Abstract
Following fusion of the human immunodeficiency virus type-1 (HIV-1) with host cells' membrane and reverse transcription of the viral RNA, the resulted cDNA is integrated into the host genome by the viral integrase enzyme (IN). Quantitative estimations have revealed that only 1–2 copies are integrated per infected cell, although many copies of the viral RNA are reverse-transcribed. The molecular mechanism that restricts the integration degree has not, so far, been elucidated. Following integration, expressed partially spliced and unspliced transcripts are exported from the nuclei by the viral Rev protein. Here, we show that in virally infected cells, the Rev interacts with the IN forming a Rev–IN complex and consequently limits the number of integration events. Disruption of the Rev–IN complex by selected IN-derived peptides or infection by a Rev-deficient virus stimulate integration resulting in large numbers of integration event/cell. Conversely, infection of Rev-expression cells blocks integration and inhibits virus production. Increased integration appears to correlate with increased cell death of infected cultures. Our results thus demonstrate a new regulatory function of Rev and probably establish a link between Rev restriction of HIV-1 integration and protection of HIV-1-infected cells from premature cell death.
Keywords: cell death, HIV-1, integrase, integration regulation, Rev
Introduction
The Rev of the human immunodeficiency virus type 1 (HIV-1) is an essential virus-encoded regulatory protein that is required for nuclear export of unspliced and single-spliced viral RNA during HIV replication (Pollard and Malim, 1998; Freed and Martin, 2001; Wolff et al., 2006). HIV replication requires the integration of a DNA copy of the viral RNA by the viral integrase protein (IN) (Freed, 2001). The IN itself is transferred into the host cells as part of the invading virus particles and eventually becomes associated with the pre-integration complex (PIC) (Freed, 2001). Following reverse transcription of the viral RNA, integration of the cDNA is carried out by two successive steps (Engelman et al., 1991). These consist of a 3′ processing step (Guiot et al., 2006) followed by the strand transfer step during which the viral genome is inserted into the host DNA double strand (Chen et al., 2006). In addition to the IN, the successful integration of the HIV DNA also requires the cellular LEDGF/p75 protein that has been suggested to tether the IN–DNA complex to the host chromatin (Maertens et al., 2003).
Quantitative estimation revealed that on the average in each infected virion ∼40–100 of viral IN molecules can be detected (Pommier et al., 2005). Also up to ∼99% of reverse-transcribed cDNA molecules remain as unintegrated within the infected cells (Chun et al., 1997). In spite of the presence of these relatively large number of free DNA molecule, only 1–2 copies are found integrated within the host genome (Butler et al., 2001). This certainly indicates a regulatory process which restricts the number of integration events in infected cells.
Recently (Rosenbluh et al., 2007), we demonstrated that IN and Rev can interact and form a stable Rev–IN complex in yeast cells. In vitro experiments showed that interaction of Rev with the IN results in inhibition of the IN enzymatic activity (Levin et al., 2009). The Rev inhibitory effect could be abrogated by specific IN derived peptides which have been shown to disrupt the Rev–IN complex (Levin et al., 2009).
Here, we demonstrate, to the best of our knowledge for the first time, an experimental evidence for the existence of Rev–IN complexes within HIV-infected cells. Our present results strongly established a direct correlation between the formation of a Rev–IN complex and inhibition of integration. Indeed, over-expression of Rev prevented integration of HIV DNA in infected cells, whereas viruses that fail to express Rev show increased numbers of integrated viral DNA copies. Multiple integration of viral cDNA led to excessive cell death. In summary, this work describes a novel and important regulatory mechanism for restriction of HIV-1 integration.
Materials and methods
Cells
Monolayer adherent HeLaP4, HEK293T and HEK293T cells over-expressing Rev (Rev10+ cells) and HeLa multinuclear activation of a galactosidase indicator (MAGI) cells (TZM-bl) (Kimpton and Emerman, 1992; Derdeyn et al., 2000) were grown in Dulbecco's modified eagle's medium (DMEM). The T-lymphocyte cell lines Sup-T1 and H9 were grown in the RPMI 1640 medium. Cells beside the Rev10+ cells were provided by the NIH Reagent, Program, Division of AIDS, NIAID, NIH, USA. Rev10+ cells were generated by transfection to HEK293T cells (Cullen, 1987). Cells were incubated at 37°C in a 5% CO2 atmosphere. All media were supplemented with 10% (v/v) fetal calf serum, 0.3 g/l l-glutamine, 100 U/ml penicillin and 100 U/ml streptomycin (Biological Industries, Beit Haemek, Israel).
Viruses
Wild-type HIV-1 (HXB2; Ratner et al., 1985) and ΔEnv (Gummuluru et al., 2000) as well as IN mutant D64N D116N (Nakajima et al., 2001) were generated by transfection to HEK293T cells (Cullen, 1987) or the virus containing plasmid or co-transected with a plasmid contain VSV-G (Levin et al., 2009). ΔRev pLAIY47H2 (Verhoef et al., 1997) and Rev M10 (Bahner et al., 1993) HIVs were generated by transfection to Rev10+ cells. Viruses were harvested and store as describe in Levin et al. (2009). The pLAIY47H2 (Verhoef et al., 1997) viruses were generous gift by Prof. Berkhout (Department of Human Retrovirology, Academic Medical Center, University of Amsterdam, The Netherlands), the IN mutant D64N D116N virus (Nakajima et al., 2001) was generous gift of Prof. Engelman (Department of Cancer Immunology and AIDS Dana-Farber Cancer Institute and Division of AIDS, Harvard Medical School, Boston, MA, USA).
Virus stock titration and normalization
Quantitative titration of HIV-1 was carried out using the MAGI assay, as described by Kimpton and Emerman (1992). Briefly, TZM-b1 cells were grown in 96-well plates at 1 × 104 cells per well. The cells were infected with 50 µl of serially diluted virus (wild-type, ΔRev or Rev M10 HIV-1) as described (Kimpton and Emerman, 1992). Two days post-infection (PI), cultured cells were fixed and β-galactosidase was estimated exactly as described previously (Kimpton and Emerman, 1992). Blue cells were counted under a light microscope at ×200 magnification. In the case of IN mutant viruses (D64N D116N), the amount of viral RNA was estimated by real-time reveres transcription PCR as described in Pizzato et al. (2009). This was also performed to the other HIV-1 viruses and was used to normalize the titer of the IN mutant virus. Virus stock concentration was preformed exactly as described in Reiser (2000). Briefly, virus stock was ultra-centrifuged (82500 g (average) at 15° for 105 min) using Beckman SW28 rotor and then virus titer was determined as described above.
Infection of cultured cells
Cultured lymphocytes were infected exactly as described in Levin et al. (2009). Cultured HEK293T, Rev10+ cells and HeLa MAGI cells (TZM-bl) were grown for 24 h before infection then medium was removed and cells were incubated at different multiplicity of infections (MOIs) with the indicated virus for 2 h at 37°C. Cells were washed three times with primer-binding site (PBS) and incubated in the DMEM. Cultured lymphocytes were infected exactly as described in Levin et al. (2009). Briefly, cultured lymphocytes (1 × 105) were centrifuged for 5 min at 2000 rpm, and after removal of the supernatant, the cells were resuspended in 0.2–0.5 ml of the RPMI 1640 medium containing virus at the indicated MOI. Following absorption for 2 h at 37°C, the cells were washed to remove unbound virus and then incubated at the same temperature in the RPMI 1640 medium.
Peptides synthesis and purification
Peptides were synthesized on an Applied Biosystems (ABI) 433A peptide synthesizer and purification was performed on a Gilson HPLC using a reverse-phase C8 semi-preparative column (ACE, Advanced Chromatography Technologies, USA) as described in Kohler et al. (1989) and Levin et al. (2009).
Isolation of cytoplasm, nuclei and PIC from infected cells
Isolation of cytoplasm, nuclei and PIC from infected cells was performed essentially as described in Zhang et al. (2007). Briefly, cells were harvested and wash twice in buffer A (20 mM Hepes pH 7.3, 150 mM KCl, 5 mM MgCl2, 1 mM DTT and 0.1 mM PMSF) and were then suspended in 200 µl of buffer A containing 0.025% digitonin. Following 10 min incubation at room temperature, cells were centrifuged 3 min at 1000g. Supernatant fraction was taken and centrifuged at 8000g and for the supernatant obtained which is the cytoplasm and pellet containing the nuclei were stored at −70°C. For PIC isolation, the cytoplasm fraction was taken and equal volume of buffer B (20 mM Hepes pH 7.4, 5 mM MgCl2, 1 mM DTT and 0.1 mM PMSF) was added. Samples were incubated for 10 min at room temperature and then centrifuge 10 min at 2000g. Supernatant was removed and pellet which is PIC aggregates was stored at −70°C.
Study of in vivo protein–protein interactions using the co-immunoprecipitation
Cells were infected with MOI of 10 of the indicated viruses, harvested at different times PI, washed three times with PBS and lysed by the addition of PBS containing 1% (v/v) Triton X100. Cytoplasm, nuclei and PICs were isolated as described above. Half of the lysate volume or of the isolated fractions was subjected to a SDS–PAGE. The samples were run in an E-PAGE™ 48 8% High-Throughput Pre-Cast Gel System (Invitrogen). Then immunoblotted with either a monoclonal anti-Rev antibody (α-Rev) (Kramer-Hammerle et al., 2005), an antiserum raised against IN amino acids 276–288 (α-IN) (NIH AIDS Research and Reference Reagent Program catalog number 758) or an anti-actin (α-actin) antibody (Santa Cruz). A complement HRP conjugated antibodies (Jackson) was used as a second antibody.
The remaining lysate or isolated fractions were incubated for 1 h at 4°C with either the α-Rev, α-IN or α-Actin antibodies. Following 3 h incubation with protein G-agarose beads (Santa Cruz) at 4°C, the samples were washed three times with PBS containing 1% Nonidet P-40. An SDS buffer was added to the samples and after boiling and running on an SDS–PAGE gel the membranes were immunoblotted with either α-Rev, α-IN or α-actin antibodies, and a complement HRP conjugated antibodies (Jackson) as second antibodies.
When peptides were used, cells were incubated with 150 µM of the indicated peptide for 2 h previous to the infection.
Quantitative analysis of the copy numbers of HIV-1 DNA integrated into cellular genome
The integration reaction and the integration events were performed exactly as described previously (Levin et al., 2009). Briefly, integrated HIV-1 sequences were amplified by two PCR replication steps using the HIV-1 LTR-specific primer (LTR-TAG-F 5′-ATGCCACGTAAGCGAAACTCTGGCTAACTAGGGAACCCACTG-3′) and Alu-targeting primers (first-Alu-F 5′-AGCCTCCCGAGTAGCTGGGA-3′ and first-Alu-R 5′-TTACAGGCATGAGCCACCG-3′) (Yamamoto et al., 2006). Alu-LTR fragments were amplified from 10 ng of total cell DNA in a 25-µl reaction mixture containing 1× PCR buffer, 3.5 mM MgCl2, 200 µM dNTPs, 300 nM primers and 0.025 units/µl of Taq-polymerase. The first-round PCR cycle conditions were as follows: a DNA denaturation and polymerase activation step of 10 min at 95°C and then 12 cycles of amplification (95°C for 15 s, 60°C for 30 s and 72°C for 5 min).
During the second-round PCR, the first-round PCR product could be specifically amplified by using the tag-specific primer (tag-F 5′-ATGCCACGTAAGCGAAACTC-3′) and the LTR primer (LTR-R 5′-AGGCAAGCTTTATTGAGGCTTAAG-3′) designed by PrimerExpress (Applied Biosystems) using default settings. The second-round PCR was performed on 1/25th of the first-round PCR product in a mixture containing 300 nM of each primer, 12.5 µl of 2X SYBR Green master mixture (Applied Biosystems) at a final volume of 25 µl, run on an ABI PRIZM 7700 (Applied Biosystems). The second-round PCR cycles began with DNA denaturation and a polymerase-activation step (95°C for 10 min), followed by 40 cycles of amplification (95°C for 15 s, 60°C for 60 s).
For generation of a standard calibration curve, the SVC21 plasmid containing the full-length HIV-1HXB2 viral DNA was used as a template. In the first-round PCR, the LTR-TAG-F and LTR-R primers were used, and the second-round PCR was performed using the tag-F and LTR-R primers. The standard linear curve was in the range of 5 ng to 0.25 fg (R = 0.99). DNA samples were assayed with quadruplets of each sample. For further experimental details, see Rosenbluh et al. (2007). The cell equivalents in the sample DNA were calculated based on amplification of the 18S gene by real-time PCR as described in Field et al. (2002).
Quantitative total viral DNA assay
Total viral DNA was estimated using SYBR green real-time quantitative PCR 12 h PI, exactly as described in Casabianca et al. (2007). Briefly, DNA samples (1 μg of DNA) were added to 95 μl containing 1× Hot-Rescue Real-Time PCR Kit-SG (Diatheva s.r.l, Fano, Italy), and 100 nM of each PBS primer: F5 (5′ primer, 5′-TAGCAGTGGCGCCCGA -3′) and R5 (3′ primer, 5′- TCTCTCTCCTTCTAGCCTCCGC -3′). All amplification reactions were carried out using an ABI Prism 7700 Sequence Detection System (Applied Biosystems): one cycle at 95°C for 10 min, followed by 45 cycles of 15 s at 95°C and 35 s at 68°C. In each PCR run, three replicates were performed.
Quantitative estimation of HIV-1 infection by determination of extracellular p24
The amount of p24 protein was estimated in the cell medium using an ELISA-based capture assay kit (SAIC, AIDS Vaccine Program, Frederick, MD, USA), according to the manufacturer's instructions and as described previously (Rosenbluh et al., 2007).
HIV-1 titration by MAGI assay
Quantitative titration of HIV-1 was carried out using the MAGI assay, as described by Kimpton and Emerman (1992). Briefly, TZMb1 cells were grown in 96-well plates at 1 × 104 cells/well and following 12 h incubation at 37°C, several dilutions of virus obtained from infected lymphocytes as described in Levin et al. (2009). Two days PI, cultured cells were fixed and β-galactosidase was estimated exactly as described previously (Levin et al., 2009).
Estimation of cell viability using the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay
H9 lymphocytes were either infected with the indicated viruses or pretreated for 2 h with the indicated peptides before infection and then medium was removed at different times PI, and the cell viability was determined in the MTT test as described in Levin et al. (2009).
Estimation of the effect of inhibitors on appearance of Rev
When inhibitors were used, they were added 1 h before infection with WT HIV-1 at MOI 10. The final concentration of each inhibitor was: AZT 2 µM; actinomycin D 5 µg/ml and cycloheximide 50 µg/ml. At 6 h PI, samples were lysed and blotted as described above.
Immunostaining
HeLaP4 cells were grown on chamber slides (Nunc), then infected with WT or ΔRev HIV-1 at an MOI of 25. Cells were fixed 6 h PI exactly as described previously (Levin et al., 2006) and immunostained essentially as described previously (Levin et al., 2006) with some modifications. Briefly, after fixation, cells were blocked with 5% IgG-free BSA (Jackson) in PBS for 60 min. For detection of HIV-1 Rev, the cells were incubated with 1:50 rat α-Rev (Kramer-Hammerle et al., 2005) at room temperature for 60 min. Cells were washed five times with PBS + 0.05% (v/v) Tween 20. Then the cells were incubated with Cy2-conjugated anti-rat secondary antibodies (Jackson) (diluted 1:100) at room temperature for 60 min, followed with five washes with PBS + 0.05% Tween 20. For detection of DNA, cells were stained with DAPI according to the manufacturer's protocol. Slides were prepared with Mounting Media (Bio-Rad) and immunofluorescent cells were detected with an Olympus confocal microscope.
Results
Co-immunoprecipitation experiments reveal the existence of a Rev–IN complex in HIV-1-infected cells
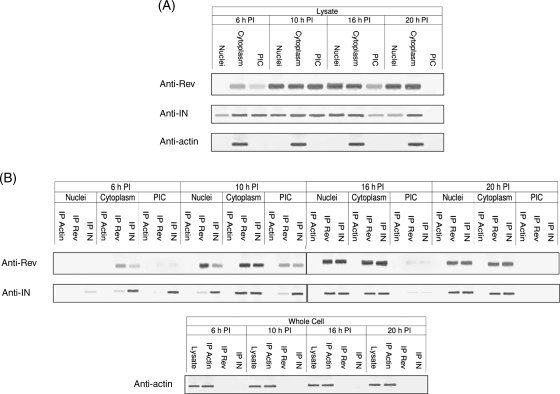
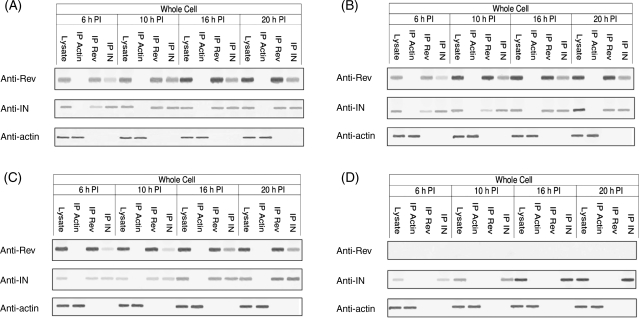
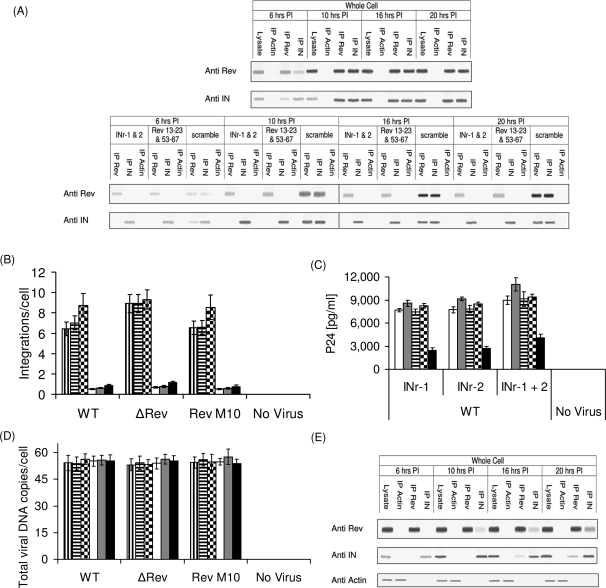
Here, we examined the occurrence of Rev and IN proteins in HIV-infected cells at various time points after exposure of cells to the virus. Western blot analysis revealed the presence of Rev and IN at 6 h PI (Fig. 1A); whereas Rev was apparent mainly in the cytoplasm, IN was detected in all three subcellular fractions tested (i.e. cytoplasm, PICs and nuclear). Starting from 10 h PI, the presence of Rev and IN coincided in all three subcellular fractions (Fig. 1A). Our observation demonstrates association of early phase Rev with the PIC. Previously, we have reported that Rev and IN interact in vitro and in cells transfected by both proteins (Rosenbluh et al., 2007). Consistent with these observations, co-IP analyses show interaction of Rev and IN in virus-infected cells (Fig. 1B). The Rev–IN complex was detected only in the cytoplasmic fraction at 6 h PI, in all three subcellular fractions at 10 h PI and in the cytoplasmic as well as the nuclear fractions but not in the PIC at 20 h PI (Fig. 1B). The Rev–IN complexes were also detected in cells infected by mutant viruses bearing either inactive IN (IN D64N D116N HIV-1; Nakajima et al., 2001) (Fig. 2A) or a nuclear export inactive Rev (Rev M10 HIV-1; Bahner et al., 1993) (Fig. 2B) suggesting that generation of these complexes did not require the known functions of these proteins. As expected, in cells infected with a virus defective for Rev-expressing (ΔRev virus; Verhoef et al., 1997), Rev–IN complexes were detected only after trans-complementation of Rev (Fig. 2C and D). Failure of anti-Rev and anti-IN antibodies to precipitate actin from the cells confirmed the specificity of these antibodies.
Fig. 1.
The HIV Rev protein interacts with the viral IN in virus-infected cells. (A) The presence of Rev, IN and of actin (control) was detected using the appropriate specific antibodies and by western blots. (B) Co-IP of Rev and IN in cells infected by WT HIV-1, co-IP experiments were performed exactly as described in the Materials and methods section.
Fig. 2.
A functional Rev or IN is not required for the formation of Rev–IN complex. Co-IP experiments were performed using the appropriate specific antibodies from lysate obtained cells infected by: (A) IN D64N D116N HIV-1, (B) Rev M10 HIV-1 or (C) ΔRev HIV-1 on Rev10+ cells, (D) ΔRev HIV-1. The presence of Rev, IN and actin was detected as described in (A). All other experimental details as described in the Material and methods section.
The presence of an early Rev is due to de novo synthesis from reverse-transcribed DNA
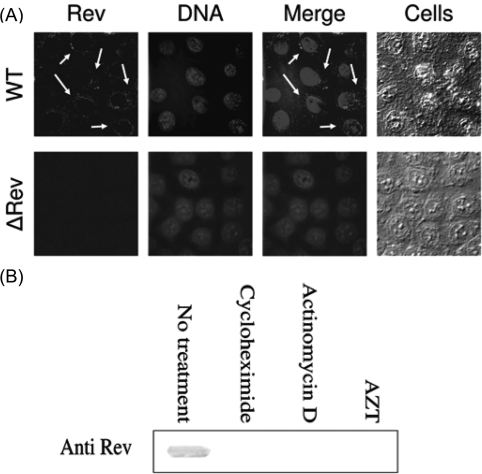
The co-IP experiments (Fig. 1) explicitly have demonstrated the presence of Rev as early as 6 h PI. Owing to the importance of this finding, it was essential to demonstrate appearance of Rev at that early time by additional experimental method. The immunofluorescence results depicted in Fig. 3A clearly demonstrate the presence of relatively low amounts of Rev in the cytoplasm (arrows in Fig. 3A), but not in the nuclei of virus-infected cells at this early time confirming the co-IP experiments (Fig. 1). Specificity of Rev expression should be inferred from the absence of any Rev specific fluorescence in ΔRev virus-infected cells (Fig. 3A). It should be added that much higher amounts of Rev—as was revealed by appearance of the fluorescence intensity—were detected at 10 h PI (not shown).
Fig. 3.
The appearance of the early Rev is due to de novo synthesis. (A) HeLa cells were infected with WT or ΔRev HIV-1 (MOI of 25) and immunostaining and observation of immunostained cells (arrows indicate the presence of Rev) as described in the Materials and methods section. (B) Treatment of cells by cycloheximide, actinomycin D and AZT as well as detection of Rev as described in the Materials and methods section.
The possibility that the Rev detected at these early times is packed within the invading viruses is ruled out by the results depicted in Fig. 3B. Both cycloheximide and actinomycin completely inhibited the appearance of early Rev indicating its de novo synthesis. Inhibition of Rev appearance by AZT (Fig. 3B) also indicates that reverse-transcription process is required to allow Rev synthesis.
Inhibition of cDNA integration by Rev and its stimulation in the absence of Rev
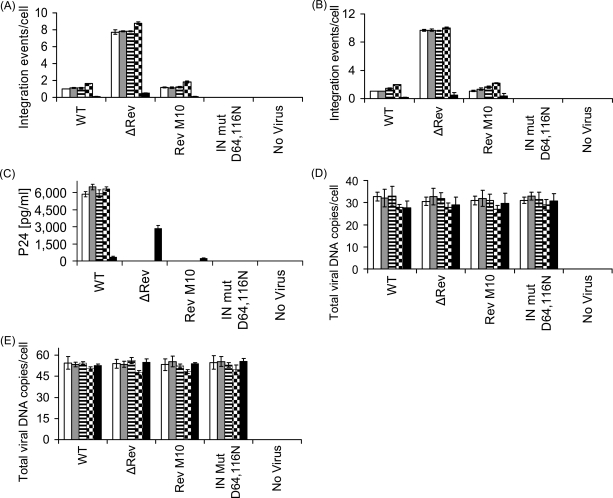
The results in Figs 1 and 2 demonstrate interaction of Rev and IN in virus-infected cells. These results necessitate studies on the influence of Rev on HIV integration events. From the results depicted in Fig. 4A and B, it is evident that Rev limits the number of integration events per cell. HIV infection of various cell lines with Rev-producing viruses led to ≤2 integration events per cell, confirming previously published results (Butler et al., 2001; Levin et al., 2009). This number was even further reduced in virus-infected Rev expressing cells (Rev10+) (Fig. 4A and B). On the other hand, the number of integration events per cell increased to ∼8–10 in cells infected with an HIV mutant that does not express Rev (ΔRev HIV) (Verhoef et al., 1997) (Fig. 4A and B and Supplementary Table S1, Supplementary data are available at PEDS online). As expected, infection with Rev M10, expressing non-functional Rev (Bahner et al., 1993), was non-productive (Fig. 4C). Also, the same amounts of total viral cDNA—which were drastically reduced in the presence of AZT (not shown)—were present under all experimental conditions indicating unimpaired virus entry and cDNA synthesis (Fig. 4D and E). Thus, both over-expression and the absence of Rev closely correlate with hypo- or hyper-integration of viral DNA, respectively. Combined with the results shown in Figs 1 and 2, these data indicate that interaction of Rev with IN restricts excessive HIV-1 DNA integration, possibly by inhibiting the IN enzymatic activity (Levin et al., 2009).
Fig. 4.
The HIV Rev protein inhibits IN-mediated integration and infection. The following cell lines; H9 lymphocytes (white bar), SupT1 lymphocytes (gray bar), TZM-bl (horizontal strips bar), 293T (black squares bar) and Rev10 cells (black bar) were infected by MOI 1.0 (A, C and E) and 2.0 (B and D) of the indicated HIVs and integration (A and B) as well as total viral cDNA (D and E) and viral p24 (C) were estimated at 24, 12 and 72 h PI, respectively. (A, B, D, E) P < 0.01, (C) P < 0.05. All others experimental conditions as well as the designation of the different virus types as described in the Materials and methods section.
IN and Rev derived peptides are able to disrupt the intracellular Rev–IN complexes and effect cDNA integration
To confirm that Rev exerts its effect on HIV-1 DNA integration through interaction with HIV-1 IN, we investigated the influence of two previously described groups of cell-permeable peptides that influence IN activity and interact with either Rev or IN in vitro (Rosenbluh et al., 2007; Hayouka et al., 2008; Levin et al., 2009). Peptides INr-1 (IN 66–80) and INr-2 (IN 118–128) are derived from the IN, interact with the Rev protein and induce dissociation of the Rev–IN complex in vitro and stimulate viral DNA integration in cultured cells (see also Levin et al., 2009). Rev 13–23 and Rev 53–67 are derived from the Rev protein, interact with the HIV-1 IN and significantly inhibit its enzymatic activity in vitro as well as integration of the viral DNA in cultured cells (Rosenbluh et al., 2007; Hayouka et al., 2008).
To examine the effect of these peptides on the formation of Rev–IN complexes in infected cells, H9 cells were pretreated with each group of the above-mentioned peptides (IN and Rev derived) or with a scrambled control peptide (Rosenbluh et al., 2007). The treated cells were exposed to HIV-1 and formation of Rev–IN complexes was analyzed by co-IP (Fig. 5A). Rev–IN complexes were detected only in cells treated with the scrambled control peptides but not in cells treated with either the Rev- or the IN-derived peptides (Fig. 5A). The view that dissociation of the Rev–IN complex by the INr peptides (Fig. 5A) stimulates viral DNA integration is inferred from the results in Fig. 5B (compare integration values to Fig. 4A and B and see also Supplementary Tables S1 and S2, Supplementary data are available at PEDS online). Consistent with increased integration, addition of INr peptides to virus-infected cells greatly stimulated production of viral p24 (Fig. 5C) as a measure of enhanced virus replication. It is of interest that the INr peptides only weakly stimulated integration in Rev10+-infected cells (Fig. 5B). This indicates that the ability of the peptides to increase virus cDNA integration could partially be reduced by the over-expressed Rev (compare results in Fig. 5B to Fig. 4A). The use of higher concentrations of peptides—to completely abrogate Rev inhibition—was avoided due to solubility limitations. The changes in the extent of integration upon pretreatment with INr peptides (Fig. 5B) cannot be explained by variations in the amounts of total viral DNA present in cells since the same amount of cDNA was detected under different experimental conditions (Fig. 5D). Scrambled peptides had no effect on the extent of HIV-1 cDNA integration (not shown and see Rosenbluh et al., 2007).
Fig. 5.
The Rev–IN complexes can be dissociated by specific cell permeable peptides and stimulate integration. (A) H9 lymphocytes were incubated for 2 h with the INr-1 and INr-2 (Levin et al., 2009) peptides as well as with the Rev 13–23 and Rev 53–67 (Rosenbluh et al., 2007; Hayouka et al., 2008) peptides and then were infected by the WT HIV-1. Rev, IN and actin were precipitated from whole cell extract and were detected by western blot analysis using the appropriate antibodies as described in Fig. 1A. (B) 293T cells and Rev10 cells were incubated for 2 h with INr-1 (vertical strips and white bars, respectively), INr-2 (horizontal strips and gray bars, respectively) and with a mixture of both peptides (black squares and black bars, respectively) and then infected with the indicated viruses. Viral cDNA integration was estimated 24 h PI. (C) The following cell lines; H9 lymphocytes (white), SupT1 lymphocytes (gray), TZM-bl (horizontal strips), 293T (black squares) and Rev10 cells (black) were incubated with the indicated peptides for 2 h and then infected with MOI 1.0 WT HIV-1 and viral p24 was estimated at 72 h PI. (D) Exactly as (B) but total viral DNA was determined 12 h PI. (E) Rev10 cells were incubated with the INr-1 and 2 peptides for 2 h, infected with the WT HIV-1 and following co-IP, Rev, IN and actin were detected as described above in A. (B and D) P < 0.01, (C) P < 0.05. Designation of the different virus types as described in Materials and methods.
The results in Fig. 4E confirm the existence of a Rev–IN complex in the INr-treated Rev10+ cells (Rev expressing). Much less stimulation of p24 production was observed in the INr-treated Rev10+ cells (compare results in Fig. 5C to those in Fig. 4C).
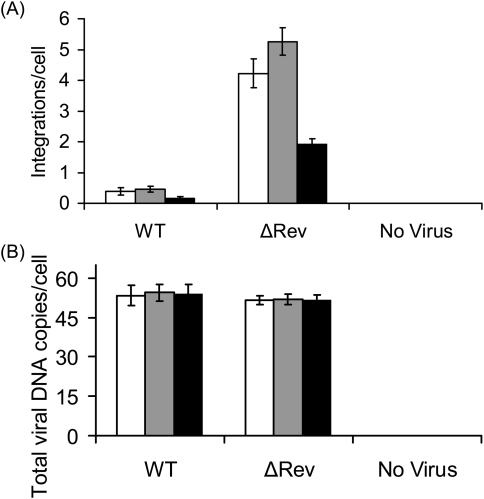
In spite of their ability to promote dissociation of the Rev–IN complex (Fig. 5A), the Rev peptides, in contrast to the INr peptides, failed to enhance the integration process in virus-infected cells (Fig. 6A; Rosenbluh et al., 2007). This result was expected since similar to the Rev protein itself (Levin et al., 2009), these peptides inhibit the enzymatic activity of IN as well as the IN-mediated integration process (Rosenbluh et al., 2007; Hayouka et al., 2008). Treatment of H9 cells with the Rev-derived peptides prior to infection with the ΔRev virus reduced the number of integration events, compared with untreated cells (compare Fig. 6A and Fig. 4A). This confirms that integration can be inhibited by the Rev-derived peptides. No changes in total cDNA amounts were observed following the addition of the Rev peptides (Fig. 6B and Supplementary Tables S1 and S2, Supplementary data are available at PEDS online).
Fig. 6.
Inhibition of integration by the Rev derived peptides. (A) Sup-T1 cells were incubated for 2 h with Rev 13–23 (white bars), Rev 53–67 (gray bars) as well as with both peptides (black bars) and were infected by the WT HIV-1 or by the DRev HIV-1. Viral cDNA integration was estimated 24 h PI. (B) Exactly as (A) but total viral DNA was determined 12 h PI, P < 0.01. Designation of the different virus types as described in Materials and methods.
Multi-integration of viral cDNA results in cell death
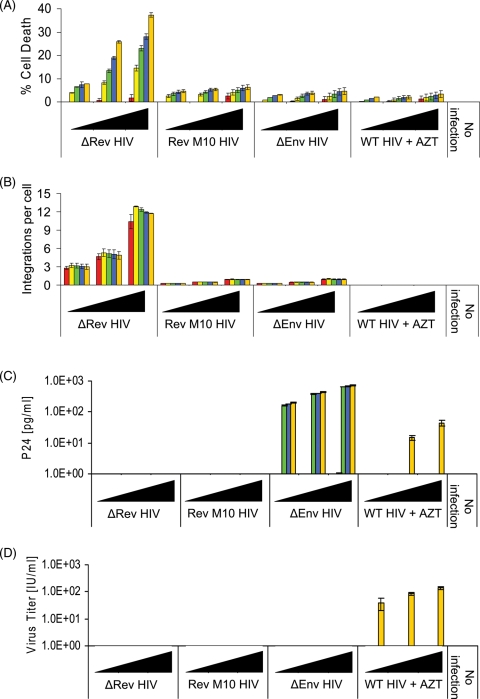
We then examined the influence of the observed increase in the integration degree on cell viability. Infection of H9 cells with ΔRev virus, which results in relatively high degree of integration (Fig. 7B, see also Fig. 4A and B), caused cell lysis of up to ∼40% (Fig. 7A). Evidently, no virus production was observed under these conditions (Fig. 7C and D) clearly indicating that the lysis observed was probably induced by the integration itself and does not result from virus replication. In contrast, limited integration, low cell lysis, but productive infection and appearance of infectious virions were observed in cells infected by viruses bearing functional Rev such as ΔEnv HIV and WT (+AZT) (AZT was added to inhibit infection by a fully functional virus), or RevM10 HIV-1 (Fig. 7A–D).
Fig. 7.
High degree of cDNA integration results in cell lysis. H9 lymphocytes were infected with increasing amounts (filled triangle, 0.01, 0.1 and 1.0 MOI) of the indicated HIVs. integration events/cell (A), cell death (B), p24 (C) and infectious particles (D) were estimated at the following different times (h) PI 12 (red), 24 (yellow), 60 (green), 108 (blue) and 156 (orange). (B) P < 0.01 and (A, C, D) P < 0.05.
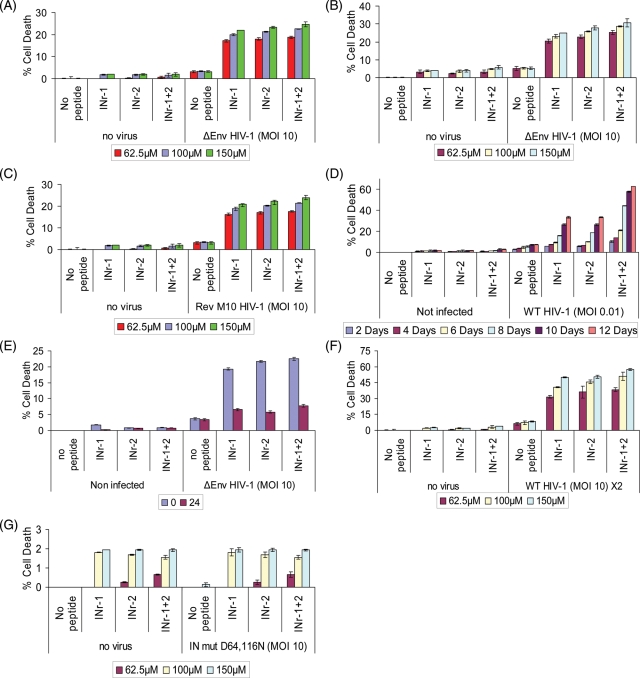
An apparent correlation between increased level of integration and induction of a certain degree of cell lysis was also observed when H9 cells were treated with the INr-1 and 2 peptides and then infected by the ΔEnv HIV (Fig. 8A and B) or by Rev M10 HIV (Fig. 8C). The use of ΔEnv and Rev M10 HIVs assured a single infection cycle again emphasizing a correlation between induction of cell death and the integration process. Cell death was also induced when WT virus-infected peptides-treated cells that were allowed to grow for a period of 12 days. Cell death reached up to ∼60% following infection with as low as 0.01 MOI (Fig. 8D). This may be due to multiple round of infection of peptide-treated cells (Fig. 8D). Addition of the peptides after completion of the integration process failed to promote cell death (Fig. 8E). When cells were double infected in the presence of the INrs, up to 60% cell death was observed (Fig. 8F) conferring the result in Fig. 5D. The INr peptides as expected did not have any effect on cell death when cells were infected by the IN mut D64116N HIV-1 (Fig. 8G).
Fig. 8.
High degree of cDNA integration results in cell lysis. Cell death was estimated 48 h PI (A) or 96 h PI (B) in peptide-treated H9 lymphocytes infected by ΔEnv HIV-1 or 48 h PI in peptide-treated H9 lymphocytes infected by Rev M10 (C) as well as at different times in cells infected by the WT virus (D). (E) as in (A) but peptides were at different times PI. (F) as in (A) but cells were double infected with ΔEnv HIV-1 (second infection was 12 h post first infection). (G) as in (A) but cells were infected by IN mut D64, 116N HIV-1. P < 0.05.
Discussion
The HIV-1 IN and Rev proteins are known to perform crucial functions at two temporal phases of the virus replication cycle, the early and the late phases, respectively (Freed, 2001). Being part of the invading virus, the IN can function immediately following infection (Freed, 2001). On the other hand, the main function attributed to Rev is nuclear export of single spliced and non-spliced viral RNA, which occurs at a later phase of the virus replication cycle (Pollard and Malim, 1998). Obviously, this function is executed by Rev molecules coded by the inserted virus genome (Pollard and Malim, 1998).
Here, we demonstrate—using co-IP experiments—that these two proteins interact and form a Rev–IN complex in HIV-infected cells. By interacting with IN, Rev blocks integration of the viral cDNA. Therefore, Rev should be present at early times PI, during the first steps of the integration process. Both, co-IP and immunofluorescence staining of virus-infected cells indeed demonstrate the presence of Rev molecules as early as 6 h PI. Rev appearance required de novo RNA and protein synthesis ruling out the possibility that Rev molecules were packed within the infectious virions. Furthermore, the requirement of a reverse-transcribed DNA for Rev expression is demonstrated by it sensitivity to AZT. It is noteworthy that our ability to detect early expressed Rev is due to the use of high titer of infectious viruses.
The ability of unintegrated viral cDNA to express viral proteins—among them Rev—at early times PI was well-established (Wu and Marsh, 2003b; Cara and Klotman, 2006; Iyer et al., 2009). Indeed, Rev-encoding HIV transcripts—expressed from unintegrated DNA molecules—were detected in the early phase of infection, prior to integration (Wu and Marsh, 2003a; Wu, 2004; Kelly et al., 2008; Iyer et al., 2009). Furthermore, using the mutant IN D64116N HIV, our results also demonstrate the ability of unintegrated cDNA to express Rev. Thus, it is conceivable that such early expressed Rev molecules are responsible for the inhibition of integration observed in the present work. If true, our findings—to the best of our knowledge—attribute, for the time, a biological function to the viral unintegrated DNA molecule. However, at this stage, the possibility that also Rev expressed from early integrated viral DNA may be involved in inhibition of integration cannot completely be excluded.
Several lines of evidences demonstrate that Rev restrict the efficiency of the integration process to 1–2 copies of integrated cDNA per infected cells. Indeed, a less number of integrated cDNA copies have been reported in various cultured cells infected by the WT HIV-1 (Butler et al., 2001) as well as in CD4+ lymphocytes of HIV-1-infected patients (Tan et al., 2006). This is in spite of the presence of numerous copies of free unintegrated DNA molecules (Chun et al., 1997). The inhibitory effect of Rev is based on the following results. (i) Augmentation of the less number of integration events in cells infected by the WT virus by peptides which either avoid the formation of or disrupt the Rev–IN complex. (ii) Overexpression of Rev reduces integration in infected cells. (iii) Multi-integration of viral cDNA in cells infected by a ΔRev virus. It certainly can be assumed that the quantity of the early expressed Rev molecules exceeds that of the invading IN molecules and thus should lead to complete inhibition of the IN activity and integration of the viral cDNA. The low number of integration observed may be due to the activity of few IN molecules that were translocated into the nuclei of infected cells before the appearance of sufficient amount of Rev molecules.
It is rather surprising that a small increase in integration resulted in a certain degree of lysis of the infected cells. Thus, it is tempting to speculate that Rev protects infected cells from premature death by restricting the number of integration events and thus assures completion of the virus replication cycle after integration. If true, it would be interesting to find out whether cell death is due to site specific integration or to expression of toxic genes. It is not surprising that cells with multiple-integrated DNA copies (>2) are only rarely observed in AIDS patients (Jung et al., 2002), since probably cells with high numbers of integrated copies of viral DNA are eliminated from the circulation of such patients by premature death. When multi-integration was observed, it was always correlated with induction of genome instability (Jung et al., 2002). The rare cells that survive the multi-integration events may produce a high titer of infectious viruses as is shown in the present work. Thus, the final outcome will be determined by the balance between the extent of cell death and virus production by the survived-infected cells.
The results of the present clearly demonstrate that in addition to its activity in nuclear export of viral RNA (Pollard and Malim, 1998), the Rev protein plays a major role in restriction of the integration degree of the viral cDNA. The question why inhibition by Rev restricts integration only to 1–2 copies is still unresolved. It is tempting to speculate that this is due to the ability of few uninhibited IN molecules to be translocated into the nuclei of the infected cells before sufficient Rev molecules are synthesized. Attempts are currently being made in our laboratory to resolve this important question.
Authors' contributions
Ab.L., Av.L., A.F., D.J.V. and R.B.-W. designed research; Av.L. preformed the immunoprecipitation experiments and cultured cells infections assays; Z.H. performs peptides synthesis and purification and Av.L. and Ab.L. wrote the paper.
Funding
This work was supported by the Israeli Science Foundation (A.L.), by a starting grant from the European Research Council (ERC) (A.F.) and D.J.V. was supported by grant DA17618 from NINDA, NIH.
Supplementary Material
Footnotes
Edited by Alan Fersht
References
- Bahner I., Zhou C., Yu X.J., Hao Q.L., Guatelli J.C., Kohn D.B. J. Virol. 1993;67:3199–3207. doi: 10.1128/jvi.67.6.3199-3207.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler S.L., Hansen M.S., Bushman F.D. Nat. Med. 2001;7:631–634. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- Cara A., Klotman M.E. J. Leukoc. Biol. 2006;80:1013–1017. doi: 10.1189/jlb.0306151. [DOI] [PubMed] [Google Scholar]
- Casabianca A., Gori C., Orlandi C., Forbici F., Federico Perno C., Magnani M. Mol. Cell. Probes. 2007;21:368–378. doi: 10.1016/j.mcp.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Chen A., Weber I.T., Harrison R.W., Leis J. J. Biol. Chem. 2006;281:4173–4182. doi: 10.1074/jbc.M510628200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun T.W., et al. Nature. 1997;387:183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- Cullen B.R. Methods Enzymol. 1987;152:684–704. doi: 10.1016/0076-6879(87)52074-2. [DOI] [PubMed] [Google Scholar]
- Derdeyn C.A., Decker J.M., Sfakianos J.N., Wu X., O'Brien W.A., Ratner L., Kappes J.C., Shaw G.M., Hunter E. J. Virol. 2000;74:8358–8367. doi: 10.1128/jvi.74.18.8358-8367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman A., Mizuuchi K., Craigie R. Cell. 1991;67:1211–1221. doi: 10.1016/0092-8674(91)90297-c. [DOI] [PubMed] [Google Scholar]
- Field F.J., Born E., Murthy S., Mathur S.N. Biochem. J. 2002;368:855–864. doi: 10.1042/BJ20020731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed E.O. Somat. Cell Mol. Genet. 2001;26:13–33. doi: 10.1023/a:1021070512287. [DOI] [PubMed] [Google Scholar]
- Freed E.O., Martin M.A. In: Fields Virology. Howley P.M., editor. Vol. 2. Philadelphia: Lippincott Williams and Wilkins; 2001. pp. 1971–2041. [Google Scholar]
- Guiot E., Carayon K., Delelis O., Simon F., Tauc P., Zubin E., Gottikh M., Mouscadet J.F., Brochon J.C., Deprez E. J. Biol. Chem. 2006;281:22707–22719. doi: 10.1074/jbc.M602198200. [DOI] [PubMed] [Google Scholar]
- Gummuluru S., Kinsey C.M., Emerman M. J. Virol. 2000;74:10882–10891. doi: 10.1128/jvi.74.23.10882-10891.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayouka Z., Rosenbluh J., Levin A., Maes M., Loyter A., Friedler A. Biopolymers. 2008;90:481–487. doi: 10.1002/bip.20930. [DOI] [PubMed] [Google Scholar]
- Iyer S.R., Yu D., Biancotto A., Margolis L.B., Wu Y. J. Virol. 2009;83:8662–8673. doi: 10.1128/JVI.00874-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung A., Maier R., Vartanian J.P., Bocharov G., Jung V., Fischer U., Meese E., Wain-Hobson S., Meyerhans A. Nature. 2002;418:144. doi: 10.1038/418144a. [DOI] [PubMed] [Google Scholar]
- Kelly J., Beddall M.H., Yu D., Iyer S.R., Marsh J.W., Wu Y. Virology. 2008;372:300–312. doi: 10.1016/j.virol.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimpton J., Emerman M. J. Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler F., Cardon G., Pohlman M., Gill R., Schider O. Plant Mol. Biol. 1989;12:189–199. doi: 10.1007/BF00020504. [DOI] [PubMed] [Google Scholar]
- Kramer-Hammerle S., Ceccherini-Silberstein F., Bickel C., Wolff H., Vincendeau M., Werner T., Erfle V., Brack-Werner R. BMC Cell Biol. 2005;6:20. doi: 10.1186/1471-2121-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin A., Kutznetova L., Kahana R., Rubinstein-Guini M., Stram Y. Virus Res. 2006;120:121–127. doi: 10.1016/j.virusres.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Levin A., Hayouka Z., Helfer M., Brack-Werner R., Friedler A., Loyter A. PLoS One. 2009;4:e4155. doi: 10.1371/journal.pone.0004155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertens G., Cherepanov P., Pluymers W., Busschots K., De Clercq E., Debyser Z., Engelborghs Y. J. Biol. Chem. 2003;278:33528–33539. doi: 10.1074/jbc.M303594200. [DOI] [PubMed] [Google Scholar]
- Nakajima N., Lu R., Engelman A. J. Virol. 2001;75:7944–7955. doi: 10.1128/JVI.75.17.7944-7955.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzato M., Erlwein O., Bonsall D., Kaye S., Muir D., McClure M.O. J. Virol. Methods. 2009;156:1–7. doi: 10.1016/j.jviromet.2008.10.012. [DOI] [PubMed] [Google Scholar]
- Pollard V.W., Malim M.H. Ann. Rev. Microbiol. 1998;52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- Pommier Y., Johnson A.A., Marchand C. Nat. Rev. Drug Discov. 2005;4:236–248. doi: 10.1038/nrd1660. [DOI] [PubMed] [Google Scholar]
- Ratner L., et al. Nature. 1985;313:277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- Reiser J. Gene Ther. 2000;7:910–913. doi: 10.1038/sj.gt.3301188. [DOI] [PubMed] [Google Scholar]
- Rosenbluh J., Hayouka Z., Loya S., Levin A., Armon-Omer A., Britan E., Hizi A., Kotler M., Friedler A., Loyter A. J. Biol. Chem. 2007;282:15743–15753. doi: 10.1074/jbc.M609864200. [DOI] [PubMed] [Google Scholar]
- Tan W., Dong Z., Wilkinson T.A., Barbas C.F., III, Chow S.A. J. Virol. 2006;80:1939–1948. doi: 10.1128/JVI.80.4.1939-1948.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoef K., Koper M., Berkhout B. Virology. 1997;237:228–236. doi: 10.1006/viro.1997.8786. [DOI] [PubMed] [Google Scholar]
- Wolff H., Hadian K., Ziegler M., Weierich C., Kramer-Hammerle S., Kleinschmidt A., Erfle V., Brack-Werner R. Exp. Cell Res. 2006;312:443–456. doi: 10.1016/j.yexcr.2005.11.020. [DOI] [PubMed] [Google Scholar]
- Wu Y. Retrovirology. 2004;1:13. doi: 10.1186/1742-4690-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Marsh J.W. J. Virol. 2003a;77:10376–10382. doi: 10.1128/JVI.77.19.10376-10382.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Marsh J.W. Microbes Infect. 2003b;5:1023–1027. doi: 10.1016/s1286-4579(03)00187-4. [DOI] [PubMed] [Google Scholar]
- Yamamoto N., Tanaka C., Wu Y., Chang M.O., Inagaki Y., Saito Y., Naito T., Ogasawara H., Sekigawa I., Hayashida Y. Virus Genes. 2006;32:105–113. doi: 10.1007/s11262-005-5851-2. [DOI] [PubMed] [Google Scholar]
- Zhang J., Scadden D.T., Crumpacker C.S. J. Clin. Invest. 2007;117:473–481. doi: 10.1172/JCI28971. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.