Abstract
In the type III secretion system (T3SS) of Aeromonas hydrophila, AcrH acts as a chaperone for translocators AopB and AopD. AcrH forms a stable 1:1 monomeric complex with AopD, whereas the 1:1 AcrH-AopB complex exists mainly as a metastable oligomeric form and only in minor amounts as a stable monomeric form. Limited protease digestion shows that these complexes contain highly exposed regions, thus allowing mapping of intact functional chaperone binding regions of AopB and AopD. AopD uses the transmembrane domain (DF1, residues 16–147) and the C-terminal amphipathic helical domain (DF2, residues 242–296) whereas AopB uses a discrete region containing the transmembrane domain and the putative N-terminal coiled coil domain (BF1, residues 33–264). Oligomerization of the AcrH-AopB complex is mainly through the C-terminal coiled coil domain of AopB, which is dispensable for chaperone binding. The three proteins, AcrH, AopB, and AopD, can be coexpressed to form an oligomeric and metastable complex. These three proteins are also oligomerized mainly through the C-terminal domain of AopB. Formation of such an oligomeric and metastable complex may be important for the proper formation of translocon of correct topology and stoichiometry on the host membrane.
Keywords: type III secretion system, translocators AopB and AopD, coexpression, oligomerization, chaperone AcrH
Introduction
Aeromonas hydrophila is an ubiquitous Gram-negative bacterium which can cause motile aeromonad septicemia in both fish and humans.1,2 Main clinical symptoms associated with Aeromonas infection are gastroenteritis, wound infections, and systemic illnesses.3 Like many other Gram-negative bacteria, a type III secretion system (T3SS) is essential for the pathogenesis of A. hydrophila.4 Two kinds of proteins, “effectors” and “translocators,” are delivered by the T3SS through the needle-shaped secretion apparatus called “injectisome.”5 Effector proteins are delivered directly from the bacterial cytoplasm into the eukaryotic cell cytosol where they interfere with signaling cascades of host cells, leading to various events, such as cytoskeleton rearrangement and apoptosis of macrophage, that aid survival of bacteria within host cells.6 Three effectors were found to be secreted by a ΔaopN mutant of A. hydrophila AH-1.7 One of these effectors shared 52% identity with ExoT of Pseudomonas aeruginosa. Recently, Vilches et al. and Sha et al. also identified ExoT homologs, AexT and AexU in strains AH-3 and SSU, respectively.8,9 Translocator proteins form a pore in the eukaryotic cell membrane, allowing passage of effectors.10 Some translocators also carry regulatory roles in the synthesis and secretion of T3SS components.11,12 The most studied T3SS translocator proteins are YopB and YopD from Yersinia sp. encoded by the lcrGVH-yopBD operon. These two proteins form a pore channel through the eukaryotic cell membrane and are essential for delivery of effectors into the cytosol of host cells.13,14 In A. hydrophila, a similar acrGVH-aopBD operon has been identified that encodes putative translocators AopB and AopD, respective homologous to YopB and YopD.4 Inactivation of AopB and AopD in A. hydrophila led to decreased cytotoxicity in carp epithelial cells, increased phagocytosis, and reduced virulence in blue gourami fish.4
As effectors and translocators are released only at the moment of host contact or under other appropriate environmental conditions, for example, low-calcium concentrations, they have to be stored inside the bacterial cytoplasm before their release.15 A class of T3SS proteins called “chaperone” is needed to stabilize these translocators within the cytoplasm and to prevent their premature association.16 The chaperone for effector proteins is called Class I chaperone and the structure of some representative members, for example, SycE from Yersinia sp., have been determined by X-ray crystallography.17,18 SycE comprises a mixture of α-β structures and functions as a dimer with the substrate YopE wrapped around this dimer in a partly unfolded form.19,20 This allows the effector to pass through the conduit of the injectisome which is too narrow for the fully folded effector.21 Relatively little is known about the molecular mechanism of the Class II chaperone which is exclusive for translocator proteins. LcrH/SycD is a well studied Yersinia sp. Class II chaperone for translocators YopB and YopD.22 It also regulates the response of T3SS to low-calcium concentrations.23 The crystal structure of Y. enterocolitica SycD has recently been determined.24 LcrH is an entirely α-helical protein comprises of three tetratricopeptide repeat (TPR) like motif.25,26 Two distinct regions on LcrH have been identified to interact separately with YopB and YopD. YopB binds at the concave peptide-binding groove, whereas YopD binds to outer convex regions of LcrH.27 In P. aeruginosa, PcrH has also been reported to bind to PopB or PopD28,29 but simultaneous binding of both PopB and PopD to PcrH has not been observed.29
The acrGVH-aopBD operon that has been identified in A. hydrophila also codes for a putative Class II chaperone AcrH that is located adjacent to its substrate translocators, AopB and AopD. In this study, by coexpressing and copurifying these proteins, we have shown that AcrH interacts with AopB or AopD. Using limited protease digestion, we found that parts of these two translocators were highly exposed even when bound onto the chaperone. A similar approach has been used to determine induced burial regions in chaperones AscE and AscG on complex formation with the needle-subunit AscF.30 Boundaries determined for intact regions of AopB and AopD binding to AcrH generally agreed with the “chaperone binding region” as determined by truncation studies with YopB and YopD on LcrH, but with clear distinctions. These findings also prompted us to identify a region in AopB that is dispensable for chaperone binding but essential for polymerization of the AcrH-AopB and AcrH-AopB-AopD complexes.
Results
The AcrH interacts with AopB or AopD to form a 1:1 complex
To investigate stabilities and oligomeric states of AcrH, AopB, and AopD, initially, we expressed these proteins separately. Both AcrH and AopD could be over-expressed but AopD could only be solubilized in a buffer containing 2% Triton X-100. On gel filtration chromatography, AcrH eluted at a volume corresponding to a dimeric form [Fig. 1(A)], whereas AopD displayed nonspecific and heterogeneous aggregation and was excluded from the column (data not shown). AopB could not be expressed at all in bacterial cells and this was likely because of its extensive hydrophobic regions. Here, we used the pETDuet-1 coexpression system to coexpress each individual translocator with the chaperone AcrH. AcrH was subcloned into the first multiple-cloning site containing an N-terminal poly-His tag. AopB or AopD were respectively subcloned into the second multiple-cloning site of the same vector. Notably, AopB or AopD could be coexpressed with AcrH to form stable AcrH-AopB or AcrH-AopD complexes that could be copurified by Ni-NTA affinity chromatography [Fig. 1(B,C)]. Homogeneity and oligomeric states of complexes were determined via gel filtration chromatography. The AcrH-AopD complex was mostly monomeric and eluted slightly earlier than the dimeric AcrH [Fig. 1(C)]. In contrast, most of the AcrH-AopB complex was oligomeric and excluded from the gel filtration column. Only small part of the complex could be purified in the monomeric form [Fig. 1(B)].
Figure 1.
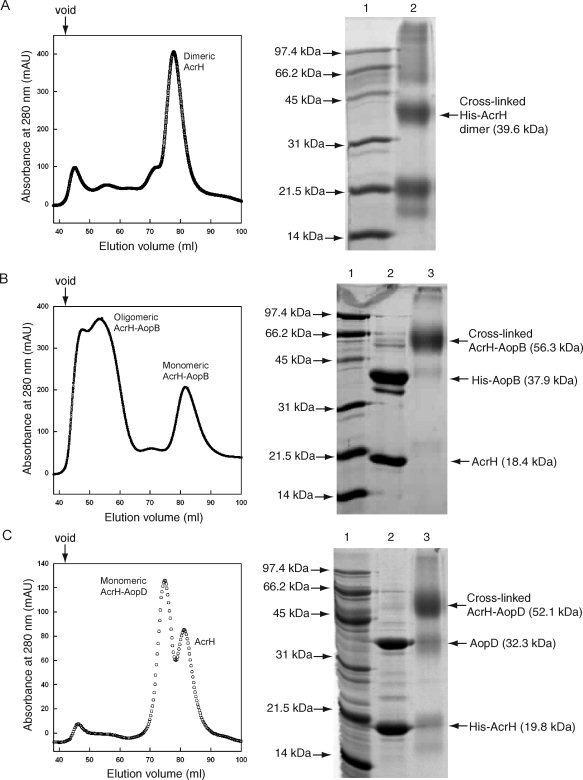
Purification and crosslinking of complexes formed between AcrH and AopB or AopD. A: HiLoad 16/60 Superdex 200 elution profile of AcrH and SDS-PAGE showing crosslinking of AcrH. Lane (1) Mw marker and lane (2) crosslinked product of AcrH. The major product is a dimeric form with a Mw of around 40 kDa. B: Elution profile of the AcrH-AopB complex. Majority of the complex exists as an oligomeric form, only part of it is in a monomeric form. SDS-PAGE shows crosslinking of the monomeric form of AcrH-AopB. Lane (1) Mw marker; lane (2) monomeric AcrH-AopB complex from gel filtration; and lane (3) crosslinked AcrH-AopB complex. The major product is a 1:1 AcrH-AopB complex with a Mw of around 55 kDa. C: Elution profile of the AcrH-AopD complex. The complex exists entirely as a monomeric form. SDS-PAGE shows crosslinking of AcrH-AopD. Lane (1) Mw marker; (2) monomeric AcrH-AopD complex from gel filtration; and (3) crosslinked AcrH-AopD complex. The major product is a 1:1 AcrH-AopD complex with a Mw of around 52 kDa. The down arrow above the elution profile indicates the void volume of the column.
To further confirm the stoichiometry of AcrH and the AcrH-AopD and AcrH-AopB complexes, we performed chemical crosslinking experiments on these proteins. Previously, using chemical crosslinking and analytical gel filtration, it had been shown that SycD/LcrH, the AcrH homolog in Yersinia enterolitica, was expressed as a homodimer.31 Similarly, based on chemical crosslinking of the purified protein, here, we demonstrated that AcrH was also expressed as a homodimer. Figure 1(A) shows that the major species after crosslinking of AcrH was a band of size ∼40 kDa, which corresponded to the molecular weight of a dimeric AcrH (monomeric Mw = 18.4 kDa). Our result agrees with the crystal structure of SycD which forms a head-to-head dimer in the absence of its cognate substrates.24 Crosslinking of the monomeric AcrH-AopB or AcrH-AopD complex [Fig. 1(B,C)] yielded a prominent band of ∼55 kDa corresponding to a 1:1 AcrH-AopB or AcrH-AopD complex. This suggested that the AcrH dimer dissociated to a monomeric state on binding to the translocator AopB (monomeric Mw = 36.5 kDa) or AopD (monomeric Mw = 32.3 kDa). The AcrH-AopD complex was shown to be monomeric, while both monomeric and oligomeric forms of the AcrH-AopB complex were observed, majority being the oligomeric form [Fig. 1(B,C)].
The chaperone binding regions of AopD
Different proteases, namely trypsin, chymotrypsin, papain, thermolysin, elastase, subtilisin, endoproteinase glu-C, and proteinase K, were tested for proteolytic cleavages of the AcrH-AopD complex. Eventually, chymotrypsin was selected for carrying out limited digestion of AcrH-AopD as it produced a stable digestion pattern over 5 h. AopD (32.3 kDa) was cleaved yielding two fragments, DF1 and DF2, that remained bound to AcrH [Fig. 2(A)]. Edman sequencing of DF1 and DF2 showed their N-terminal sequences to be VAPAD and SQANA, respectively. Further analysis by mass spectrometry identified DF1 as the fragment consisting of residues Val16 to Leu147 (132 residues, Mw = 13.9 kDa) and DF2 as the fragment consisting of residues Ser242 to Phe296 (55 residues, Mw = 6.4 kDa) [Fig. 2(B)]. AcrH in the AcrH-AopD complex was resistant to digestion by the protease. The DF1 fragment included the predicted transmembrane domain, whereas the DF2 fragment contained a C-terminal amphipathic domain of AopD.
Figure 2.
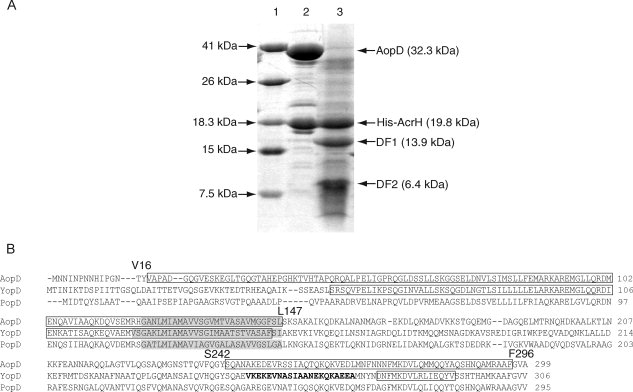
Limited protease digestion of the AcrH-AopD complex. A: SDS-PAGE showing partial digestion of AcrH-AopD with chymotrypsin. Lane (1) Mw marker; lane (2) monomeric AcrH-AopD from gel filtration before digestion; and lane (3) AopD is being digested into two separate fragments, DF1 (residues 16–147) and DF2 (residues 242–296), whereas AcrH remains intact. B: Sequence alignment of AopD from Aeromonas hydrophila (AAR26342) with YopD from Yersinia enterolitica (AAD16812) and PopD from Pseudomonas aeruginosa (AAC45938). Predicted transmembrane domains (TMHMM Server v. 2.0, http://www.cbs.dtu.dk/services/TMHMM-2.0/) are highlighted in gray. Residues that are determined to be involved in chaperone binding are boxed (this work and Francis et al.32). Residues from the coiled coil regions predicted by the program COILS33 are boldfaced.
The chaperone binding region of AopB
To determine the chaperone binding region of AopB, the protease elastase was used to partially cleave the AcrH-AopB complex. AopB (Mw = 36.5 kDa) was reduced to a band of around 24.1 kDa after 5 min of elastase digestion [Fig. 3(A)]. This 24.1 kDa fragment, BF1, was tightly bound to AcrH and eluted together with AcrH through gel filtration chromatography and this was further confirmed by SDS-PAGE. Mass spectrometry and N-terminal sequencing confirmed the identity of this intact chaperone binding BF1 fragment to consist of residues Ala33 to Leu264 of AopB (232 residues, 24.1 kDa) indicating that 32 N-terminal residues and 83 C-terminal residues had been digested away. This BF1 fragment contained the predicted transmembrane domains and a putative N-terminal coiled coil domain [Fig. 3(C)].
Figure 3.
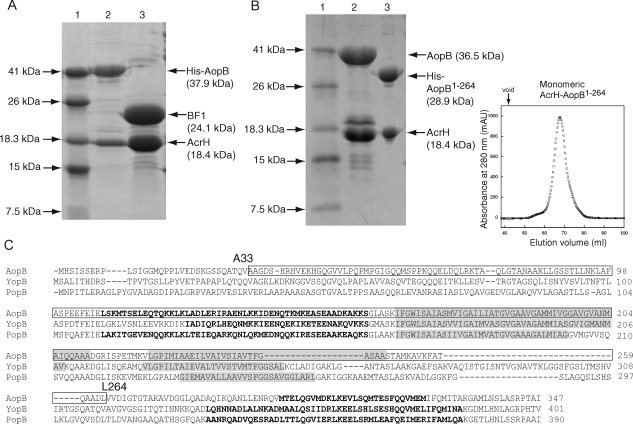
Limited protease digestion of the AcrH-AopB complex. A: SDS-PAGE showing partial digestion of AcrH-AopB with elastase. Lane (1) Mw marker; lane (2) monomeric AcrH-AopB from gel filtration before digestion; and lane (3) AopB is being digested into a single fragment, BF1 (residues 33–264), whereas AcrH remains intact. B: Coexpression and purification of AcrH-AopB1–264 complex. Lane (1) Mw marker; lane (2) AcrH-AopB complex; lane (3) AcrH-AopB1–264 complex. The gel filtration elution profile shows that the AcrH-AopB1–264 complex exists entirely as a monomeric form. The down arrow above the elution profile indicates the void volume of the column. C: Sequence alignment of AopB from Aeromonoas hydrophila (AAR26341) with YopB from Yersinia enterolitica (AAK69211) and PopB from Pseudomonas aeruginosa (AAG05097). Predicted transmembrane domains (TMHMM Server v. 2.0, http://www.cbs.dtu.dk/services/TMHMM-2.0/) are highlighted in gray. Residues that are determined to be involved in chaperone binding are boxed. Residues from the predicted coiled coil regions by the program COILS33 are boldfaced.
Subsequently, we coexpressed AcrH with this BF1 fragment. The BF1 fragment failed to express even in the presence of AcrH (data not shown), suggesting that either N-terminal or C-terminal residues were essential for folding of AopB although they were dispensable for formation of the AcrH-AopB complex. Further, we tried to combine the BF1 and the N-terminus residues and found that this fragment containing residues 1–264 of AopB could be coexpressed with AcrH to form a soluble complex that could be copurified on a Ni-NTA column [Fig. 3(B)]. However, 32 N-terminal residues of this AcrH-AopB1–264 complex could be digested away with protease leaving the BF1 fragment intact (data not shown) indicating that they still remained exposed and the BF1 fragment represented a functional chaperone binding region of AopB.
In addition to mapping the chaperone binding region of AopB, surprisingly, we also found that this AcrH-AopB1–264 complex existed entirely as a monomeric form as determined from the gel filtration profile [Fig. 3(B)]. This result suggested that although the 83 C-terminal residues of AopB, which contained a putative coiled coil domain, were not involved in chaperone binding, they may have a role in the formation of the oligomeric form of the AcrH-AopB complex.
AopB and AopD interact with AcrH to form an oligomeric AcrH-AopB-AopD complex
LcrH, the homolog of AcrH, contains three tandem TPRs. A site-directed mutation study suggested separate surfaces on the protein for binding YopB and YopD.27 Based on the sequence alignment with LcrH, AcrH is also comprised of three tandem TPRs. To determine whether AcrH can form a complex with both AopB and AopD, we coexpressed three proteins concomitantly from an acrHaopBaopD tricistronic system with the His-tag on AopD. AcrH could be coexpressed with both AopB and AopD to form a soluble complex that could be copurified on a Ni-NTA column [Fig. 4(A)]. However, the AcrH-AopB-AopD complex was excluded from the gel filtration column indicating that it mostly existed as an oligomeric form [Fig. 4(A)]. So far, no monomeric species of the AcrH-AopB-AopD complex can be obtained.
Figure 4.
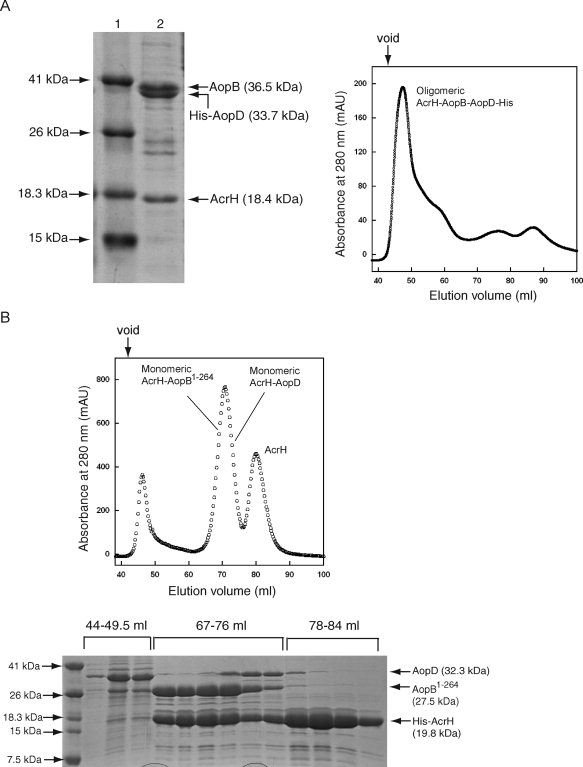
Purification of the AcrH-AopB-AopD and the AcrH-AopB1–264-AopD complexes. A: Purification of the AcrH-AopB-AopD complex. Lane (1) Mw marker and lane (2) purified AcrH-AopB-AopD complex from Ni-NTA column with His-tag placed on AopD. The gel filtration elution profile shows that the AcrH-AopB-AopD complex exists as an oligomeric form. B: Purification of the AcrH-AopB1–264-AopD complex from Ni-NTA column with His-tag placed on AcrH. The gel filtration elution profile shows that a mixture of monomeric AcrH-AopB1–264 and AcrH-AopD complexes are obtained from 67 to 76 mL. The down arrow above the elution profiles indicates the void volume of the column.
As the removal of the 83 C-terminal residues of AopB was found to disrupt the oligomeric state of the AcrH-AopB complex, we attempted to coexpress AcrH, AopB1–264, and AopD together with His-tag on AcrH. Majority of the complexes formed were found to elute from the gel filtration column as a mixture of monomeric AcrH-AopB1–264 and AcrH-AopD complexes [Fig. 4(B)] suggesting that 83 C-terminal residues of AopB were not only responsible for oligomerization of the AcrH-AopB complex but also for oligomerization of the AcrH-AopB-AopD complex.
The oligomeric AcrH-AopB-AopD complex is metastable
The stability of a chaperone-substrate complex determines how easily a substrate can be dissociated from its chaperone. The metastable state of a protein has lower stability and less interaction between amino acid residues. To compare relative stabilities of AcrH and its various complexes, we performed thermal denaturation studies on these complexes using Far-UV circular dichroism (CD) and the data was curve fitted using a two-state equation that also accounts for the drift of CD signal at the base line regions.34 The dimeric AcrH was soluble and stable by itself with a Tm of 51.92 ± 0.02°C (Fig. 5), which was comparable with that observed at 48°C for PcrH.29 The thermostability of the AcrH-AopD complex (Tm of 57.76 ± 0.05°C) was higher than that of AcrH alone (Fig. 5). This stability is comparable with that of the 1:1 PcrH-PopD complex at neutral pH (Tm of 55°C), but is much higher than that of the oligomeric and metastable PopD at acidic pH (Tm of 42°C).29
Figure 5.
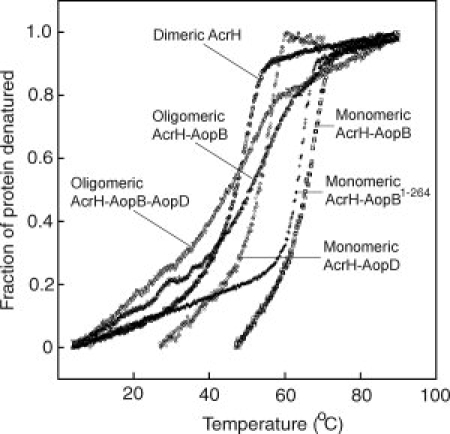
Thermal denaturation of AcrH and various complexes as monitored by FarUV-CD at 222 nm. The fraction of protein denatured is plotted against temperature (°C). Legends for different proteins are: dimeric AcrH (open circle); monomeric AcrH-AopB (open square); monomeric AcrH-AopD (open rhombus); oligomeric AcrH-AopB-AopD (open inverted triangle); monomeric AcrH-AopB1–264 (cross); and oligomeric AcrH-AopB (open triangle).
The heterodimeric AcrH-AopB complex isolated showed exceptionally high-thermal stability of Tm = 70.40 ± 0.04°C (Fig. 5), which was 18.5 and 12.6°C higher than that of AcrH and AcrH-AopD, respectively, indicating that either a much larger portion of AopB was structurally folded, or that it exhibited more extensive interactions with AcrH in the complex. Both explanations agreed with our observation that a large and discrete fragment of AopB remained intact in the AcrH-AopB complex after a limited proteolysis. The thermal stability of the monomeric AcrH-AopB1–264 complex, with a Tm of 66.44 ± 0.01°C, was comparable with that of the monomeric AcrH-AopB complex. Interestingly, the stability of the oligomeric form of AcrH-AopB complex was very low and the accurate Tm could not be obtained due to poor curve fitting (Fig. 5). The low stability of the oligomeric AcrH-AopB complex agreed with stability findings for the metastable and oligomeric PcrH-PopB at acidic (Tm of 32°C) or neutral pH (Tm of 40°C).29 Although the accurate Tm for the oligomeric AcrH-AopB-AopD complex could not be obtained also due to a very gentle slope of transition, we found that the oligomeric AcrH-AopB-AopD complex was least stable, with its Tm of around 5°C lower than that of the oligomeric AcrH-AopB complex (Fig. 5).
Discussion
The role of the AcrH chaperone is to keep translocators AopB and AopD in soluble and stable forms in the cytoplasm, likely by burial of their hydrophobic transmembrane helices. But at the same time, the chaperone also should keep translocators in conformations that are suitable for secretion through the conduit of the needle complex and insertion into the host membrane. Coexpression experiments showed that AcrH formed a stable and soluble 1:1 complex with either AopB or AopD. These findings correspond to those previously observed in Yersinia sp. that YopB or YopD alone could not be expressed as monomeric and stable protein.13,22 YopD and YopB, however, could be stabilized on binding to the chaperone LcrH/SycD in Yersinia pseudotuberculosis.13,23 It has also been reported that the translocator has a molten globule conformation both in its free and chaperone bound forms.35 To speculate that exposed regions may be present in the complexes formed between AcrH and AopB or AopD, we carried out limited protease digestions on these complexes. Intact chaperone binding regions mapped this way represent actual functional binding domains and are different from arbitrary boundaries determined by assessing loss of chaperone binding by use of deletion mutants. The deletion mutation method cannot give proper information about regions that are exposed and not involved in chaperone binding.13 It is also possible that deletion of a large fragment of a protein could lead to protein unfolding, thus preventing it from binding to the chaperone properly.
Using limited proteolysis, we discovered that both AcrH-AopB and AcrH-AopD complexes had extensive exposed regions that were not involved in chaperone binding. Functional and intact chaperone binding regions of AopD were mapped to two separate regions, DF1 (residues 16–147) and DF2 (residues 242–296). These findings agree with mapped regions of YopD binding to the chaperone LcrH32,36 but with slight differences. Residues 53–149 (transmembrane domain) and 278–292 (amphipathic domain) of YopD were found to be important for LcrH attachment.32,37 In PopD, the putative transmembrane fragment was buried within the PcrH-PopD complex.35 However, unlike in the cases of AopD and YopD, the C-terminal amphipathic helix of PopD was not bound to PcrH and was susceptible to trypsin digestion. The region (residues 20–49) at the N-terminus of DF1 is aligned to a region in IpaC (residues 50–80), a homolog of AopD from Shigella flexneri, which had been reported to be critical for its invasion function and also essential for chaperone binding.38 The region (residues 246–266) at the N-terminus of DF2 corresponded to a putative amphipathic coiled coil domain predicted in YopD showing a similar pattern of hydrophobic residues [Fig. 2(B)]. This coiled coil domain, however, has not been located in AopD and PopD and the biological significance remains to be determined.
The BF1 fragment of AopB that bound to the chaperone was a single large fragment comprising a putative N-terminal coiled coil domain33 and two predicted transmembrane domains39 but not the putative C-terminal coiled coil domain [Fig. 3(C)]. Transmembrane helices are highly hydrophobic and burial of this region through chaperone binding or protein folding can highly increase its stability and avoid their nonspecific interactions. As the size of the chaperone binding region of AopB is relatively large, it is reasonable to expect that multiple regions within this single fragment of BF1 may be involved directly in chaperone binding. Our results agree with previous findings that SycD/LcrH did not bind to a unique clearly defined segment of YopB, but rather bind at different sites of the protein.13 On the basis of our findings, we can conclude that these sites are located within the BF1 fragment and not at the N- or C-terminal regions of the protein. It should also be noted that the first 32 N-terminal residues of AopB, was not identified as a chaperone binding region by proteolytic assay but found to be essential for coexpression of the AcrH-AopB complex.
It had been determined that PopB and PopD can only be inserted into a host membrane in an oligomeric and metastable form.29 Studies on the T3SS of P. aeruginosa had also shown that PcrH formed a heterodimeric complex with PopD and a hetero-oligomeric (and/or possibly heterodimeric) complex with PopB on coexpression in the bacterial cytoplasm.29 In our study, we found that the AcrH-AopD complex existed as a monomer, whereas the AcrH-AopB complex was found mainly as an oligomeric form with a small portion as the monomeric form. The AcrH-AopB-AopD complex exists entirely as an oligomeric form when these three proteins are coexpressed. The monomeric AcrH-AopB and AcrH-AopD complexes are relatively stable and dissociation of such complexes inside the cell is unlikely. Indeed, the oligomeric AcrH-AopB and AcrH-AopB-AopD complexes have significantly lower stabilities. This suggested that extensive conformational changes or dissociation must have taken place during oligomerization of these complexes. It has been shown that InvC, the ATPase of the Salmonella typhi T3SS, is responsible to recognize, disassemble, and unfold protein substrate of the SicP-SptP chaperone-effector complex. The dissociation and unfolding process require hydrolysis of ATP and are essential for recycling of the chaperone and secretion through the narrow conduit of the needle complex.40 The importance of a metastable substrate for T3SS secretion was shown by the experiment that only a mutant of DHFR with a folding defect, but not the wild type DHFR, could be secreted when fused to the secretion signal of YopE.41 It is the least stable complex that allows effective dissociation of translocators from the chaperone and unfolding of the substrate before secretion.
During the mapping of the chaperone binding region of AopB, we found that oligomerization of the AcrH-AopB complex was mainly mediated through the exposed C-terminal domain (residues 265–347) of AopB and that truncation of this domain led to the formation of an entirely monomeric AcrH-AopB complex. The ability of AopB to oligomerize is also essential for the formation of the AcrH-AopB-AopD complex as coexpression of AopB1–264 with both AcrH and AopD resulted in a mixture of monomeric AcrH-AopB and AcrH-AopD instead of the oligomeric AcrH-AopB-AopD complex. In addition, oligomerization of the AcrH-AopB-AopD complex likely to occur only in vivo, as in vitro mixing of monomeric or oligomeric AcrH-AopB with AcrH-AopD did not form the AcrH-AopB-AopD complex (data not shown). To investigate the importance of oligomerization of the AcrH-AopB-AopD complex on the secretion of translocators, an in-frame deletion mutant ΔaopNB was constructed by deleting aopB from a negative regulator mutant (ΔaopN) of A. hydrophila which is competent for T3SS secretion.7 Our result showed that AopD could be secreted from this ΔaopN mutant of A. hydrophila even in the absence of AopB, suggesting that secretion of AopD is independent of AopB and oligomerization of the AcrH-AopB-AopD complex (Supporting Information Figure 1). Rather than affecting secretion of the translocators, we speculate that oligomerization of the metastable AcrH-AopB-AopD complex may be important for proper translocation and formation of translocon on the host cell with the correct topology and stoichiometry. The exact role of oligomerization of the AcrH-AopB-AopD complex remains to be elucidated.
Materials and Methods
Cloning of AcrH, AcrH-AopB, AcrH-AopD, and AcrH-AopB-AopD
AcrH, AopB, or AopD DNA constructs were subcloned separately into a modified pET-32a vector (with thioredoxin and S-tag removed) using restriction enzymes BamH I and EcoR I for expression of these three proteins as their respective His-tag fusion proteins. For coexpression of AcrH-AopB or AcrH-AopD complexes, full length AcrH was subcloned into the first multiple-cloning site of the pETDuet-1 coexpression vector (Novagen) using restriction enzymes BamH I and EcoR I. A full-length AopD was subsequently subcloned at the second multiple-cloning site of the same vector using Nde I and Xho III allowing coexpression of both proteins. The (His)6 tag was encoded upstream of AcrH. A similar subcloning was done for the full-length AopB and the C-terminal truncated mutant AopB1–264 at the first multiple-cloning site of the same vector by using restriction enzymes EcoR I and Hind III. While AcrH was subcloned into the second multiple-cloning site by using restriction enzymes Nde I and Bgl II. The His-tag was attached at the N-termini of AopB constructs, allowing the pull-down of AcrH when an AcrH-AopB interaction occurred. Sequences of constructs were verified by big dye DNA sequencing using 3100 Genetic Analyzer automated DNA sequencer (Applied Biosystems). For coexpression of the AcrH-AopB-AopD complex, genes encoding the AcrHAopBD part of the acrGVHaopBD operon from the genomic DNA of A. hydrophila were amplified by standard PCR procedures and subcloned between the Xba I and Hind III restriction sites of a modified pET-32a vector (Novagen) which had its S-Tag and the thioredoxin tag removed. The His-tag was attached at the C-terminus of AopD. As for the expression of AcrH-AopB1–264-AopD, the construct was generated by the PCR-based overlap extension method to remove residues 264–347 of AopB on the acrGVHaopBD operon before acrHaopB1–264D was amplified for cloning. The His-tag was attached at the N-terminus of AcrH.
Expression and purification of AcrH, AcrH-AopB, AcrH-AopD, and AcrH-AopB-AopD
A single ampicillin-resistant colony of Escherichia coli BL21 (DE3) cells transformed with the suitable plasmid was used to inoculate 50 mL of Luria broth supplemented with 100 μg/mL of ampicillin. The culture was grown overnight at 37°C with shaking at a speed of 200 rpm. Ten milliliter of the overnight culture was inoculated to 1 L of fresh Luria Broth and the cells were grown to early log phase (OD600 = 0.6). IPTG was added to a final concentration of 0.3 mM to induce expression and cells were grown at 25°C for additional 6 h. Cells were harvested by centrifugation at 6891g for 15 min. The cell pellet obtained from 1 L of culture was resuspended in 30 mL of Ni-binding buffer (20 mM Tris-HCl pH 7.5, 200 mM NaCl, 5% glycerol, 10 mM β-mercaptoethanol, and 5 mM imidazole) along with one tablet of Complete EDTA-free cocktail protease-inhibitor (Roche). Cells were lyzed by sonication and cell debris were removed by centrifugation at 26,581g for 30 min. The supernatant was purified using Ni-NTA beads (Qiagen). Ni-binding buffer with 0.4M of imidazole were used to elute proteins from the beads. They were further purified on a prepacked HiLoad 16/60 Superdex 200 prep grade (GE Healthcare) gel filtration chromatography column on the AKTA fast protein liquid chromatography system (GE Healthcare) using gel filtration buffer (20 mM Tris-Cl, pH 7.5, 200 mM NaCl, 5% glycerol, and 10 mM β-mercaptoethanol). The eluted protein was then stored at −80°C for subsequent analysis.
Limited protease digestion
All proteases used for limited digestion studies were from Sigma. Small-scale digestions using different amounts of proteases for different periods of time were performed before scaling up the reaction. Limited protease digestion was performed on the monomeric AcrH-AopB and AcrH-AopD complexes in gel filtration buffer (20 mM Tris-Cl, pH 7.5, 200 mM NaCl, 5% glycerol, and 10 mM β-mercaptoethanol). The purified AcrH-AopB complex was concentrated to 30 mg/mL and subsequently incubated with elastase (E7885, from porcine pancreas) at a molar ratio of 1:200 at 25°C for 5 min before loading onto the HiLoad 16/60 Superdex 75 prep grade (GE Healthcare) gel filtration chromatography column to remove the protease and obtain the digested complex. The purified AcrH-AopD complex was incubated with chymotrypsin (C4129, Type II α-chymotrypsin from bovine pancreas) at a molar ratio of 1:100 at 25°C for 5 h. The reaction was terminated by passing digested proteins through a gel filtration chromatography column as described for AcrH-AopB.
Mass spectrometry and N-terminal sequencing
Protease digested AcrHB and AcrHD complexes containing intact domains still bound to AcrH were collected after gel filtration chromatography. These samples were spotted on a MALDI target plate, with sinapinic acid as a matrix, and the molecular weight of each component in the complex was determined using Voyager-DE STR mass spectrometer (Applied Biosystems) with a voltage of 2100 V and laser intensity of 2500. As for the N-terminal sequencing analysis, protease digested samples were separated on a 15% SDS-PAGE gel and then blotted onto a PVDF membrane using a Mini Trans-Blot Electrophoretic Transfer Cell (Bio-Rad). Transblotting was performed at 4°C with a voltage of 100 V for 1 h. Bands were visualized with Coomassie staining and membrane-bound proteins were excised and analyzed on an Applied Biosystems 494A Procise protein sequencer.
Chemical crosslinking
Crosslinking was performed using glutaraldehyde purchased from Sigma. 0.1% glutaraldehyde was added to 100 μL of AcrH, AcrH-AopD, or monomeric AcrH-AopB in PBS with a protein concentration of 1 mg/mL. The reaction was performed at 4°C for a period of 30 min. Ten microliter of the reaction mixture was sampled every 5 min to monitor progress of the crosslinking process. Equal volume of SDS-PAGE sample loading buffer containing 5% β-mercaptoethanol was added to the 10 μL of reaction mixture and sample was boiled for 5 min at 95°C before loading on a 15% SDS-PAGE gel for electrophoresis.
Circular dichroism
All protein samples used for heat denaturation studies were dialyzed in PBS buffer and concentrated to 20 μM each. Thermodynamic stability experiments were conducted on a Jasco J-810 spectropolarimeter using a wavelength of 222 nm in a 1 mm path-length quartz cuvette (Hellma). The CD signal was recorded for a range of temperature from 4 to 90°C with a scan rate of 1°C/min and readings were recorded every 0.1°C. The measured ellipticity was converted to the fraction of protein denatured before curve fitting to obtain the Tm.
Acknowledgments
K.Y.L., J.S., and Y.-K.M. are members of the Structural Biology and Proteomics Research group of the Office of Life Sciences, NUS. The authors thank Mr. Shashikant Joshi for proof reading the manuscript.
References
- 1.Thune RL, Stanley LA, Cooper K. Pathogenesis of gram-negative bacterial infections in warmwater fish. Annu Rev Fish Dis. 1993;3:37–68. [Google Scholar]
- 2.Austin B, Adams C. Fish pathogens. In: Austin B, Altwegg M, Gosling PJ, Joseph SW, editors. The genus Aeromonas. New York: Wiley; 1996. pp. 197–229. [Google Scholar]
- 3.Janda JM. Aeromonas and Plesiomonas. In: Sussman M, editor. Molecular medical microbiology. San Diego: Academic Press; 2001. pp. 1237–1270. [Google Scholar]
- 4.Yu HB, Srinivasa Rao PS, Lee HC, Vilches S, Merino S, Tomas JM, Leung KY. A type III secretion system is required for Aeromonas hydrophila AH-1 pathogenesis. Infect Immun. 2004;72:1248–1256. doi: 10.1128/IAI.72.3.1248-1256.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marenne MN, Journet L, Mota LJ, Cornelis GR. Genetic analysis of the formation of the Ysc-Yop translocation pore in macrophages by Yersinia enterocolitica: role of LcrV, YscF and YopN. Microb Pathog. 2003;35:243–258. doi: 10.1016/s0882-4010(03)00154-2. [DOI] [PubMed] [Google Scholar]
- 6.Zaharik ML, Gruenheid S, Perrin AJ, Finlay BB. Delivery of dangerous goods: type III secretion in enteric pathogens. Int J Med Microbiol. 2002;291:593–603. doi: 10.1078/1438-4221-00179. [DOI] [PubMed] [Google Scholar]
- 7.Yu HB, Kaur R, Lim S, Wang XH, Leung KY. Characterization of extracellular proteins produced by Aeromonas hydrophila AH-1. Proteomics. 2007;7:436–449. doi: 10.1002/pmic.200600396. [DOI] [PubMed] [Google Scholar]
- 8.Vilches S, Wilhelms M, Yu HB, Leung KY, Tomás JM, Merino S. Aeromonas hydrophila AH-3 AexT is an ADP-ribosylating toxin secreted through the type III secretion system. Microb Pathog. 2008;44:1–12. doi: 10.1016/j.micpath.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 9.Sha J, Wang SF, Suarez G, Sierra JC, Fadl AA, Erova TE, Foltz SM, Khajanchi BK, Silver AC, Graf J, Schein CH, Chopra AK. Further characterization of a type III secretion system (T3SS) and of a new effector protein from a clinical isolate of Aeromonas hydrophila—Part I. Microb Pathog. 2007;43:127–146. doi: 10.1016/j.micpath.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 10.Neyt C, Cornelis GR. Insertion of a Yop translocation pore into the macrophage plasma membrane by Yersinia enterocolitica: requirement for translocators YopB and YopD, but not LcrG. Mol Microbiol. 1999;33:971–981. doi: 10.1046/j.1365-2958.1999.01537.x. [DOI] [PubMed] [Google Scholar]
- 11.Williams AW, Straley SC. YopD of Yersinia pestis plays a role in negative regulation of the low-calcium response in addition to its role in translocation of Yops. J Bacteriol. 1998;180:350–358. doi: 10.1128/jb.180.2.350-358.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson DM, Ramamurthi KS, Tam C, Schneewind O. YopD and LcrH regulate expression of Yersinia enterocolitica YopQ by a posttranscriptional mechanism and bind to yopQ RNA. J Bacteriol. 2002;184:1287–1295. doi: 10.1128/JB.184.5.1287-1295.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neyt C, Cornelis GR. Role of SycD, the chaperone of the Yersinia Yop translocators YopB and YopD. Mol Microbiol. 1999;31:143–156. doi: 10.1046/j.1365-2958.1999.01154.x. [DOI] [PubMed] [Google Scholar]
- 14.Ryndak MB, Chung H, London E, Bliska JB. Role of predicted transmembrane domains for type III translocation, pore formation, and signaling by the Yersinia pseudotuberculosis YopB protein. Infect Immun. 2005;73:2433–2443. doi: 10.1128/IAI.73.4.2433-2443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parsot C, Hamiaux C, Page A-L. The various and varying roles of specific chaperones in type III secretion systems. Curr Opin Microbiol. 2003;6:7–14. doi: 10.1016/s1369-5274(02)00002-4. [DOI] [PubMed] [Google Scholar]
- 16.Feldman MF, Cornelis GR. The multitalented type III chaperones: all you can do with 15 kDa. FEMS Microbiol Lett. 2003;219:151–158. doi: 10.1016/S0378-1097(03)00042-9. [DOI] [PubMed] [Google Scholar]
- 17.Birtalan S, Ghosh P. Structure of the Yersinia type III secretory system chaperone SycE. Nat Struct Mol Biol. 2001;8:974–978. doi: 10.1038/nsb1101-974. [DOI] [PubMed] [Google Scholar]
- 18.Evdokimov AG, Tropea JE, Routzahn KM, Waugh DS. Three-dimensional structure of the type III secretion chaperone SycE from Yersinia pestis. Acta Crystallogr D. 2002;58:398–406. doi: 10.1107/s090744490200015x. [DOI] [PubMed] [Google Scholar]
- 19.Birtalan SC, Phillips RM, Ghosh P. Three-dimensional secretion signals in chaperone-effector complexes of bacterial pathogens. Mol Cell. 2002;9:971–980. doi: 10.1016/s1097-2765(02)00529-4. [DOI] [PubMed] [Google Scholar]
- 20.Phan J, Tropea JE, Waugh DS. Structure of the Yersinia pestis type III secretion chaperone SycH in complex with a stable fragment of YscM2. Acta Crystallogr D. 2004;60:1591–1599. doi: 10.1107/S0907444904017597. [DOI] [PubMed] [Google Scholar]
- 21.Stebbins CE, Galán JE. Priming virulence factors for delivery into the host. Nat Rev Mol Cell Biol. 2003;4:738–743. doi: 10.1038/nrm1201. [DOI] [PubMed] [Google Scholar]
- 22.Wattiau P, Bernier B, Deslée P, Michiels T, Cornelis GR. Individual chaperones required for Yop secretion by Yersinia. Proc Natl Acad Sci USA. 1994;91:10493–10497. doi: 10.1073/pnas.91.22.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Francis MS, Lloyd SA, Wolf-Watz H. The type III scretion chaperone LcrH co-operates with YopD to establish a negative, regulatory loop for control of Yop synthesis in Yersinia pseudotuberculosis. Mol Microbiol. 2001;42:1075–1093. doi: 10.1046/j.1365-2958.2001.02702.x. [DOI] [PubMed] [Google Scholar]
- 24.Büttner CR, Sorg I, Cornelis GR, Heinz DW, Niemann HH. Structure of the Yersinia enterocolitica type III secretion translocator chaperone SycD. J Mol Biol. 2008;375:997–1012. doi: 10.1016/j.jmb.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 25.Pallen MJ, Francis MS, Fütterer K. Tetratricopeptide-like repeats in type-III-secretion chaperones and regulators. FEMS Microbiol Lett. 2003;223:53–60. doi: 10.1016/S0378-1097(03)00344-6. [DOI] [PubMed] [Google Scholar]
- 26.D'Andrea LD, Regan L. TPR proteins: the versatile helix. Trends Biochem Sci. 2003;28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 27.Edqvist PJ, Bröms JE, Betts HJ, Forsberg Å, Pallen MJ, Francis MS. Tetratricopeptide repeats in the type III secretion chaperone, LcrH: their role in substrate binding and secretion. Mol Microbiol. 2006;59:31–44. doi: 10.1111/j.1365-2958.2005.04923.x. [DOI] [PubMed] [Google Scholar]
- 28.Allmond LR, Karaca TJ, Nguyen VN, Nguyen T, Wiener-Kronish JP, Sawa T. Protein binding between PcrG-PcrV and PcrH-PopB/PopD encoded by the pcrGVH-popBD operon of the Pseudomonas aeruginosa type III secretion system. Infect Immun. 2003;71:2230–2233. doi: 10.1128/IAI.71.4.2230-2233.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schoehn G, Di Guilmi AM, Lemaire D, Attree I, Weissenhorn W, Dessen A. Oligomerization of type III secretion proteins PopB and PopD precedes pore formation in Pseudomonas. EMBO J. 2003;22:4957–4967. doi: 10.1093/emboj/cdg499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan YW, Yu HB, Leung KY, Sivaraman J, Mok Y-K. Structure of AscE and induced burial regions in AscE and AscG upon formation of the chaperone needle-subunit complex of type III secretion system in Aeromonas hydrophila. Protein Sci. 2008;17:1748–1760. doi: 10.1110/ps.036798.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmid A, Dittmann S, Grimminger V, Walter S, Heesemann J, Wilharm G. Yersinia enterocolitica type III secretion chaperone SycD: recombinant expression, purification and characterization of a homodimer. Protein Expr Purif. 2006;49:176–182. doi: 10.1016/j.pep.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 32.Francis MS, Aili M, Wiklund M-L, Wolf-Watz H. A study of the YopD-LcrH interaction from Yersinia pseudotuberculosis reveals a role for hydrophobic residues within the amphipathic domain of YopD. Mol Microbiol. 2000;38:85–102. doi: 10.1046/j.1365-2958.2000.02112.x. [DOI] [PubMed] [Google Scholar]
- 33.Lupas A, van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 34.Ruiz-Sanz J, de Prat Gay G, Otzen DE, Fersht AR. Protein fragments as models for events in protein folding pathways: protein engineering analysis of the association of two complementary fragments of the barley chymotrypsin inhibitor 2 (CI-2) Biochemistry. 1995;34:1695–1701. doi: 10.1021/bi00005a026. [DOI] [PubMed] [Google Scholar]
- 35.Faudry E, Job V, Dessen A, Attree I, Forge V. Type III secretion system translocator has a molten globule conformation both in its free and chaperone-bound forms. FEBS J. 2007;274:3601–3610. doi: 10.1111/j.1742-4658.2007.05893.x. [DOI] [PubMed] [Google Scholar]
- 36.Olsson J, Edqvist PJ, Bröms JE, Forsberg Å, Wolf-Watz H, Francis MS. The YpoD translocator of Yersinia pseudotuberculosis is a multifunctional protein comprised of discrete domains. J Bacteriol. 2004;186:4110–4123. doi: 10.1128/JB.186.13.4110-4123.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tengel T, Sethson I, Francis MS. Conformational analysis by CD and NMR spectroscopy of a peptide encompassing the amphipathic domain of YopD from Yersinia. Eur J Biochem. 2002;269:3659–3668. doi: 10.1046/j.1432-1033.2002.03051.x. [DOI] [PubMed] [Google Scholar]
- 38.Harrington AT, Hearn PD, Picking WL, Barker JR, Wessel A, Picking WD. Structural characterization of the N terminus of IpaC from Shigella flexneri. Infect Immun. 2003;71:1255–1264. doi: 10.1128/IAI.71.3.1255-1264.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 40.Akeda Y, Galán JE. Chaperone release and unfolding of substrates in type III secretion. Nature. 2005;437:911–915. doi: 10.1038/nature03992. [DOI] [PubMed] [Google Scholar]
- 41.Feldman MF, Müller S, Wüest E, Cornelis GR. SycE allows secretion of YopE-DHFR hybrids by the Yersinia enterocolitica type III Ysc system. Mol Microbiol. 2002;46:1183–1197. doi: 10.1046/j.1365-2958.2002.03241.x. [DOI] [PubMed] [Google Scholar]


