The invertase from Schwanniomyces occidentalis has been expressed in Saccharomyces cerevisiae, purified and crystallized. The wild-type enzyme was also purified and crystallized and diffraction data were collected to 2.9 Å resolution.
Keywords: yeast invertases, fructofuranosidases, glycoside hydrolase family 32
Abstract
Schwanniomyces occidentalis invertase is an extracellular enzyme that releases β-fructose from the nonreducing termini of various β-d-fructofuranoside substrates. Its ability to produce 6-kestose by transglycosylation makes this enzyme an interesting research target for applications in industrial biotechnology. The enzyme has been expressed in Saccharomyces cerevisiae. Recombinant and wild-type forms, which showed different glycosylation patterns, were crystallized by vapour-diffusion methods. Although crystallization trials were conducted on both forms of the protein, crystals suitable for X-ray crystallographic analyses were only obtained from the wild-type enzyme. The crystals belonged to space group P212121, with unit-cell parameters a = 105.78, b = 119.49, c = 137.68 Å. A diffraction data set was collected using a synchrotron source. Self-rotation function and sedimentation-velocity experiments suggested that the enzyme was dimeric with twofold symmetry.
1. Introduction
Enzymes that hydrolyse sucrose are collectively referred to as invertases or β-fructofuranosidases (EC 3.2.1.26) and catalyse the release of β-fructose from the nonreducing termini of various β-d-fructofuranoside substrates. Based on amino-acid sequence (Coutinho & Henrissat, 1999 ▶), they are classified into family 32 of the glycosyl hydrolases (GHs), which is included in the GH-J clan together with the GH68 (inulosucrase) family. To date, the three-dimensional structures of a β-fructofuranosidase from Thermotoga maritima (Alberto et al., 2004 ▶), an exoinulinase from Aspergillus niger (var. awamori; Nagem et al., 2004 ▶), a plant FEH from Chicorium intybus (Verhaest et al., 2005 ▶) and a cell-wall invertase from Arabidopsis thaliana (Lammens et al., 2008 ▶), all of which are members of the GH32 family, have been solved by X-ray crystallography. However, no member from the yeast family has been structurally analyzed to date. This is particularly remarkable taking into account that different oligomerization states have been described in yeast enzymes and this seems to regulate the in vivo enzymatic activity (Reddy et al., 1990 ▶). Therefore, the structural basis of this unique property will be of great biological interest.
In addition to releasing d-glucose and d-fructose from sucrose, microbial β-fructofuranosidases may catalyze the synthesis of short-chain fructooligosaccharides (FOS) in which one to three fructosyl moieties are linked to the sucrose by different glycosidic bonds depending on the enzyme source (Sangeetha et al., 2005 ▶). FOS containing β-(2→1) bonds (1-kestose, nystose and 1F-fructofuranosylnystose) are produced commercially in vast quantities owing to their prebiotic properties; they exert a beneficial effect on human health, being selectively fermented by colonic flora (Ghazi et al., 2005 ▶). Consequently, there is a great interest in the development of novel FOS with improved prebiotic and physiological properties. In this context, β-(2→6)-linked FOS (6-kestose being the first in the series) exhibit enhanced prebiotic properties compared with those of commercial FOS (Marx et al., 2000 ▶).
Schwanniomyces (Sw.) occidentalis (formerly Debaryomyces occidentalis) efficiently utilizes a wide variety of carbon compounds by employing a number of extracellular enzymes. This yeast has been used in biotechnology, where it has a high potential for enzyme production. An extracellular β-fructofuranosidase (SoInv) from Sw. occidentalis has been characterized (Álvaro-Benito et al., 2007 ▶). The enzyme shows broad substrate specificity, hydrolyzing sucrose, 1-kestose, nystose and raffinose, and it also produces several FOS by transfructosylation, the major product being the prebiotic trisaccharide 6-kestose. Accordingly, structural analysis of this protein is necessary in order to fully understand and improve its biological function and biotechnological potential. In this study, we describe the purification, crystallization and preliminary X-ray crystallographic analysis of the fructofuranosidase from Sw. occidentalis.
2. Experimental
2.1. Expression in an heterologous host and purification
The SoINV gene (X17604) encoding the invertase from Sw. occidentalis, a 75 kDa protein, was amplified by polymerase chain reaction using the forward primer 5′-TAGGATCCAACATGGTACAAGTTTTAAGTGTATTAG-3′ and the reverse primer 5′-TATCTTCATCAATAGACTATCTAGACTTATTTAGTTCCCT-3′. Restriction sites for BamHI and XbaI (shown in bold) were included in these primers for cloning of the PCR product into the pYES2.0/CT shuttle vector (Invitrogen). The resulting fusion protein including a 6×His tag was expressed in Saccharomyces cerevisiae EUROSCARF Y02321 (BY4741; Mat a; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; YIL162w::kanMX4) and was purified using Ni–NTA affinity resin (Qiagen) in accordance with the manufacturer’s recommendations. The purification of the recombinant cell-associated enzyme yields a final fraction of 3 mg ml−1. The homogeneity of the samples was confirmed by SDS–PAGE (Fig. 1 ▶). Fructofuranosidase activity was measured using sucrose as described previously (Álvaro-Benito et al., 2007 ▶).
Figure 1.
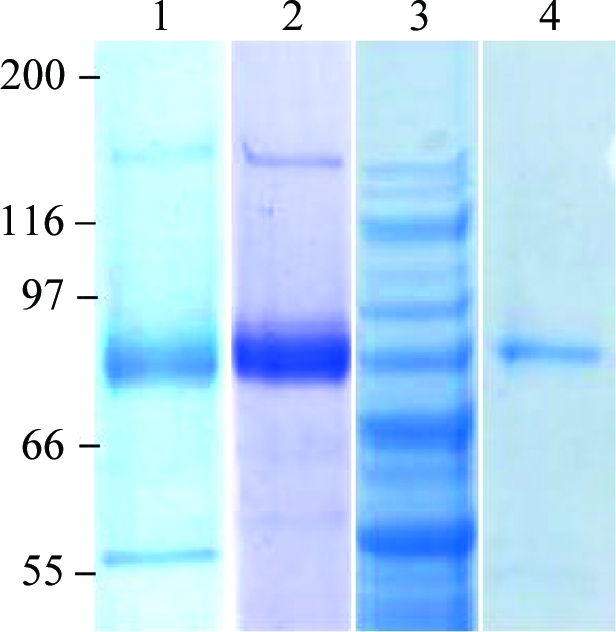
SDS–PAGE analysis of the fructofuranosidase from Sw. occidentalis. Lane 1, culture filtrate expressing fructofuranosidase activity before DEAE-Sephacel column chromatography pH 7; lane 2, after DEAE-Sephacel column chromatography. Lane 3, S. cerevisiae cellular extract expressing SoInv; lane 4, the purified enzyme from this organism. The positions of molecular-mass markers are indicated (in kDa) on the left.
Sw. occidentalis ATCC26077 was grown at 301–303 K on inulin-based medium [2%(w/v) yeast extract, 1.5%(w/v) inulin]. Growth was monitored spectrophotometrically at 600 nm (A 600nm). The fructofuranosidase (14.5 U ml−1) secreted in a 2.4 l culture (A 600nm = 8.9) was concentrated, fractionated and dialyzed (against 20 mM Tris–HCl pH 7.0) by tangential filtration through a 30 000 molecular-weight cutoff HY Viva Flow 50 system (Vivascience). The resultant fraction (53 ml, 607.45 U ml−1) was applied onto DEAE-Sephacel (25 ml). The protein was eluted with a 0–0.5 M NaCl gradient at a flow rate of 1.25 ml min−1 and the active fractions eluting at 0.15 M NaCl (20 ml, 1058.23 U ml−1) were pooled and concentrated to 7 mg ml−1 by ultracentrifugation using YM-30 membranes (Amicon, Millipore Corporation, Bedford, Massachusetts, USA) in a solution containing 10 mM Tris–HCl pH 7 (Fig. 1 ▶). All procedures were performed at room temperature (295 K).
2.2. Analysis of the molecular weight
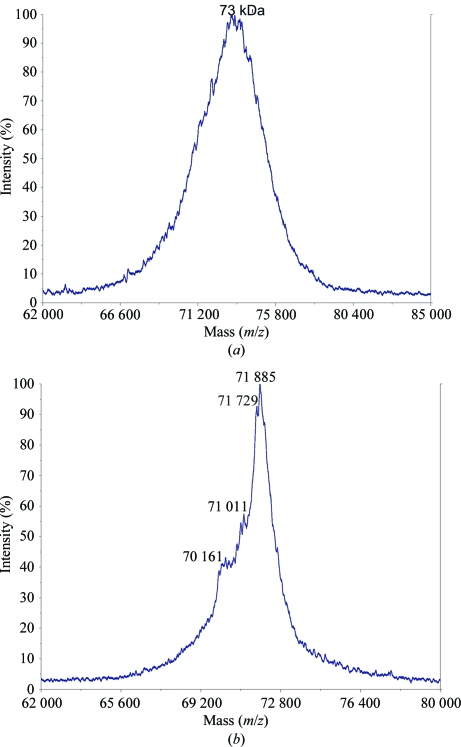
Matrix-assisted laser desorption ionization–time-of-flight (MALDI–TOF) mass spectra were recorded using an Applied Biosystems Voyager System 6214 mass spectrometer. Data were visualized and analyzed using the software supplied by Applied Biosystems (Fig. 2 ▶).
Figure 2.
MALDI–TOF mass spectra of (a) recombinant SoInv and (b) wild-type SoInv. The mass/charge ratios of the principal peaks are given. Note that in the MALDI–TOF technique dimers are separated into monomers.
2.3. Determination of the oligomeric state
Sedimentation-equilibrium experiments were performed in a Beckman Optima XL-A ultracentrifuge using a Ti 50 rotor and six-channel centrepieces of Epon charcoal (optical path length 12 mm). Samples of purified SoInv in the concentration range 0.2–0.5 mg ml−1 were equilibrated against 20 mM HEPES pH 7 and were centrifuged at 8000, 10 000 and 14 000 rev min−1 and 293 K. Radial scans at 280 nm were taken at 12, 14 and 16 h. The three scans were identical (equilibrium conditions were reached). The weight-average molecular mass (M w) of SoInv was determined using the program EQASSOC with the partial specific volume of SoInv set to 0.71 at 293 K as calculated from its amino-acid composition.
2.4. Crystallization
Initial crystallization conditions of the purified protein samples were investigated by high-throughput techniques with a NanoDrop robot (Innovadyne Technologies Inc.) using the commercially available Crystal Screens I, II and Lite, Index Screen and SaltRx from Hampton Research and PACT Suite and JCSG+ Suite from Qiagen. The assays were carried out by the sitting-drop vapour-diffusion method at 291 K on Innovaplate SD-2 microplates (Innovadyne Technologies Inc.) by mixing 250 nl protein solution with 250 nl precipitant solution and equilibrating against 60 µl well solution. Many PEG 3350-containing solutions gave needle-like crystals and these conditions were optimized through further sitting-drop experiments by mixing 1 µl protein solution with 1 µl precipitant solution and equilibrating against 500 µl well solution on Cryschem plates (Hampton Research).
2.5. Data collection and processing
All crystals were soaked in precipitant solutions additionally containing 30% glycerol before being flash-cooled to 100 K. Crystals were tested using in-house facilities or synchrotron radiation at various beamlines. Diffraction data sets were collected at ESRF (Grenoble, France). The data sets were processed using the program MOSFLM (Leslie, 1990 ▶) and the CCP4 suite (Collaborative Computational Project, Number 4, 1994 ▶).
3. Results and discussion
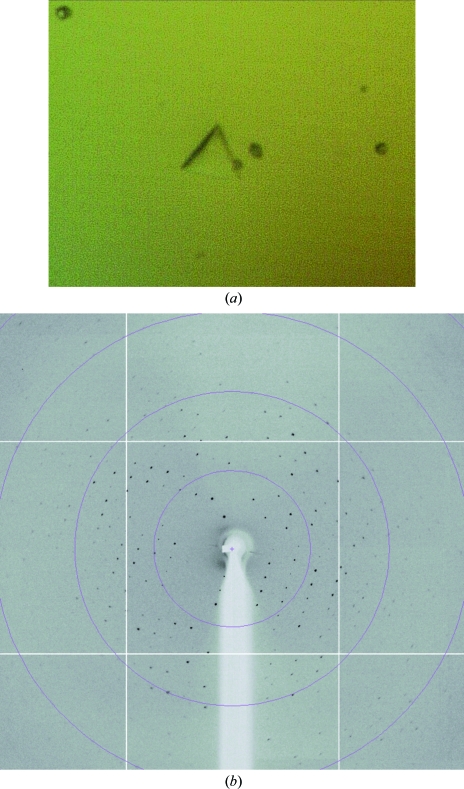
Recombinant SoInv shows a different glycosylation pattern to that obtained from the wild-type enzyme, which presents a broad peak in the mass spectrum with a maximum of 73 kDa. A preliminary search for crystallization conditions with the recombinant enzyme led to needle-like crystals that were subsequently refined by varying several parameters, e.g. the buffer type, pH, temperature, sample size and the type and concentration of precipitant agent. Other crystal habits such as plates, rods or even crystals with rhombohedral shape were then observed under different conditions with a PEG 3350 concentration of between 18 and 25% and acidic pH values from 4 to 5.5, most of which grew to very small sizes and failed to diffract on various synchrotron beamlines. Screening of many additives (Additive Screens from Hampton Research) was also attempted but did not improve the crystal quality. Only one crystal (Fig. 3 ▶) grown from 21% PEG 3350, 3% 2-propanol and 0.1 M citrate buffer pH 4.2 reached dimensions that allowed a data set to be collected to 3 Å on the ESRF (Grenoble, France) ID14-4 beamline. This data set indexed in the apparent space group H32, with unit-cell parameters a = b = 300, c = 112 Å and two molecules in the asymmetric unit. However, the very poor quality of the data at resolution higher than 4.5 Å prevented further analysis.
Figure 3.
(a) Crystal of recombinant SoInv with dimensions of 0.10 × 0.10 × 0.02 mm. (b) X-ray diffraction pattern of this crystal obtained using a synchrotron source. The outer resolution shell is 5.3 Å.
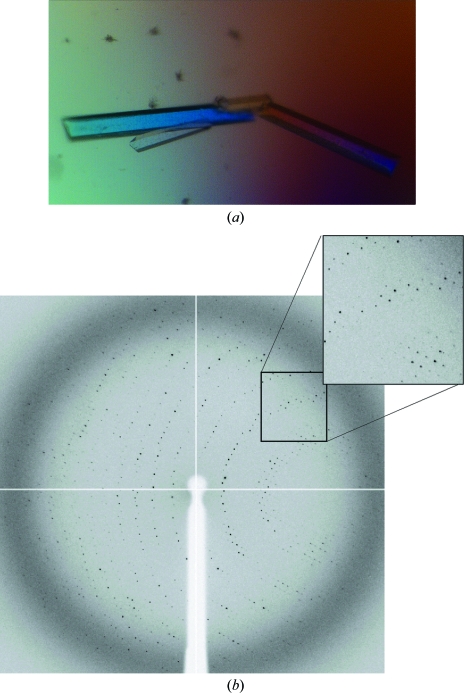
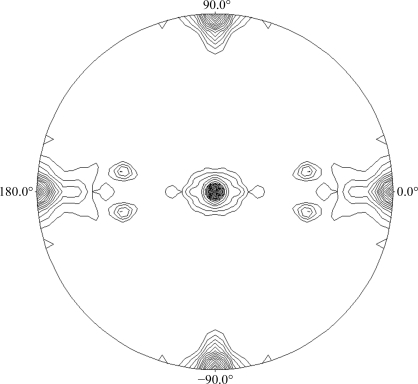
As the wild-type sample became available, new conditions were screened and best results were again obtained with PEG 3350 as precipitant agent, but at lower concentrations and a higher pH, ranging from 7 to 8. Long regularly shaped crystals were obtained using 14–16% PEG 3350 and 0.1 M HEPES pH 7, reaching maximum dimensions of 0.6 × 0.2 × 0.1 mm (Fig. 4 ▶) in 10–15 d. A full data set was collected to 2.9 Å resolution on the ESRF ID14-1 beamline. The data-collection statistics are given in Table 1 ▶. The unit-cell parameters were a = 105, b = 119, c = 134 Å in space group P212121. As seen in the mass-spectrometric analysis, the molecular weight of the monomer is 72 kDa and therefore, assuming the presence of two molecules in the asymmetric unit, the Matthews coefficient (Matthews, 1968 ▶) of 2.98 Å3 Da−1 leads to a 60% solvent content within the cell. We have investigated the local symmetry relating the units in the asymmetric unit using POLARRFN (Kabsch, 1976 ▶) from the CCP4 package. Several self-rotation functions were computed in the resolution range 15–3 Å, with Patterson vectors from 25 to 40 Å radius of integration. Analysis of the self-rotation peaks revealed the presence of noncrystallographic twofold symmetry near the ac plane. The stereographic projection (κ = 180° section) of the self-rotation is shown in Fig. 5 ▶. As the analytical centrifugation experiments gave an estimated molecular weight of 140 000, which indicates a dimeric aggregation state for SoInv, a functional dimer is expected to form the asymmetric unit of the crystals. Structure determination was initiated using the exo-inulinase from Aspergillus awamori (PDB code 1y4w), which shows 35% sequence identity, as a model. Molecular replacement was performed using the MOLREP program (Vagin & Teplyakov, 1997 ▶) with a corrected model using the SoInv sequence and data to 3.5 Å resolution. The rotation and translation functions yielded two clear positions for the model, with a final correlation coefficient of 0.49 and an R factor of 0.49. Structural refinement of the SoInv model is currently in progress.
Figure 4.
(a) Crystals of wild-type SoInv with maximum dimensions of 0.60 × 0.20 × 0.10 mm. (b) X-ray diffraction pattern for a crystal of wild-type SoInv obtained using a synchrotron source. The maximum observed resolution is 2.9 Å.
Table 1. Data-collection statistics for the SoInv crystal.
Values in parentheses are for the highest resolution shell.
| Wavelength (Å) | 0.9334 |
| Source | ESRF |
| Beamline | ID14-1 |
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 105.78, b = 119.49, c = 137.68 |
| Limiting resolution (Å) | 2.9 (2.95–2.90) |
| Unique reflections | 36939 |
| Rmerge† | 0.16 (0.48) |
| Completeness (%) | 99.7 (99.8) |
| Mean multiplicity | 6.9 (6.7) |
| Mean I/σ(I) | 9.7 (2.9) |
| Wilson B factor (Å2) | 52.9 |
R
merge(I) = 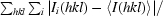
 , where Ii(hkl) is the ith observed amplitude of reflection hkl and 〈I(hkl)〉 is the mean amplitude for measurements of reflection hkl.
, where Ii(hkl) is the ith observed amplitude of reflection hkl and 〈I(hkl)〉 is the mean amplitude for measurements of reflection hkl.
Figure 5.
Plot of the self-rotation function of SoInv crystals using data between 15 and 3 Å resolution and a 25 Å radius of integration in the κ = 180° section. The view is down the c axis. ϕ = 0° and ϕ = 90° correspond to the a and b* axes, respectively.
Acknowledgments
This work was supported by grants BIO2007-67708-C04-04 and BIO2007-67708-C04-03 from Dirección General de Investigación and by an institutional grant from the Fundación Ramón Areces to the Centro de Biología Molecular Severo Ochoa. This is a product of the Project ‘Factoría Española de Cristalización’ Ingenio/Consolider 2010.
References
- Alberto, F., Bignon, C., Sulzenbacher, G., Henrissat, B. & Czjzek, M. (2004). J. Biol. Chem.279, 18903–18910. [DOI] [PubMed]
- Álvaro-Benito, M., de Abreu, M., Fernandez-Arrojo, L., Plou, F. J., Jiménez-Barbero, J., Ballesteros, A., Polaina, J. & Fernández-Lobato, M. (2007). J. Biotechnol.132, 75–81. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Coutinho, P. M. & Henrissat, B. (1999). Carbohydrate-Active Enzymes Server. http://afmb.cnrs-mrs.fr/CAZY/.
- Ghazi, I., Gómez de Segura, A., Fernández-Arrojo, L., Alcalde, M., Yates, M., Rojas-Cervantes, M. L., Plou, F. J. & Ballesteros, A. (2005). J. Mol. Catal. B. Enzym.35, 19–27.
- Kabsch, W. (1976). Acta Cryst. A32, 922–923.
- Lammens, W., Le Roy, K., Van Laere, A., Rabijns, A. & Van den Ende, W. (2008). J. Mol. Biol.377, 378–385. [DOI] [PubMed]
- Leslie, A. G. W. (1990). Crystallographic Computing 5: From Chemistry To Biology, edited by D. Moras, A. D. Podjarny & J. C. Thierry. Oxford University Press.
- Marx, S. P., Winkler, S. & Hartmeier, W. (2000). FEMS Microbiol. Lett.182, 163–169. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Nagem, R. A. P., Rojas, A. L., Golubev, A. M., Korneeva, O. S., Eneyskaya, E. V., Kulminskaya, A. A., Neustroev, K. N. & Polikarpov, I. (2004). J. Mol. Biol.120, 621–623. [DOI] [PubMed]
- Reddy, A. V., MacColl, R. & Maley, F. (1990). Biochemistry, 29, 2482–2487. [DOI] [PubMed]
- Sangeetha, P. T., Ramesh, M. N. & Prapulla, S. G. (2005). Process. Biochem.40, 1085–1088.
- Vagin, A. & Teplyakov, A. (1997). J. Appl. Cryst.30, 1022–1025.
- Verhaest, M., Van den Ende, W., Le Roy, K., De Ranter, C. J., Van Laere, A. & Rabijns, A. (2005). Plant J.41, 401–411. [DOI] [PubMed]