Mutations in the human AAA+ protein p97 cause a disease in humans called IBMPFD. How these mutations affect the structure and function of p97 is unknown. Here, the crystallization of three disease-related mutants of p97 in the presence of ATPγS are reported.
Keywords: p97, AAA+ proteins, disease-related mutants
Abstract
The human type II AAA+ protein p97 participates in various cellular activities, presumably through its involvement in the ubiquitin–proteasome degradation pathway. Mutations in p97 have been implicated in patients with inclusion-body myopathy associated with Paget’s disease of the bone and frontotemporal dementia (IBMPFD). In this work, three mutant p97 N-D1 fragments, R86A, R95G and R155H, were crystallized in the presence of ATPγS with PEG 3350 as a main precipitant, yielding two different crystal forms. The R155H mutant crystal belonged to space group R3, with unit-cell parameters in the hexagonal setting of a = b = 134.2, c = 182.9 Å, and was merohedrally twinned, with an estimated twin fraction of 0.34. The crystals of the R86A and R95G mutants belonged to space group P1, with similar unit-cell parameters of a = 90.89, b = 102.6, c = 107.2 Å, α = 97.5, β = 90.6, γ = 91.5° and a = 92.76, b = 103.7, c = 107.7 Å, α = 97.7, β = 91.9, γ = 89.7°, respectively.
1. Introduction
Human p97 belongs to the AAA+ (extended family of ATPases associated with a variety of cellular activities) family of proteins, which hydrolyze ATP to induce conformational changes and use the resulting mechanical energy for substrate remodeling (Neuwald et al., 1999 ▶). p97 belongs to the type II AAA+ proteins, featuring two AAA ATPase domains in tandem. It is highly abundant in the cytoplasm (Peters et al., 1990 ▶) and takes part in various cellular pathways such as ER-associated degradation, homotypic membrane fusion and cell-cycle control, often with the assistance of various adaptor proteins such as p47 and Ufd1-Npl4 (Halawani & Latterich, 2006 ▶; Ye, 2006 ▶; Uchiyama & Kondo, 2005 ▶). Despite its apparent multivalent activities, p97 presumably carries out these functions through the ubiquitin–proteasome degradation pathway, in which p97 recognizes and shuttles polyubiquitinated proteins to proteasomes for degradation (Wang et al., 2004 ▶). Autosomal inclusion-body myopathy associated with Paget’s disease of the bone and frontotemporal dementia (IBMPFD) has been linked to mutations in p97 (Kimonis et al., 2000 ▶). To date, mutations of p97 at positions Arg93, Arg95, Arg155, Gly157, Arg159, Arg191, Leu198, Ala232, Thr262 or Asn387 have been identified in patients with IBMPFD (Kimonis et al., 2008 ▶; Djamshidian et al., 2009 ▶). Inclusion bodies containing polyubiquitinated proteins and p97 have been found in tissues from IBMPFD patients, a fact that is consistent with the role of p97 in the ubiquitin–proteasome degradation pathway (Schroder et al., 2005 ▶).
The structure of p97 consists of three domains. The N-terminal domain (N-domain) interacts with different protein partners such as p47 (Dreveny et al., 2004 ▶) and Ufd1-Npl4 (Pye et al., 2007 ▶), which allows p97 to function in different pathways. Following the N-domain are two tandem AAA ATPase domains, the D1-domain and the D2-domain, with linker regions between them. In the wild-type structures that have been determined of p97 (Zhang et al., 2000 ▶; DeLaBarre & Brunger, 2003 ▶; Huyton et al., 2003 ▶), the D1-domains and D2-domains form two stacked hexameric rings, with the N-domains radiating outward in the same plane as the D1-domain ring. Interestingly, in all the p97 structures published to date the D1-domains are always occupied by ADP, while the D2-domains are either bound with ADP–AlFx or AMP-PNP or are nucleotide-free (Zhang et al., 2000 ▶; DeLaBarre & Brunger, 2003 ▶, 2005 ▶; Huyton et al., 2003 ▶). To understand how IBMPFD mutations might affect the structure and function of p97, we purified three N-D1 fragments of p97 bearing the IBMPFD mutations R86A, R95G and R155H, and crystallized each in the presence of ATPγS.
2. Materials and methods
2.1. Construction of wild-type p97 and IBMPFD mutant p97 N-D1 expression plasmids
The DNA fragment encoding full-length p97 was amplified from a p97 clone (accession No. BC110913) available from Thermo Scientific (Huntsville, Alabama, USA) using the first primer pair listed in Table 1 ▶ (Nos. 1 and 2). The primers contain restriction sites for BamHI and BglII, respectively. The PCR product was cloned into a pQE60 expression vector (Qiagen, Valencia, California, USA) through the BamHI/BglII sites, generating the expression plasmid pQE-p97. A hexahistidine-affinity tag was attached to the C-terminus of p97 via a two-residue linker Arg-Ser (RSH6 tag). The p97 DNA fragment encoding Met1–Gly481 (N-D1 fragment) was then generated by deletion mutagenesis, removing residues 482–806 from the plasmid pQE-p97, by using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, California, USA) with the second primer pair (Nos. 3 and 4) listed in Table 1 ▶. The resulting PCR product is the expression plasmid pQE-p97ND1 containing the wild-type p97 N-D1 fragment inserted between BamHI/BglII sites in the pQE60 vector with a C-terminal RSH6 affinity tag. The N-D1 fragments for mutants R86A, R95G and R155H were obtained using the QuikChange site-directed mutagenesis kit with the primer pairs listed in Table 1 ▶. Constructs were confirmed by DNA sequencing.
Table 1. Primers used in cloning and mutagenesis.
| No. | Mutations | Direction | Sequence | Restriction site |
|---|---|---|---|---|
| 1 | Wt FL† | Forward | TGAGGATCCATGGCTTCTGGAGCCGATTC | BamHI |
| 2 | Wt FL† | Reverse | TCAAGATCTGCCATACAGGTCATCATCATTGTC | BglII |
| 3 | Wt N-D1 | Forward | ACCTGGGAAGACATCGGGGGCAGATCTCATCACCATCACCATC | BglII |
| 4 | Wt N-D1 | Reverse | GATGGTGATGGTGATGAGATCTGCCCCCGATGTCTTCCCAGGT | BglII |
| 5 | R53A | Forward | GAATTGCAGTTGTTCCGCGGTGACACAGTGTTGC | — |
| 6 | R53A | Reverse | GCAACACTGTGTCACCGCGGAACAACTGCAATTC | — |
| 7 | R86A | Forward | CTTGTTCTGATGAGAAGATTCGGATGAATGCTGTTGTTCGGAATAACCTTCG | — |
| 8 | R86A | Reverse | CGAAGGTTATTCCGAACAACAGCATTCATCCGAATCTTCTCATCAGAACAAG | — |
| 9 | R95G | Forward | GTTCGGAATAACCTTCGTGTAGGTCTAGGGGATGTCATCAGCATCC | — |
| 10 | R95G | Reverse | GGATGCTGATGACATCCCCTAGACCTACACGAAGGTTATTCCGAAC | — |
| 11 | R155H | Forward | GGAGACATTTTTCTTGTCCATGGTGGGATGCGTGCTGTG | — |
| 12 | R155H | Reverse | CACAGCACGCATCCCACCATGGACAAGAAAAATGTCTCC | — |
Wild-type full-length p97.
2.2. Expression and purification of p97 N-D1 fragments
Expression plasmids were transformed into Escherichia coli strain M15 (QIAexpression system, Qiagen). Transformed cells were grown at 310 K in Terrific Broth supplemented with 100 µg ml−1 ampicillin and 50 µg ml−1 kanamycin and were induced by 1 mM isopropyl β-d-1-thiogalactopyranoside at an OD600 of 1.0. Cells were harvested by centrifugation and the pellet was resuspended in buffer A (25 mM Tris–HCl pH 7.6, 0.3 mM NaCl, 10 mM imidazole and 0.25 mM DTT) supplemented with a protease-inhibitor cocktail (Santa Cruz Biotechnology, Santa Cruz, California, USA). Cells were lysed by sonication followed by centrifugation to remove cell debris at 15 000g for 40 min at 277 K. Subsequently, the supernatant was incubated with Ni–NTA resin (Qiagen) pre-equilibrated with buffer A for 1 h at 277 K. Unbound materials were washed away in two steps with buffer A supplemented with 50 mM and then 100 mM imidazole and bound proteins were eluted with 250 mM imidazole in buffer A. After elution, proteins were concentrated, dialyzed against a buffer containing 20 mM Tris–HCl pH 8.0, 10% glycerol and stored at 193 K until use. All proteins were purified using the same procedure. The hexahistidine tag remained uncleaved at the C-terminus in all purified proteins. The protein concentration was determined using BCA methods (Smith et al., 1985 ▶).
2.3. Oligomeric state of p97 N-D1 fragments
The oligomeric states of the p97 variants were analyzed by both native gel electrophoresis using blue native PAGE (Invitrogen, Carlsbad, California, USA) and gel-filtration chromatography using a Superdex 200 column (GE Healthcare, Piscataway, New Jersey, USA) in a buffer containing 20 mM Tris–HCl pH 8.0 and 10% glycerol.
2.4. Crystallization of mutant p97 N-D1 fragments
Prior to crystallization trials, all protein samples were first treated with 5 mM DTT followed by the addition of MgCl2 and ATPγS to final concentrations of 40 and 4 mM, respectively. The mixture was centrifuged at 12 600g for 1 h and the supernatant was used for crystallization. Mutant p97 N-D1 fragments were crystallized by the hanging-drop vapor-diffusion method at 288 K. Typically, for each mutant an initial crystallization screening was performed robotically with a Mosquito automated solution dispenser (TTP LabTech, Royston, England) using commercially available high-throughput screening kits (from Sigma, Hampton Research and Molecular Dimensions) in hanging-drop format. Each droplet consisted of a mixture of 300 nl protein solution and 300 nl reservoir solution and was equilibrated over 50 µl reservoir solution. The conditions that yielded initial hits were repeated and confirmed with solutions prepared in-house.
2.5. Cryoprotection and X-ray diffraction data collection
The crystals of the p97 N-D1 mutants were fragile and difficult to freeze cryogenically. A large number of cryoprotection agents such as glycerol, various ethylene glycols, Paratone oil mixtures etc. were screened to search for the best conditions. Only a few crystals survived the treatment while retaining sufficient diffraction resolution for the collection of complete data sets. Mutant p97 crystals consistently performed better on soaking them for 3 min in buffer solutions containing 10 mM Tris–HCl pH 8.0, 2% benzamidine, 50 mM citrate buffer pH 5.8, 20 mM MgCl2, 5 mM DTT, 20% PEG 3350 with increasing amounts of glycerol. This stepwise process (typically 15 steps) continued until a final concentration of 28% glycerol was reached. The crystals were then flash-frozen in liquid propane.
X-ray diffraction experiments were carried out at 100 K on the SER-CAT (ID-22) beamline at the Advanced Photon Source at Argonne National Laboratory. Diffraction images were collected with a MAR 225 CCD detector and processed and scaled with the HKL-2000 package (Otwinowski & Minor, 1997 ▶). Statistics of X-ray diffraction data collection and for the qualities of data sets are given in Table 2 ▶.
Table 2. Statistics for the quality of diffraction data sets for mutant p97.
All data sets were collected on SERCAT beamline ID22 at Argonne National Laboratory. Values in parentheses are for the last shell.
| Data set | R155H | R95G | R86A |
|---|---|---|---|
| Space group | R3† | P1 | P1 |
| Bound nucleotide | ATPγS | ATPγS | ATPγS |
| Unit-cell parameters (Å, °) | a = b = 134.2, c = 182.9† | a = 92.76, b = 103.3, c = 107.7, α = 97.7, β = 91.9, γ = 89.7 | a = 90.89, b = 102.6, c = 107.2, α = 97.5, β = 90.6, γ = 91.5 |
| Resolution (Å) | 50–2.1 (2.23–2.10) | 50–2.8 (2.90–2.80) | 50–2.85 (2.95–2.85) |
| Mosaicity (°) | 0.76 | 0.82 | 0.96 |
| No. of observations | 235711 | 151406 | 200205 |
| No. of unique reflections | 66557 | 87240 | 85163 |
| Completeness (%) | 99.5 (95.6) | 90.5 (71.5) | 91.8 (67.8) |
| Rmerge‡ | 0.057 (0.524) | 0.050 (0.493) | 0.049 (0.502) |
| I/σ(I) | 19.7 (1.7) | 12.3 (0.8) | 13.8 (1.02) |
| Phasing | |||
| Model used | PDB entry 1e32 | R155H N-D1 fragment | R155H N-D1 fragment |
| Log-likelihood gain | 446§ | 5792¶ | 6717¶ |
| R in initial rigid-body refinement | 0.43 | 0.38 | 0.37 |
| FOM | 0.36 | 0.59 | 0.62 |
R3 space group in hexagonal setting.
R
merge is defined as  −
− 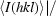
 , where I
i(hkl) is the intensity of the ith observation of a reflection with Miller indices hkl and 〈I(hkl)〉 is the mean intensity for all measured I(hkl) and Friedel pairs.
, where I
i(hkl) is the intensity of the ith observation of a reflection with Miller indices hkl and 〈I(hkl)〉 is the mean intensity for all measured I(hkl) and Friedel pairs.
MR was performed with Phaser using the automatic MR option. Two PDB files were provided: one for the D1-domain and another for the N-domain. We asked for two copies each of the D1-domain and N-domain. A single solution was obtained. A search with an intact N-D1 from PDB entry 1e32 produced a wrong solution with a log-likelihood gain of −42.
A monomeric N-D1 fragment from the R155H mutant structure was used as a phasing model, asking for six N-D1 subunits in a crystallographic asymmetric unit. A single solution was obtained.
2.6. Structure determination of the N-D1 mutants
The structure of the R155H mutant with bound ATPγS was determined by molecular replacement (MR) using the program Phaser (Storoni et al., 2004 ▶). Merohedral twinning in the data set was detected using the twinning server (http://nihserver.mbi.ucla.edu/Twinning; Yeates, 1997 ▶) and the data set was detwinned using the program DETWIN in the CCP4 program package (Collaborative Computational Project, Number 4, 1994 ▶). The structure was further refined with the program REFMAC (Murshudov et al., 1997 ▶) using the detwinned data set. Bound ligands were inserted manually with the graphical modeling program O (Jones et al., 1991 ▶). The structures of both the R95G and R86A mutants in the ATPγS-bound form were also solved with Phaser using a monomeric template from the R155H mutant as a search model.
3. Results and discussion
Three wild-type structures of p97 are available and all are hexameric: one is an N-D1 fragment (Zhang et al., 2000 ▶) and the other two are full-length proteins (DeLaBarre & Brunger, 2003 ▶; Huyton et al., 2003 ▶). Interestingly, ADP molecules are bound to the D1-domains in all of them, regardless of the nucleotide form used for crystallization. In the present study, three N-D1 fragments of IBMPFD mutant p97 (R86A, R95G and R155H) were crystallized in the presence of ATPγS and their structures were determined, with a highest resolution of 2.1 Å.
3.1. Mutant p97 N-D1 fragments form hexamers in solution
Purified mutant proteins were subjected to blue native PAGE analysis (Ma & Xia, 2008 ▶; Wittig et al., 2006 ▶). All mutants formed hexamers in solution, with an estimated molecular weight of 333.6 kDa (Fig. 1 ▶ a), suggesting that they behave just like the wild-type N-D1 fragment. When subjected to SDS–PAGE analysis, the same protein migrated as monomers with a molecular weight of 54.5 kDa (Fig. 1 ▶ b).
Figure 1.
Oligomeric states of purified N-D1 fragments of p97 mutant R155H. (a) Blue native PAGE analysis with NativeMarker (Invitrogen) as molecular-weight standards. (b) SDS–PAGE analysis with SeeBlue-Plus2 (Invitrogen) as molecular-weight standards.
3.2. p97 mutants crystallize in two different crystal forms
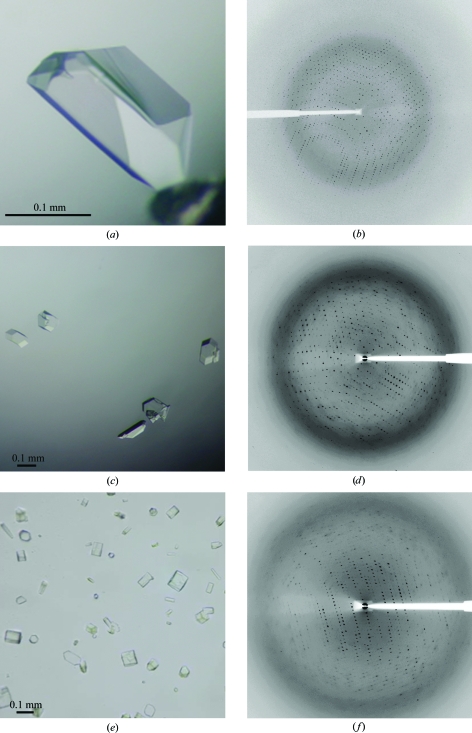
Our robotic screens began with the R155H mutant N-D1 at a protein concentration of 10 mg ml−1 in a buffer containing 20 mM Tris–HCl pH 8.0 and 20% glycerol. An initial set of conditions was identified from the Sigma HT screen kit, which gave rise to small crystals (condition A9; 0.2 M ammonium acetate, 0.1 mM sodium citrate pH 5.6 and 30% PEG 4000). This set of conditions was subsequently refined with various PEG and additive screens. The best crystals for the R155H mutant were obtained by mixing the protein (7 mg ml−1 in 20 mM Tris–HCl pH 8.0, 40 mM MgCl2, 4 mM ATPγS, 5 mM DTT and 10% glycerol) in a 1:1 ratio with a reservoir solution containing 0.1 M citrate buffer pH 5.6, 6% benzamidine, 7% PEG 3350 and 20% glycerol. Similarly, crystals of the R95G mutant were obtained by mixing the protein (5 mg ml−1 in 10 mM Tris–HCl pH 8.0, 40 mM MgCl2, 4 mM ATPγS, 5 mM DTT and 10% glycerol) in an equal ratio with a reservoir solution consisting of 0.1 M citrate buffer pH 5.7, 0.1 M NaCl, 13.5% PEG 3350 and 20% glycerol. The R86A mutant (5 mg ml−1 in 20 mM Tris–HCl pH 8.0, 40 mM MgCl2, 4 mM ATPγS, 5 mM DTT and 10% glycerol) was crystallized with a well solution containing 0.1 M citrate buffer pH 5.8, 0.1 M NaCl, 4% benzamidine, 16.5% PEG 3350 and 20% glycerol. Crystals appeared a few days after setup and reached full size (0.1–0.2 mm) in about 2–3 weeks in hanging drops.
After treatment with cryoprotectant, the mutant p97 crystals were frozen in liquid propane and subjected to an X-ray diffraction screen. Most crystals were not protected well by the procedures described in §2 and diffracted X-rays poorly. Occasionally, crystals diffracted to 3 Å resolution or better and complete data sets were collected. R155H mutant crystals could grow to substantial sizes (Fig. 2 ▶ a) and displayed the symmetry of space group R3. The best R155H mutant crystal we obtained diffracted X-rays to 2.1 Å resolution (Table 2 ▶). There are two copies of N-D1 in the asymmetric unit, giving rise to a Matthews coefficient (V M) of 2.96 Å3 Da−1, corresponding to a solvent content of 58% (Matthews, 1968 ▶). Both the R95G and R86A mutant crystals behaved much more poorly than those of R155H in diffraction experiments, diffracting X-rays to only 2.8 and 2.85 Å resolution, respectively. They have very similar unit-cell parameters with one hexamer in the unit cell, corresponding to a V M value of 3.19 Å3 Da−1 and a solvent content of 61%. Statistics of data collection and data quality are given in Table 2 ▶.
Figure 2.
Crystals and X-ray diffraction patterns for N-D1 fragments of p97 mutants. (a) Typical crystals of the R155H mutant grown in the presence of ATPγS. (b) X-ray diffraction pattern of an R155H mutant crystal. (c) Crystals of the R95G mutant grown in the presence of ATPγS. (d) Diffraction image of an R95G mutant crystal. (e) Crystals of the R86A mutant grown in the presence of ATPγS. (f) Diffraction pattern of an R86A mutant crystal.
3.3. Structure solution of the R155H mutant in the presence of merohedral twinning
We began our structure determination with the data set from the R155H mutant crystal because it had the highest resolution. Molecular replacement (MR) using a model template based on the wild-type N-D1 fragment with bound ADP (PDB code 1e32; Zhang et al., 2000 ▶) failed to yield successful solutions for the expected two NCS-related copies. Assuming that the mutations and/or the presence of ATPγS might have changed the relative positions of the N-domain and D1-domain, we carried out MR using separate templates for the N-domain and D1-domain and found two solutions for each domain. The structure was subsequently refined and rebuilt in many cycles with the addition of new features, but the free R value remained high at approximately 38%. The existence of merohedral twinning in the R3 crystal was detected; the data set was detwinned with an initial estimated twin fraction of 0.34 and a twin operator of k, −h, l (in the hexagonal setting). Structure refinement against the detwinned data set is in progress.
3.4. Structure determination of the R86A and R95G mutants by molecular replacement
The structures of both the R95G and R86A mutants in the ATPγS-bound form were solved by molecular replacement with Phaser using a monomeric template from the high-resolution R155H mutant as a search model. Six copies of the p97 N-D1 protomer are present in the unit cell. Structure refinement and analysis of the two mutants of p97 with ATPγS bound are in progress.
Acknowledgments
The authors wish to thank the staff members of the SER-CAT beamline (ID-22) at Advanced Photon Source, Argonne National Laboratory for their assistance in data collection. We also thank George Leiman for editorial assistance. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
References
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- DeLaBarre, B. & Brunger, A. T. (2003). Nature Struct. Biol.10, 856–863. [DOI] [PubMed]
- DeLaBarre, B. & Brunger, A. T. (2005). J. Mol. Biol.347, 437–452. [DOI] [PubMed]
- Djamshidian, A., Schaefer, J., Haubenberger, D., Stogmann, E., Zimprich, F., Auff, E. & Zimprich, A. (2009). Muscle Nerve, 39, 389–391. [DOI] [PubMed]
- Dreveny, I., Kondo, H., Uchiyama, K., Shaw, A., Zhang, X. & Freemont, P. S. (2004). EMBO J.23, 1030–1039. [DOI] [PMC free article] [PubMed]
- Halawani, D. & Latterich, M. (2006). Mol. Cell, 22, 713–717. [DOI] [PubMed]
- Huyton, T., Pye, V. E., Briggs, L. C., Flynn, T. C., Beuron, F., Kondo, H., Ma, J., Zhang, X. & Freemont, P. S. (2003). J. Struct. Biol.144, 337–348. [DOI] [PubMed]
- Jones, T. A., Zou, J.-Y., Cowan, S. W. & Kjeldgaard, M. (1991). Acta Cryst. A47, 110–119. [DOI] [PubMed]
- Kimonis, V. E., Fulchiero, E., Vesa, J. & Watts, G. (2008). Biochim. Biophys. Acta, 1782, 744–748. [DOI] [PubMed]
- Kimonis, V. E., Kovach, M. J., Waggoner, B., Leal, S., Salam, A., Rimer, L., Davis, K., Khardori, R. & Gelber, D. (2000). Genet. Med.2, 232–241. [DOI] [PMC free article] [PubMed]
- Ma, J. & Xia, D. (2008). J. Appl. Cryst.41, 1150–1160. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Neuwald, A. F., Aravind, L., Spouge, J. L. & Koonin, E. V. (1999). Genome Res.9, 27–43. [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Peters, J. M., Walsh, M. J. & Franke, W. W. (1990). EMBO J.9, 1757–1767. [DOI] [PMC free article] [PubMed]
- Pye, V. E., Beuron, F., Keetch, C. A., McKeown, C., Robinson, C. V., Meyeer, H. H., Zhang, X. & Freemont, P. S. (2007). Proc. Natl Acad. Sci. USA, 104, 467–472. [DOI] [PMC free article] [PubMed]
- Schroder, R., Watts, G. D., Mehta, S. G., Evert, B. O., Broich, P., Fliessbach, K., Pauls, K., Hans, V. H., Kimonis, V. & Thal, D. R. (2005). Ann. Neurol.57, 457–461. [DOI] [PubMed]
- Smith, P. K., Krohn, R. I., Hermanson, G. T., Mallia, A. K., Gartner, F. H., Provenzano, M. D., Fujimoto, E. K., Goeke, N. M., Olson, B. J. & Klenk, D. C. (1985). Anal. Biochem.150, 76–85. [DOI] [PubMed]
- Storoni, L. C., McCoy, A. J. & Read, R. J. (2004). Acta Cryst. D60, 432–438. [DOI] [PubMed]
- Uchiyama, K. & Kondo, H. (2005). J. Biochem.137, 115–119. [DOI] [PubMed]
- Wang, Q., Song, C. & Li, C. (2004). J. Struct. Biol.146, 44–57. [DOI] [PubMed]
- Wittig, I., Braun, H. P. & Schagger, H. (2006). Nature Protoc.1, 418–428. [DOI] [PubMed]
- Ye, Y. (2006). J. Struct. Biol.156, 29–40. [DOI] [PubMed]
- Yeates, T. O. (1997). Methods Enzymol.276, 344–358. [PubMed]
- Zhang, X., Shaw, A., Bates, P. A., Newman, R. H., Gowen, B., Orlova, E., Gorman, M. A., Kondo, H., Dokurno, P., Lally, J., Leonard, G., Meyer, H., van Heel, M. & Freemont, P. S. (2000). Mol. Cell, 6, 1473–1484. [DOI] [PubMed]