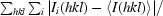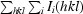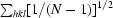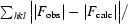The 1.45 Å structure of E. coli N-acetyl-d-neuraminic acid lyase in complex with pyruvate in space group P212121 is reported from new low-salt crystallization conditions that will facilitate soaking experiments with substrates and inhibitors.
Keywords: N-acetyl-d-neuraminic acid lyase, directed evolution, Schiff base, aldolases
Abstract
The structure of a mutant variant of Escherichia coli N-acetyl-d-neuraminic acid lyase (NAL), E192N, in complex with pyruvate has been determined in a new crystal form. It crystallized in space group P212121, with unit-cell parameters a = 78.3, b = 108.5, c = 148.3 Å. Pyruvate has been trapped in the active site as a Schiff base with the catalytic lysine (Lys165) without the need for reduction. Unlike the previously published crystallization conditions for the wild-type enzyme, in which a mother-liquor-derived sulfate ion is strongly bound in the catalytic pocket, the low-salt conditions described here will facilitate the determination of further E. coli NAL structures in complex with other active-site ligands.
1. Introduction
N-Acetyl-d-neuraminic acid lyase (NAL; EC 4.1.3.3) catalyses the reversible aldol condensation of pyruvate and N-acetyl-d-mannosamine to yield N-acetyl-d-neuraminic acid (Brunetti et al., 1962 ▶; Schauer et al., 1999 ▶). NAL belongs to the aldolase family, which can be subdivided into two classes on the basis of the reaction mechanism. In class I aldolases, including NAL, the reaction proceeds through the formation of a Schiff base between a protein lysine residue and the substrate pyruvate, whereas in class II aldolases the intermediates are stabilized by a metal cofactor (e.g. Zn+) (Plater et al., 1999 ▶). Many structures of class I aldolases have been reported and all share the same TIM-barrel fold (Izard et al., 1994 ▶; Wymer et al., 2001 ▶; Theodossis et al., 2004 ▶; Pauluhn et al., 2008 ▶). The structure of NAL from Escherichia coli has previously been determined in space group P3221 crystallized from a solution containing saturated ammonium sulfate (Izard et al., 1994 ▶). The presence of a sulfate ion strongly bound in the catalytic pocket of this structure has been considered to be the likely cause of the failure of substrate-soaking experiments. Crystals of a mutant E. coli NAL from low-salt conditions have been reported in complex with the inhibitor β-hydroxypyruvate (Joerger et al., 2003 ▶), but no substrate-complex structures are available. In contrast, NAL from Haemophilus influenzae crystallizes readily under low-salt conditions and several complexes with inhibitors or substrate analogues have been reported (Barbosa et al., 2000 ▶), although the pyruvate Schiff-base complex could only be trapped after reduction by sodium borohydride (Izard et al., 1994 ▶).
A range of analogues of N-acetyl-d-neuraminic acid are well established drugs for the treatment of influenza, targeting the sialidase (von Itzstein, 2007 ▶). Some potent inhibitors of influenza A sialidase have been discovered in which the glycerol side chain of N-acetyl-d-neuraminic acid has been replaced by a dialkylaminocarbonyl substituent (Smith et al., 1998 ▶; Taylor et al., 1998 ▶). Unfortunately, these modified ligands may not readily be prepared using wild-type E. coli NAL as the catalyst and require complex chemical synthesis. The enzyme has therefore been subjected to directed evolution (Williams et al., 2005 ▶) in order to broaden its substrate specificity. This programme resulted in the discovery of the mutant variant E192N, which has a sixfold higher specificity (k cat/K m) for dipropylaminocarbonyl-substituted derivatives than wild-type NAL has for its natural substrate sialic acid (Williams et al., 2005 ▶). We are pursuing X-ray structures of complexes of this NAL variant with substrates and inhibitors in order to identify the structural basis of the improved specificity. Here, we report a new crystal form of E. coli NAL obtained under low-salt conditions, which will allow us to carry out substrate-soaking experiments. We present the high-resolution structure (1.45 Å) of the variant E192N–pyruvate complex obtained by cocrystallization with pyruvate without the need for trapping by sodium borohydride reduction.
2. Experimental
The E. coli NAL variant E192N was isolated and purified as previously described (Williams et al., 2005 ▶). The protein was concentrated to 10 mg ml−1 and incubated for 1 h at 310 K with 100 mM sodium pyruvate before setting up crystallization trials.
Initial sitting-drop trials were performed by screening different commercial conditions from Hampton Research (Index, SaltRx, Crystal Screen and MembFac) and Emerald BioSystems (Wizard I and II) using an Oryx 6 robot (Douglas Instruments). The best crystals grew from 100 mM Tris–HCl pH 8.2, 200 mM ammonium acetate, 18% PEG 3350. Plate-shaped crystals appeared after 10 d at 277 K. Data were collected to a resolution of 1.45 Å from a single crystal at 100 K at the Diamond Synchrotron Light Source (station I04), were indexed using MOSFLM (Leslie, 2006 ▶) and were scaled and merged using SCALA (Evans, 2006 ▶) (Table 1 ▶). 5% of the reflections were excluded from the refinement and constituted the R free set. The structure was solved by molecular replacement using Phaser (McCoy, 2007 ▶) with the structure of the E. coli NAL wild-type protein as the search model (PDB code 1nal). Refinement was carried out with REFMAC (Murshudov et al., 1997 ▶). Stereochemical restraints for the ligand pyruvate bound to Lys165 were obtained from PRODRG (http://davapc1.bioch.dundee.ac.uk/prodrg/index.html), while TLS refinement was applied following determination of TLS domains using the TLSMD server (http://skuld.bmsc.washington.edu/~tlsmd/). Superposition of structures and calculation of their r.m.s. deviation were performed with LSQKAB (Collaborative Computational Project, Number 4, 1994 ▶). Water molecules were added in Coot (Emsley & Cowtan, 2004 ▶) for peaks over 3.0σ in the F o − F c map and structure validation was carried out with MolProbity (Davis et al., 2007 ▶).
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the outermost shell of the resolution range.
| Data collection | |
| Space group | P212121 |
| Unit-cell parameters () | a = 78.3, b = 108.5, c = 148.3 |
| Resolution () | 63.411.45 (1.531.45) |
| R merge † | 0.077 (0.375) |
| R p.i.m. ‡ | 0.036 (0.172) |
| I/sdI | 11.6 (3.4) |
| Completeness (%) | 96.2 (89.5) |
| Redundancy | 5.1 (4.9) |
| Refinement | |
| No. of reflections | 1095072 (138933) |
| No. of unique reflections | 213226 (28642) |
| R work § | 0.167 (0.280) |
| R free | 0.189 (0.290) |
| No. of atoms | 10677 |
| Protein | 9336 |
| Ligands | 34 |
| Water | 1307 |
| Average B factors (2) | |
| Protein | 14.4 |
| Covalently bound pyruvate | 10.8 |
| Noncovalently bound pyruvate | 28.7 |
| PEG 400 | 50.0 |
| Waters | 16.5 |
| R.m.s. deviations¶ | |
| Bond lengths () | 0.012 |
| Bond angles () | 1.346 |
| Ramachandran statistics†† (%) | |
| Most favoured | 100 |
| Outliers | 0 |
| PDB code | 2wkj |
3. Results and discussion
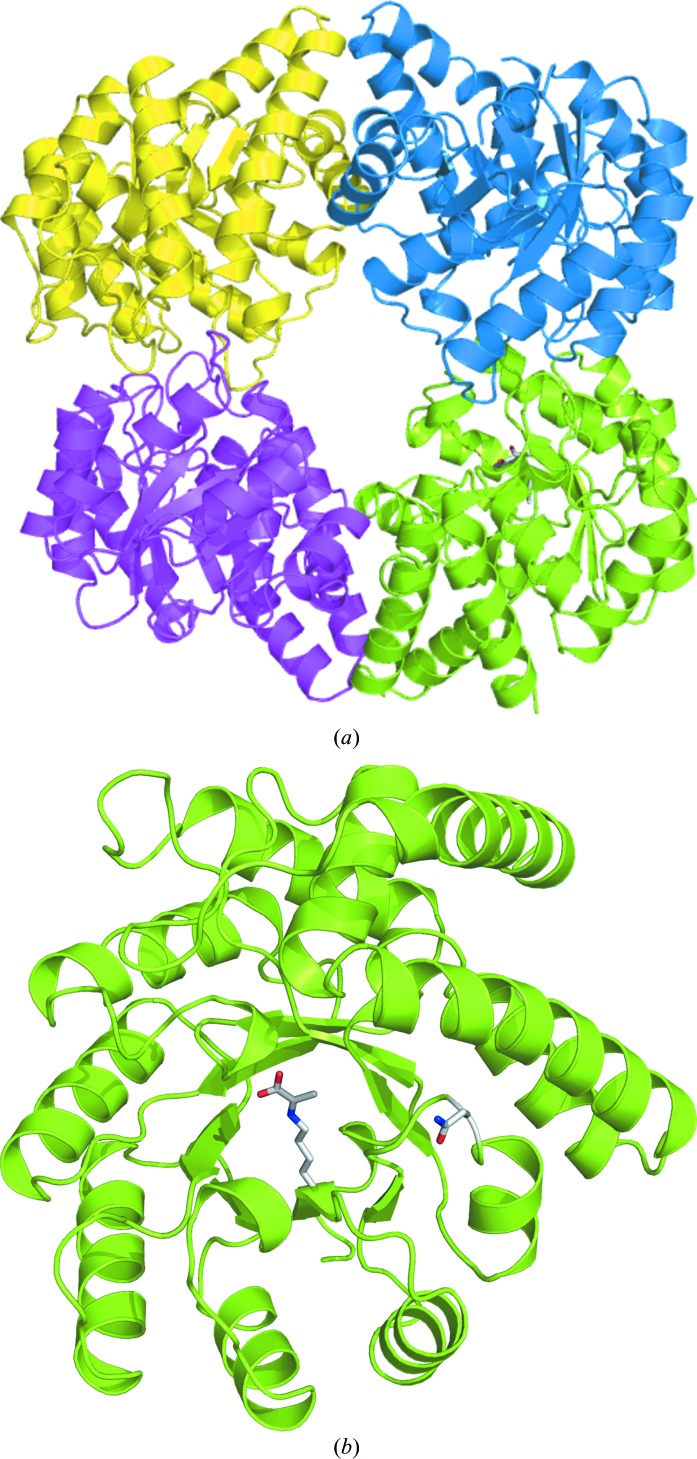
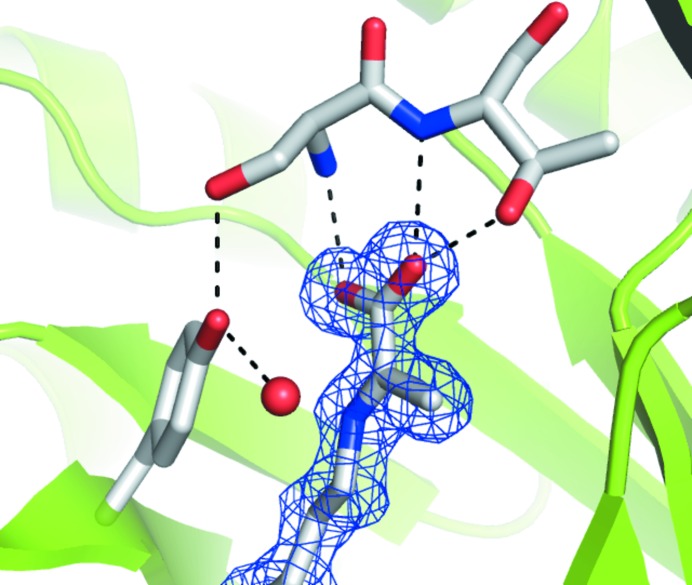
E192N NAL in space group P212121 has one noncrystallographic tetramer per asymmetric unit, with a very similar structure to that of the wild-type enzyme (PDB code 1nal) determined in space group P3221 (Fig. 1 ▶). The r.m.s.d. between these structures is only 0.5 Å for all atoms, despite the difference in space group and crystallization conditions. The structure is also very similar to that of the L142R β-hydroxypyruvate inhibitor-complex structure (PDB code 1hl2; 0.8 Å r.m.s.d. for all atoms; Joerger et al., 2003 ▶), which was determined under different low-salt conditions in space group P21 (100 mM sodium acetate pH 4.6 and 8% PEG 4000). For the first time for E. coli NAL we were able to trap the Schiff-base complex between pyruvate and Lys165 without the need for reduction with sodium borohydride. Schiff-base formation is confirmed by the planarity of the electron-density map for the conjugated pyruvate (Fig. 2 ▶). The hydrogen-bond network between pyruvate and Ser47 and Thr48, and between Tyr137 and Ser47 and a water (here water 48) is well conserved in all the NAL family members (Fig. 2 ▶). One additional noncovalently bound pyruvate molecule is present in the catalytic pocket of subunits C and D, probably owing to the high concentration of sodium pyruvate in the crystallization buffer. One molecule of cryoprotecting agent PEG 400 is also present in both chain A and chain C, lying on the twofold axis. We have also been able to obtain diffracting crystals of wild-type E. coli NAL under the same conditions (data not shown). These new low-salt crystallization conditions for E. coli NAL are particularly exciting as they will allow ligand- and inhibitor-soaking studies that were not possible under the previously published high-salt crystallization conditions. These will enable us to probe the origin of the broadened substrate specificity of the E192N variant.
Figure 1.
(a) Overall structure of the NAL variant E192N homotetramer showing each chain of the NAL variant E192N as a coloured ribbon. In the green-coloured monomer both the Schiff-base complex between Lys165 and pyruvate and the mutated residue E192N are shown as sticks coloured by atom type. (b) View of a single NAL monomer, showing the location of the lysine–pyruvate Schiff base and the E192N mutation within the α/β barrel. This figure was generated using PyMOL v.0.99 (DeLano, 2002 ▶).
Figure 2.
Electron density for the Schiff base between Lys165 and pyruvate. The planarity of the 2F o − F c map (contoured at 1.0 r.m.s.) is consistent with the formation of a Schiff base between Lys165 and pyruvate. The hydrogen-bond network between pyruvate and Thr48 and Ser47 and between Tyr137, Ser47 and a water is well conserved in all members of the NAL family.
Supplementary Material
PDB reference: N-acetyl-d-neuraminic acid lyase, 2wkj, r2wkjsf
Acknowledgments
IC is supported by a Wellcome Trust PhD Studentship. ARP is supported by a RCUK Fellowship. This work was supported by the BBSRC (BB/E000622/1). We thank the MX beamline staff at Diamond Light Source for assistance with data collection.
References
- Barbosa, J. A., Smith, B. J., DeGori, R., Ooi, H. C., Marcuccio, S. M., Campi, E. M., Jackson, W. R., Brossmer, R., Sommer, M. & Lawrence, M. C. (2000). J. Mol. Biol. 303, 405–421. [DOI] [PubMed]
- Brunetti, P., Jourdian, G. W. & Roseman, S. (1962). J. Biol. Chem. 237, 2447–2453. [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Davis, I. W., Leaver-Fay, A., Chen, V. B., Block, J. N., Kapral, G. J., Wang, X., Murray, L. W., Arendall, W. B. III, Snoeyink, J., Richardson, J. S. & Richardson, D. C. (2007). Nucleic Acids Res. 35, W375–W383. [DOI] [PMC free article] [PubMed]
- DeLano, W. L. (2002). The PyMOL Molecular Graphics System. http://www.pymol.org.
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Engh, R. A. & Huber, R. (1991). Acta Cryst. A47, 392–400.
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Itzstein, M. von (2007). Nature Rev. Drug Discov. 6, 967–974. [DOI] [PubMed]
- Izard, T., Lawrence, M. C., Malby, R. L., Lilley, G. G. & Colman, P. M. (1994). Structure, 2, 361–369. [DOI] [PubMed]
- Joerger, A. C., Mayer, S. & Fersht, A. R. (2003). Proc. Natl Acad. Sci. USA, 100, 5694–5699. [DOI] [PMC free article] [PubMed]
- Leslie, A. G. W. (2006). Acta Cryst. D62, 48–57. [DOI] [PubMed]
- McCoy, A. J. (2007). Acta Cryst. D63, 32–41. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Pauluhn, A., Ahmed, H., Lorentzen, E., Buchinger, S., Schomburg, D., Siebers, B. & Pohl, E. (2008). Proteins, 72, 35–43. [DOI] [PubMed]
- Plater, A. R., Zgiby, S. M., Thomson, G. J., Qamar, S., Wharton, C. W. & Berry, A. (1999). J. Mol. Biol. 285, 843–855. [DOI] [PubMed]
- Schauer, R., Sommer, U., Krueger, D., van Unen, H. & Traving, C. (1999). Biosci. Rep. 19, 373–383. [DOI] [PubMed]
- Smith, P. W. et al. (1998). J. Med. Chem. 41, 787–797. [DOI] [PubMed]
- Taylor, N. R., Cleasby, A., Singh, O., Skarzynski, T., Wonacott, A. J., Smith, P. W., Sollis, S. L., Howes, P. D., Cherry, P. C., Bethell, R., Colman, P. & Varghese, J. (1998). J. Med. Chem. 41, 798–807. [DOI] [PubMed]
- Theodossis, A., Walden, H., Westwick, E. J., Connaris, H., Lamble, H. J., Hough, D. W., Danson, M. J. & Taylor, G. L. (2004). J. Biol. Chem. 279, 43886–43892. [DOI] [PubMed]
- Williams, G. J., Woodhall, T., Nelson, A. & Berry, A. (2005). Protein Eng. Des. Sel. 18, 239–246. [DOI] [PubMed]
- Wymer, N., Buchanan, L. V., Henderson, A., Mehta, N., Botting, C. H., Pocivasvsek, L., Fierke, C. A., Toon, E. J. & Naismith, J. H. (2001). Structure, 9, 1–9. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: N-acetyl-d-neuraminic acid lyase, 2wkj, r2wkjsf