The structure of N-terminally truncated DR2204, a Nudix hydrolase, is presented at 2.0–Å resolution.
Keywords: ADP-ribose pyrophosphatases, Deinococcus radiodurans
Abstract
Nudix pyrophosphatases are a well represented protein family in the Deinococcus radiodurans genome. These hydrolases, which are known to be enzymatically active towards nucleoside diphosphate derivatives, play a role in cleansing the cell pool of potentially deleterious damage products. Here, the structure of DR2204, the only ADP-ribose pyrophosphatase in the D. radiodurans genome that is known to be active towards flavin adenosine dinucleotide (FAD), is presented at 2.0 Å resolution.
1. Introduction
Deinococcus radiodurans is a Gram-positive bacterium that has been shown to have remarkable resistance to ionizing radiation (Battista, 1997 ▶), in addition to an extraordinary resistance to desiccation. Analysis of the complete genome of D. radiodurans showed that there is a specific expansion of certain protein families; such an expansion may be connected to the organism’s response to stress and damage resistance. One of the most highly represented protein families are hydrolases, amongst which are the Nudix pyrophosphatases, calcineurin-like phosphoesterases, lipase/epoxidase-like (α/β) hydrolases, subtilisin-like proteases and sugar deacetylases (Makarova et al., 2000 ▶). D. radiodurans has 24 open reading frames (ORFs) annotated as MutT-like/Nudix hydrolases, a reasonably large number considering its proteome size (3187 protein-coding genes) and more than double the number identified for Thermus thermophilus (ten ORFs), an extremophile belonging to the same bacterial clade (Omelchenko et al., 2005 ▶).
Nudix hydrolases are a group of phosphoanhydrases that catalyse the hydrolysis of a nucleoside diphosphate linked to another moiety X. Members of the Nudix family contain the consensus sequence GX 5EX 7REUXEEXGU (known as the Nudix box, where U may be Ile, Leu or Val and X represents any amino acid), a unique loop–helix–loop motif which forms the versatile catalytic site for diphosphate hydrolysis. The Nudix motif has been identified in protein sequences from species from all three kingdoms. Outside the conserved sequence box, Nudix-family proteins have low overall sequence identity. However, the core fold is conserved and is composed of a highly conserved α/β/α sandwich, with the Nudix box sitting within a helix of the β-grasp motif. Catalytic activity takes place in the Nudix box, in which both the substrate and metal ions which are essential for activity are bound. Beyond the central fold, other regions of the sequence such as loops and the C-terminal and N-terminal regions account for substrate specificity, thus separating catalytic activity from substrate recognition and accommodation (for reviews, see Mildvan et al., 2005 ▶; McLennan, 2006 ▶).
Nudix hydrolases have been termed house-cleaning enzymes, together with other enzymes such as dUTPases, ITPases and all-α nucleoside triphosphate (NTP) pyrophosphatases (Bessman et al., 1996 ▶; Galperin et al., 2006 ▶). They play a role in cleansing the cell pool of potentially deleterious damage products. These substrates, cell signalling molecules, regulators of cellular metabolism and metabolic intermediates, are potentially toxic compounds whose concentrations require fine modulation at different stages of the cell cycle. Typically, each enzyme may be active towards more than one nucleoside diphosphate derivative, rendering identification of the true biological substrate a difficult task. Nudix hydrolases can process a wide variety of substrates including various ribonucleoside and deoxyribonucleoside triphosphates, nucleoside diphosphate sugar molecules, dinucleoside polyphosphates and several coenzymes including NADH, FAD and CoA. In addition, some of these enzymes have also been reported to be active towards noncanonical nucleoside triphosphates, as is the case for Escherichia coli MutT. The natural substrate of this Nudix hydrolase is 8-oxo-dGTP (Bhatnagar & Bessman, 1988 ▶; Akiyama et al., 1989 ▶; Maki & Sekiguchi, 1992 ▶; Saraswat et al., 2002 ▶), but the enzyme is also active towards non-nucleoside diphosphate derivatives such as diphosphoinositol (Safrany et al., 1998 ▶) and 5-phosphoribosyl-1-pyrophosphate (Fisher et al., 2002 ▶).
To date, the crystal structures of three other Nudix hydrolases from D. radiodurans have been reported: DR1184, DR1025 and DR0079 (PDB entries 1nqy, 1su2 and 2o5f, respectively). All three contain the conserved elements of the Nudix fold; nevertheless, substantial differences in substrate binding have been reported. DR1184 is known to be active towards CoA and has additional structural elements characteristic of the CoA-hydrolysing Nudix subfamily (Kang et al., 2003 ▶). The DR1025 crystal structure showed the pivotal role of the N-terminal extension in ATP, GTP or P1,P4-diadenosine-5′-tetraphosphate binding and selectivity (Ranatunga et al., 2004 ▶). While the biological substrate of DR0079 remains unknown, it has been proven that it is not a nucleotide triphosphate (Buchko et al., 2004 ▶). Of the Nudix enzymes belonging to the D. radiodurans genome, DR2204 was reported to have the widest substrate specificity, being active towards ADP-ribose, GDP-mannose, FAD and NADH (Xu et al., 2001 ▶) and moderately active towards dATP and dGTP. Here, we report the crystal structure of an N-terminally truncated form of DR2204 at 2.0 Å resolution.
2. Experimental
2.1. Cloning, expression and purification
The open reading frame annotated as DR2204 was amplified by PCR from the genomic DNA of D. radiodurans. The PCR product comprising the 5′-CACC overhang was inserted into the expression pET151/D-TOPO directional vector (Invitrogen) following the manufacturer’s recommended procedure. The resulting expression vector was used to transform E. coli BL21 (DE3) competent cells (Invitrogen). Transformed cells carrying pET151-DR2204 were grown in Luria–Bertani enriched growth medium containing 100 µg ml−1 ampicillin; the medium was inoculated with a sample of an overnight culture and grown at 310 K until an optical density of approximately 0.6–0.7 at 600 nm was reached, at which point IPTG (isopropyl β-d-1-thiogalactopyranoside) was added to a final concentration of 0.3 mM, thus inducing overexpression. Cultures were allowed to grow for a further 4 h, after which the cells were harvested at 7000g for 20 min at 277 K and resuspended in lysis buffer [50 mM Tris–HCl pH 7.0, 300 mM NaCl, 2 mM β-mercaptoethanol, 5%(v/v) glycerol]. The cells were frozen at 193 K, thawed and lysed by two passages through a French press. DNase I was added to a final concentration of 20 µg ml−1 together with an EDTA-free protease-inhibitor tablet (Roche). The lysed culture was centrifuged at 40 000g for 30 min at 277 K and the soluble fraction was loaded onto a 5 ml His-Trap column (GE Healthcare) equilibrated with buffer A [50 mM Tris–HCl pH 7.5, 300 mM NaCl, 5%(v/v) glycerol]. The first wash consisted of a step elution to 12% buffer B (buffer A supplemented with 500 mM imidazole) followed by a gradient elution from 12 to 100% buffer B. DR2204 was eluted and pooled near 250 mM imidazole, as confirmed by SDS–PAGE. The pooled fractions were exchanged into a suitable buffer for His-tag cleavage by enzymatic digestion with tobacco etch virus (TEV) protease. The tag-free protein was again loaded onto a 5 ml His-Trap column and the flowthrough was collected. Further polishing was performed and oligomerization states were confirmed using a Superdex S200 HR HiPrep 16/60 column (GE Healthcare) equilibrated with 25 mM Tris–HCl pH 7.6, 150 mM NaCl, 1 mM DTT and 1 mM MgCl2. The oligomerization state and monodispersity of DR2204 in solution were estimated by dynamic light scattering (Malvern Instruments, Zetasizer, Nano series).
2.2. Protein crystallization and data collection
DR2204 was successfully crystallized in two crystallization conditions using the hanging-drop vapour-diffusion method. The protein was concentrated to 16 mg ml−1 and mixed with an equal volume of reservoir solution containing either 0.2 M LiNO3, 20%(w/v) polyethylene glycol 3350 (PEG/Ion Screen, Hampton Research; crystal type A) or 0.2 M KSCN, 0.1 M bis-tris propane pH 6.5, 20%(w/v) polyethylene glycol 3350 (PACT2 Screen, Molecular Dimensions; crystal type B) and allowed to equilibrate at 294 K. Crystals of tetragonal bipyramidal shape grew in 10–15 d using both crystallization conditions.
Diffraction data were collected at 100 K on beamline ID14-4 at the European Synchrotron Radiation Facility (Grenoble, France) at a wavelength of 0.98 Å. For both crystallization conditions, crystals were flash-cooled in liquid nitrogen (100 K) after soaking them for a few seconds in a solution containing a 2% increment in the precipitant and 18%(v/v) glycerol. For de novo phasing, a heavy-atom derivative crystal was prepared by soaking crystals of type B in 1 mM GdCl3 for 18 h. Native crystals of type A and derivative crystals diffracted to 2.0 and 2.1 Å resolution, respectively. The solvent content was estimated as 50% based on a Matthews coefficient of 2.46 Å3 Da−1 for two molecules in the crystallographic asymmetric unit and a unit-cell volume of 622 859 Å3. Diffraction data were processed with MOSFLM (Powell, 1999 ▶) and further data analysis was carried out using the CCP4 suite v.6.0.2 (Collaborative Computational Project, Number 4, 1994 ▶). Data-collection statistics are summarized in Table 1 ▶.
Table 1. Data-collection statistics.
Values in parentheses are the highest resolution shell.
| Gd-SAD | Native | |
|---|---|---|
| Beamline | ID14-4, ESRF | ID14-4, ESRF |
| Space group | P43212 | P43212 |
| Unit-cell parameters (Å) | ||
| a | 60.9 | 61.3 |
| b | 60.9 | 61.3 |
| c | 166.5 | 165.7 |
| Resolution (Å) | 50–2.1 (2.21–2.10) | 50–2.0 (2.11–2.00) |
| Wavelength (Å) | 0.98 | 0.98 |
| No. of unique reflections | 18230 (2068) | 22215 (3200) |
| Multiplicity | 15.4 (7.3) | 4.4 (4.6) |
| Completeness (%) | 95.3 (77.3) | 99.7 (100) |
| 〈I/σ(I)〉 | 24.4 (5.5) | 11 (3.6) |
| Rmerge† (%) | 7.6 (24.9) | 8.0 (39.1) |
| Ranom‡ (%) | 2.5 (10.6) | |
| FOM§ | 0.31 | |
| FOM (after DM) | 0.79 | |
| Phasing power | 1.20 | |
| RCullis | 0.78 |
R
merge = 
 .
.
R
anom = 
 .
.
Figure of merit.
2.3. Structure determination and refinement
The structure of DR2204 was solved by single-wavelength anomalous dispersion (SAD) from highly redundant data collected to 2.1 Å resolution from the gadolinium derivative at a wavelength of 0.98 Å. After analysing the structure factors using SHELXC and SHELXD from HKL2MAP v.0.2 (Sheldrick, 2008 ▶; Pape & Schneider, 2004 ▶), the position of one gadolinium site per monomer could be determined. The coordinates were then refined and phase calculations were performed using the SHARP suite (Bricogne et al., 2003 ▶). The phases were further improved by density modification using SOLOMON (Abrahams & Leslie, 1996 ▶) and DM (Cowtan, 1999 ▶). A partial starting model was obtained by automatic model building with the program ARP/wARP (Perrakis et al., 1999 ▶). The initial model was then completed by rounds of manual building with Coot (Emsley & Cowtan, 2004 ▶) and refinement with REFMAC. The structure of the native protein was obtained by molecular replacement with the program Phaser (McCoy et al., 2007 ▶) using the gadolinium-derivative structure as a starting model. The native structure was refined against data collected from a crystal of type A, which were of better quality than the data collected from crystals of type B. Refinement using REFMAC (Murshudov et al., 1997 ▶) and further cycles of manual model building using Coot gave a final model consisting of an N-terminally truncated form of the protein which lacked the first 55 residues in the sequence. Mass-spectrometric analysis and sequencing performed on protein crystals confirmed the protein to be truncated at this position. In both monomers the electron density is poor for a short loop between residues 149–158 and 148–159 in monomers A and B, respectively. No model could therefore be built in this region; this problem is likely to arise from disorder associated with this part of the protein. The structure was refined using bulk-solvent correction and a maximum-likelihood target function. All the data measured, apart from a 5% test set for R free calculation, were used in the refinement. Side-chain atoms without visible electron density at 1.0σ were removed from the model. The quality of the structures was assessed using PROCHECK (Laskowski et al., 1993 ▶), WHAT IF (Vriend, 1990 ▶) and MolProbity (Davis et al., 2004 ▶). All residues fall within the allowed regions of the Ramachandran plot; refinement statistics and geometry analysis of the structures are summarized in Table 2 ▶.
Table 2. Crystallographic refinement statistics.
| Rcryst† (%) | 20.84 |
| Rfree‡ (%) | 24.69 |
| R.m.s.d.§ bond lengths (Å) | 0.0163 |
| R.m.s.d. angles (°) | 1.583 |
| Mean B (Å2) | 33.1 |
| MolProbity statistics | |
| Ramachandran | |
| Most favoured (%) | 96.2 |
| Allowed (%) | 3.8 |
| Outliers (%) | 0.0 |
| Rotamer outliers (%) | 1.94 |
| Clashscore¶ | 1.97 (100th percentile) |
| Overall score¶ | 1.18 (100th percentile) |
| Overall G factor†† | 0.02 |
R
cryst = 
 .
.
R free was calculated with a small fraction (5%) of randomly selected reflections.
Root-mean-square deviation.
The scores are ranked according to structures of similar resolution as formulated in MolProbity.
The overall measure of structure quality from PROCHECK (Laskowski et al., 1993 ▶).
3. Results and discussion
Full-length recombinant DR2204 was overexpressed in E. coli and purified to homogeneity as confirmed by SDS–PAGE. The gene product corresponding to DR2204 encodes a protein of 200 amino acids with a calculated molecular mass of 21.8 kDa and a theoretical pI of 4.9. Size-exclusion chromatography indicated the presence of a homodimer in solution, a feature common to Nudix hydrolases. Dynamic light-scattering analysis confirmed the monodispersity of the purified protein.
We have determined the crystal structure of DR2204 at 2.0 Å resolution using the SAD phasing method (for data-collection and refinement statistics, see Tables 1 ▶ and 2 ▶, respectively). Both crystal types contained a homodimer in the asymmetric unit. Upon inspection of the map after density modification, no electron density was observed for about 55 residues. Inspection of maps before density modification also confirmed this observation. Further analysis of the crystal packing also suggested a lack of space to accommodate this number of N-terminal residues. Direct loading of a crystallization drop containing crystals showed a single band migrating to a molecular weight lower than 21.8 kDa. Identical drops were analysed by mass spectrometry, resulting in a spectrum dominated by a polypeptide of 15.9 kDa. N-terminal protein sequencing identified three polypeptides starting at Thr51, Tyr53 and Tyr55. The latter corresponds to a polypeptide with a calculated molecular weight of 15.8 kDa.
The crystal structure described here corresponds to an unexpected truncation of the first 55 amino acids of the N-terminus of DR2204. Based on partial proteolysis, human NUDT9, an ADP-ribose pyrophosphatase (ADPRase), was reported to have a resistant C-terminal domain which retains specific enzymatic activity while the proteolytically labile N-terminal domain enhances substrate affinity (Perraud et al., 2003 ▶; Shen et al., 2003 ▶). Stability studies of concentrated DR2204 in solution using SDS–PAGE analysis showed that the protein retains its molecular weight with no degradation at room temperature for over two weeks (result not shown).
The refined crystal structure of DR2204 consists of the conserved α/β/α sandwich fold typical of Nudix hydrolases: the Nudix box (residues 94–115) holding the α1 helix sits between two loops accessible to the solvent, while the other two helices, α2 and α3, lie on the other side of the central β-sheet and participate in dimer-interface formation. Sequence alignment (Fig. 1 ▶) shows that DR2204 shares common characteristics with other members of the ADP-ribose pyrophosphatase (EC 3.6.1.13) subfamily, with sequence identities of as high as 24% to E. coli ADPRaseII (ORF186) and 21% to E. coli ADPRaseI (ORF209). More significantly, the overall structure of truncated DR2204 has a high identity to that of the recently solved Thermus thermophilus HB8 Ndx2 crystal structure (PDB code 2yvm; Wakamatsu et al., 2008 ▶), with a root-mean-square deviation (r.m.s.d) of 0.73 Å for equivalent Cα atoms (Fig. 2 ▶ c) and a sequence identity of 50% (40% for the full-length protein). DR2204 was found to exist as a dimer both in solution and in the crystal form (Fig. 2 ▶ a), in contrast to T. thermophilus Ndx2 which crystallized with a monomer in the asymmetric unit. Despite lacking the N-terminal β-strand, which is known to domain-swap between two subunits and to offer additional interfacial area and structural stabilization in other Nudix structures (Gabelli et al., 2001 ▶; Yoshiba et al., 2004 ▶; Wakamatsu et al., 2008 ▶), DR2204 dimerized with the same orientation between the monomers, burying ∼1100 Å2 of solvent-accessible surface (Fig. 2 ▶ a).
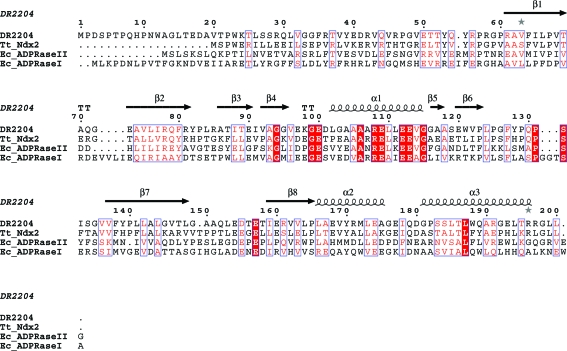
Figure 1.
Sequence alignment of DR2204 with the Tt_Ndx2 (Thermus thermophilus HB8; 40% identity), Ec_ADPRaseII (E. coli ORF186; 24% identity) and Ec_ADPRaseI (E. coli ORF209; 21% sequence identity) proteins. Identical residues are shown in white on a red background and similar residues are shown in red on a white background. For the secondary-structure assignment, α-helices are represented as helices, β-strands are represented as arrows and β-turns are marked ‘TT’. This figure was prepared with ESPript (Gouet et al., 1999 ▶). The multiple sequence alignment was performed with CLUSTALW (v1.83) (Thompson et al., 1994 ▶)
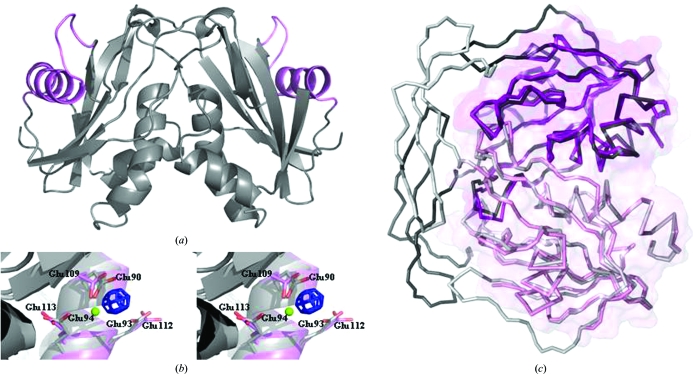
Figure 2.
(a) The overall structure of the DR2204 dimer as observed in the asymmetric unit. The protein in grey is shown in cartoon representation, with the Nudix box highlighted in magenta. (b) Stereo picture showing a close-up of the Nudix box. The experimental map (blue; contoured at 10.0σ) calculated from data collected from the gadolinium-derivative crystal used for phasing is overlaid on the final model refined from data collected from the native crystal (magenta). A significant positive peak for the Gd3+ ion is observed in the region where Mg2+ is expected to bind during catalysis, between the side chains of Glu109 and Glu112. Both residues are located in the Nudix-box region, a versatile divalent metal-binding site characteristic of this family of enzymes. For clarity, the model of the ADP-ribose pyrophosphatase (Ndx2) from T. thermophilus (grey; PDB code 2yvm) including the Mg2+ 1002 ion (in green) bound to residues Glu90 and Glu94 is shown. (c) Superposition of DR2204 (magenta) on the structures of the ADP-ribose pyrophosphatase from T. thermophilus (grey; 0.73 Å r.m.s.d.; PDB code 2yvm). The surface of DR2204 is overlaid on the proteins represented in ribbon; the dimer formation buries ∼1100 Å2 of solvent-accessible surface. This figure was prepared with PyMOL (DeLano, 2003 ▶).
Although the protein was purified in the presence of MgCl2, the native structure of DR2204 was found to be in the apo form; no bound magnesium ions were found. However, using the gadolinium data and by analogy with the T. thermophilus Ndx2 structure, the Mg2+-binding site can be identified in our structure (Fig. 2 ▶ b). The Gd3+ ion is bound between the side chains of Glu109 and Glu112, thus corresponding to the magnesium ion labelled Mg2 in the T. thermophilus structure. In this structure Mg2 has an octahedral coordination to three glutamate residues (Glu90, Glu94 and Glu139), the α-phosphate of the bound nucleotide and two water molecules. The first two coordinating residues Glu90 and Glu94 correspond by sequence alignment to Glu109 and Glu113, respectively, in the DR2204 structure. In both human NUDT9 and T. thermophilus Ndx4 the metal-binding residues involve the second and fourth glutamates from the Nudix box. However, E. coli ADP-ribose pyrophosphatase (ADPRaseI; ORF209) also has the metal ion bound to the same corresponding glutamates as DR2204. This suggests versatility in ion binding within the Nudix box, accounting for the ability to bind both divalent and trivalent cations.
According to Xu et al. (2001 ▶), DR2204 is the only Nudix hydrolase in D. radiodurans with enzymatic activity towards FAD and NADH as well as ADP-ribose. In the D. radiodurans genome the Nudix hydrolase with the highest sequence identity to DR2204 is DR1007 (26% identity); however, this enzyme exhibits activity towards ADP-ribose alone, qualifying it as an ADPRaseI according to Wakamatsu et al. (2008 ▶). We propose DR2204 to be an ADPRaseII or rather an FAD pyrophosphatase (EC 3.6.1.18), as is T. thermophilus Ndx2, and possibly the only one in the entire D. radiodurans genome. Attempts to cocrystallize DR2204 with ADP-ribose and FAD are under way.
Supplementary Material
PDB reference: DR2204, 2w4e, r2w4esf
Acknowledgments
Mass-spectrometric analysis was performed by I. Bérard from the Laboratoire de Spectrométrie de Masse des Protéines and N-terminal protein sequencing was performed by J.-P. Andrieu from the Laboratoire d’Enzymologie Moléculaire at the Institut de Biologie Structurale, 41 Rue Jules Horowitz, 38027 Grenoble, France. AMDG acknowledges a postdoctoral research grant from Fundação para a Ciência e Tecnologia (SFRH/BEST/15939/2005).
References
- Abrahams, J. P. & Leslie, A. G. W. (1996). Acta Cryst. D52, 30–42. [DOI] [PubMed]
- Akiyama, M., Maki, H., Sekiguchi, M. & Horiuchi, T. (1989). Proc. Natl Acad. Sci. USA, 86, 3949–3952. [DOI] [PMC free article] [PubMed]
- Battista, J. R. (1997). Annu. Rev. Microbiol.51, 203–224. [DOI] [PubMed]
- Bessman, M. J., Frick, D. N. & O’Handley, S. F. (1996). J. Biol. Chem.271, 25059–25062. [DOI] [PubMed]
- Bhatnagar, S. K. & Bessman, M. J. (1988). J. Biol. Chem.263, 8953–8957. [PubMed]
- Bricogne, G., Vonrhein, C., Flensburg, C., Schiltz, M. & Paciorek, W. (2003). Acta Cryst. D59, 2023–2030. [DOI] [PubMed]
- Buchko, G. W., Shuisong, N., Holbrook, S. R. & Kennedy, M. A. (2004). Proteins, 56, 28–39. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Cowtan, K. (1999). Acta Cryst. D55, 1555–1567. [DOI] [PubMed]
- Davis, I. W., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2004). Nucleic Acids Res.32, W615–W619. [DOI] [PMC free article] [PubMed]
- DeLano, W. L. (2003). The PyMOL Molecular Graphics System. DeLano Scientific LLC, San Carlos, California, USA.
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Fisher, D. I., Safrany, S. T., Strike, P., McLennan, A. G. & Cartwright, J. L. (2002). J. Biol. Chem.277, 47313–47317. [DOI] [PubMed]
- Gabelli, S. B., Bianchet, M. A., Bessman, M. J. & Amzel, L. M. (2001). Nature (London), 8, 467–472. [DOI] [PubMed]
- Galperin, M. Y., Moroz, O. V., Wilson, K. S. & Murzin, A. G. (2006). Mol. Microbiol.59, 5–19. [DOI] [PubMed]
- Gouet, P., Courcelle, E., Stuart, D. I. & Métoz, F. (1999). Bioinformatics, 15, 305–308. [DOI] [PubMed]
- Kang, L.-W., Gabelli, S. B., Bianchet, M. A., Xu, W. L., Bessman, M. J. & Amzel, L. M. (2003). J. Bacteriol.185, 4110–4118. [DOI] [PMC free article] [PubMed]
- Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl. Cryst.26, 283–291.
- Makarova, K. S., Aravind, L., Daly, M. J. & Koonin, E. V. (2000). Genetica, 108, 25–34. [DOI] [PubMed]
- Maki, H. & Sekiguchi, M. (1992). Nature (London), 355, 273–275. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst.40, 658–674. [DOI] [PMC free article] [PubMed]
- McLennan, A. G. (2006). Cell. Mol. Life Sci.63, 123–143. [DOI] [PMC free article] [PubMed]
- Mildvan, A. S., Xia, Z., Azurmendi, H. F., Saraswat, V., Legler, P. M., Massiah, M. A., Gabelli, M. A., Bianchet, M. A., Kang, L.-W. & Amzel, L. M. (2005). Arch. Biochem. Biophys.433, 129–143. [DOI] [PubMed]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Omelchenko, M. V., Wolf, Y. I., Gaidamakova, E. K., Matrosova, V. Y., Vasilenko, A., Zhai, M., Daly, M. J., Koonin, E. V. & Makarova, K. S. (2005). BMC Evol. Biol.5, 57. [DOI] [PMC free article] [PubMed]
- Pape, T. & Schneider, T. R. (2004). J. Appl. Cryst.37, 843–844.
- Perrakis, A., Morris, R. & Lamzin, V. S. (1999). Nature Struct. Biol.6, 458–463. [DOI] [PubMed]
- Perraud, A.-L., Shen, B., Dunn, C. A., Rippe, K., Smith, M. K., Bessman, M. J., Stoddard, B. L. & Scharenberg, A. M. (2003). J. Biol. Chem.278, 1794–1801. [DOI] [PubMed]
- Powell, H. R. (1999). Acta Cryst. D55, 1690–1695. [DOI] [PubMed]
- Ranatunga, W., Hill, E. E., Mooster, J. L., Holbrook, E. L., Schulze-Gahmen, U., Xu, W. L., Bessman, M. J., Brenner, S. E. & Holbrook, S. R. (2004). J. Mol. Biol.339, 103–116. [DOI] [PubMed]
- Safrany, S. T., Caffrey, J. J., Yang, X., Bembenek, M. E., Moyer, M. B., Burkhart, W. A. & Shears, S. B. (1998). EMBO J.17, 6599–6607. [DOI] [PMC free article] [PubMed]
- Saraswat, V., Massiah, M. A., Lopez, G., Amzel, L. M. & Mildvan, A. S. (2002). Biochemistry, 41, 15566–15577. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Shen, B. W., Perraud, A.-L., Scharenberg, A. & Stoddard, B. L. (2003). J. Mol. Biol.332, 385–398. [DOI] [PubMed]
- Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994). Nucleic Acids Res.22, 4673–4680. [DOI] [PMC free article] [PubMed]
- Vriend, G. (1990). J. Mol. Graph.8, 52–56. [DOI] [PubMed]
- Wakamatsu, T., Nakagawa, N., Kuramitsu, S. & Masui, R. (2008). J. Bacteriol.190, 1108–1117. [DOI] [PMC free article] [PubMed]
- Xu, W. L., Shen, J. Y., Dunn, C. A., Desai, S. & Bessman, M. J. (2001). Mol. Microbiol.39, 286–290. [DOI] [PubMed]
- Yoshiba, S., Ooga, T., Nakagawa, N., Shibata, T., Inoue, Y., Yokoyama, S., Kuramitsu, S. & Masui, R. (2004). J. Biol. Chem.35, 37163–37174. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: DR2204, 2w4e, r2w4esf
PDB reference: DR2204, 2w4e, r2w4esf