in the subunit FG loop. J Mol Biol. 1996;256:330–339. doi: 10.1006/jmbi.1996.0089. [DOI] [PubMed] [Google Scholar]
- 8.Hill HR, Stonehouse NJ, Fonseca SA, Stockley PG. Analysis of phage MS2 coat protein mutants expressed from a reconstituted phagemid reveals that proline 78 is essential for viral infectivity. J Mol Biol. 1997;266:1–7. doi: 10.1006/jmbi.1996.0786. [DOI] [PubMed] [Google Scholar]
- 9.Axblom C, Tars K, Fridborg K, Orna L, Bundule M, Liljas L. Structure of phage fr capsids with a deletion in the FG loop: implications for viral assembly. Virology. 1998;249:80–88. doi: 10.1006/viro.1998.9279. [DOI] [PubMed] [Google Scholar]
- 10.Peabody DS, Lim F. Complementation of RNA binding site mutations in MS2 coat protein heterodimers. Nucleic Acids Res. 1996;24:2352–2359. doi: 10.1093/nar/24.12.2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plevka P, Tars K, Liljas L. Crystal packing of a bacteriophage MS2 coat protein mutant corresponds to octahedral particles. Protein Sci. 2008;17:1731–1739. doi: 10.1110/ps.036905.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kleywegt GJ, Brunger AT. Checking your imagination: applications of the free R value. Structure. 1996;4:897–904. doi: 10.1016/s0969-2126(96)00097-4. [DOI] [PubMed] [Google Scholar]
- 13.Peabody DS. A viral platform for chemical modification and multivalent display. J Nanobiotechnol. 2003;1:5. doi: 10.1186/1477-3155-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Penin F, Geourjon C, Montserret R, Bockmann A, Lesage A, Yang YS, Bonod BC, Cortay JC, Negre D, Cozzone AJ, Deleage G. Three-dimensional structure of the DNA-binding domain of the fructose repressor from Escherichia coli by H-1 and N-15 NMR. J Mol Biol. 1997;270:496–510. doi: 10.1006/jmbi.1997.1123. [DOI] [PubMed] [Google Scholar]
- 15.Mastico RA, Talbot SJ, Stockley PG. Multiple presentation of foreign peptides on the surface of an RNA-free spherical bacteriophage capsid. J Gen Virol. 1993;74:541–548. doi: 10.1099/0022-1317-74-4-541. [DOI] [PubMed] [Google Scholar]
- 16.Brown WL, Mastico RA, Wu M, Heal KG, Adams CJ, Murray JB, Simpson JC, Lord JM, Taylor-Robinson AW, Stockley PG. RNA bacteriophage capsid-mediated drug delivery and epitope presentation. Intervirology. 2002;45:371–380. doi: 10.1159/000067930. [DOI] [PubMed] [Google Scholar]
- 17.Peabody DS, Manifold-Wheeler B, Medford A, Jordan SK, Caldeira JD, Chackerian B. Immunogenic display of diverse peptides on virus-like particles of RNA phage MS2. J Mol Biol. 2008;380:252–263. doi: 10.1016/j.jmb.2008.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valegård K, Unge T, Montelius I, Strandberg B, Fiers W. Purification, crystallization and preliminary X-ray data of the bacteriophage MS2. J Mol Biol. 1986;190:587–591. doi: 10.1016/0022-2836(86)90244-5. [DOI] [PubMed] [Google Scholar]
- 19.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter CW Jr, Sweet RM, editors. Methods in enzymology. New York: Academic Press; 1996. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 20.Tong L, Rossmann MG. Rotation function calculations with GLRF program. Methods Enzymol. 1997;276:594–611. [PubMed] [Google Scholar]
- 21.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr Sect D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 23.Kleywegt GJ, Read RJ. Not your average density. Structure. 1997;5:1557–1569. doi: 10.1016/s0969-2126(97)00305-5. [DOI] [PubMed] [Google Scholar]
- 24.Read RJ. Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallogr Sect A. 1986;42:140–149. [Google Scholar]
- 25.Rowsell S, Stonehouse NJ, Convery MA, Adams CJ, Ellington AD, Hirao I, Peabody DS, Stockley PG, Phillips SEV. Crystal structure of a series of RNA aptamers complexed to the same protein target. Nat Struct Biol. 1998;5:970–975. doi: 10.1038/2946. [DOI] [PubMed] [Google Scholar]


