Abstract
This study employed cultured human primary hepatocytes to investigate the ability of the putative chemopreventive phytochemicals curcumin (CUR), 3,3′-diindolylmethane (DIM), isoxanthohumol (IXN), or 8-prenylnaringenin (8PN) to reduce DNA adduct formation of the hepatocarcinogen aflatoxin B1 (AFB). Following 48 h of pretreatment, DIM and 8PN significantly increased AFB-DNA adduct levels, whereas CUR and IXN had no effect. DIM greatly enhanced the transcriptional expression of cytochrome P450 (CYP) 1A1 and CYP1A2 mRNA. Glutathione S-transferase mRNAs were not increased by any of the treatments. In vitro enzyme activity assays demonstrated that 8PN and DIM, but not CUR or IXN, inhibited human CYP1A1, CYP1A2, and CYP3A4 activities. To distinguish between treatment effects on transcription versus direct effects on enzyme activity for DIM, we evaluated the effects of pretreatment alone (transcriptional activation) versus cotreatment alone (enzyme inhibition). The results demonstrated that effects on gene expression, but not catalytic activity, are responsible for the observed effects of DIM on AFB-DNA adduct formation. The increase in AFB-DNA damage following DIM treatment may be explained through its substantial induction of CYP1A2 and/or its downregulation of GSTM1, both of which were significant. The increase in DNA damage by DIM raises potential safety risks for dietary supplements of DIM and its precursor indole-3-carbinol.
Keywords: phytochemicals, biotransformation, hepatocytes, human, CYP
Numerous studies in laboratory animals, and limited human epidemiological data, suggest that a variety of plant-derived compounds (phytochemicals) can lower the risk for certain types of cancer (Kelloff et al., 2000). Widely consumed cruciferous vegetables such as broccoli, cabbage, cauliflower, and Brussels sprouts are rich in sulfur-containing glycosides (glucosinolates). Glucobrassicin is one of the most common glucosinolate compounds in cruciferous vegetables (Ciska et al., 2000). Once the vegetables are mechanically damaged, the biologically active compound, indole-3-carbinol (I3C), is released from glucobrassicin by the plant-derived enzyme myrosinase (Shertzer and Senft, 2000). The acidic environment of the stomach fosters the non-enzymatic self-condensation of two I3C molecules to form 3,3′-diindolylmethane (DIM). DIM and I3C have demonstrated anticancer effects in vitro and in vivo, are currently used as dietary supplements, and have been the subject of clinical studies (Higdon et al., 2007; Reed et al., 2006, 2008). Isoxanthohumol (IXN) and 8-prenylnaringenin (8PN) are derived from xanthohumol (XN), a major flavanone in hops (Stevens et al., 1999b). XN is converted to IXN and other prenylated naringenins such as 8PN during brewing, thus making the latter ones more relevant candidates than XN (Stevens et al., 1999a). Curcumin (CUR) is a major yellow pigment found in turmeric and curry and has putative chemopreventive properties (Singh and Khar, 2006).
I3C, DIM, and CUR protect animals against experimentally induced tumors from a variety of chemical carcinogens (Rogan, 2006). Numerous in vitro studies have demonstrated protective effects of DIM, CUR, IXN, and 8PN toward (geno)toxicological end points of several carcinogens/mutagens that require metabolic activation to exert their genotoxic effects. Suggested mechanisms for these chemoprotective effects of phytochemicals include inhibition of bioactivation of procarcinogens (Henderson et al., 2000), enhanced detoxification of reactive intermediates (Kensler et al., 2000; Keum et al., 2005), modulation of oxidative stress (Kwak et al., 2001; Singh and Khar, 2006), and alterations in cell cycle regulation or apoptosis (Kong et al., 2001; Singh and Khar, 2006).
The four phytochemicals utilized in the present study may act as modulators of expression and/or catalytic activity of phase I and II biotransformation enzymes that play key roles in the bioactivation of the hepatocarcinogenic mycotoxin aflatoxin B1 (AFB). In particular, cytochromes P450 (CYP) 1A2 and CYP3A4/5 are involved in the activation of AFB to the genotoxic aflatoxin B1-8,9-oxide (AFBO) (Gallagher et al., 1996). The detoxification of AFBO is mediated by certain glutathione S-transferases (GSTs), but in a species-specific manner. Alpha-class GSTs in rats (rGSTA5-5) and mice (mGSTA3-3) are highly effective in detoxifying AFBO (Buetler et al., 1992) and are inducible by diet (Hayes et al., 1998), but human alpha class GSTs (hGSTA1, hGSTA2) lack any measurable activity toward AFBO (Eaton et al., 2001). However, in the absence of a high-activity alpha-class GST, the low but measurable activity of hGSTM1 may afford some protection against AFBO (Deng et al., 2005; Kirk et al., 2005; Long et al., 2006; McGlynn et al., 1995). Alternatively, human microsomal epoxide hydrolase (mEH) may also participate in the detoxification of AFBO in the absence of significant GST activity (Eaton et al., 2001; Kelly et al., 2002; Kirk et al., 2005; McGlynn et al., 1995).
Despite a large body of evidence that these phytochemicals act as modulators of gene expression and/or enzymatic activities of biotransformation enzymes involved in AFB activation and detoxification, few studies have utilized intact human liver cells to evaluate the effects of putative chemoprotective agents on AFB genotoxicity. Using a human-derived test system is important because there are large species differences in susceptibility toward genotoxic effects of AFB, and much of this is attributed to a wide variation in expression, regulation, and substrate specificity of biotransformation enzymes involved in AFB bioactivation and detoxification (Eaton et al., 2001). Indeed, a previous study in our laboratory demonstrated that effects of phytochemicals on gene expression in human hepatocytes are quite different from those observed in rodent-derived test models as well as in human tumor-derived cell cultures (e.g., HepG2 cells), emphasizing the importance of utilizing primary human-derived test systems in the evaluation of potential chemoprotective mechanisms of relevance to humans (Gross-Steinmeyer et al., 2004). Thus, the present study utilizes freshly isolated human hepatocytes in primary culture, derived from livers rejected for transplantation, to evaluate the putative chemoprotective effects of CUR, DIM, and two XNs on AFB-DNA adduct formation and to elucidate potential mechanisms responsible for modulation of AFB-mediated genotoxicity in human liver.
MATERIALS AND METHODS
Phytochemicals and AFB.
CUR was purchased from Sigma-Aldrich (St. Paul, MN). DIM was purchased from LKT Laboratories; IXN and 8PN were isolated and purified, as described previously (Miranda et al., 2000). All phytochemicals were at > 98% purity as determined via high performance liquid chromatography. 3H-AFB was obtained from Moravek Biochemicals (Brea, CA). Specific activities of different 3H-AFB product lots ranged from 16 to 28 mCi/mmol.
Isolation of human hepatocytes, culture, and treatment.
Human hepatocytes were isolated from viable human livers that were rejected for transplantation for various reasons. All human subjects’ protocols were reviewed and approved by the University of Washington and the University of Pittsburgh Institutional Review Boards. Hepatocyte isolation was performed as described previously (Strom et al., 1996). Hepatocyte cultures were maintained at 37°C under 5% CO2/95% humidified air on a rigid collagen substratum overlaid with Matrigel (Collaborative Biochemicals, Bedford, MA) in supplemented William's E media as described previously, as previous work in our laboratory demonstrated that Matrigel overlays facilitate the maintenance and induction of xenobiotic metabolizing enzymes in primary human hepatocytes (Gross-Steinmeyer et al., 2005). Following a minimum 48 h recovery period, hepatocytes were treated with phytochemicals, and different end points were evaluated following treatments, as outlined below. During the treatment periods, incubation media–containing compounds were changed after 24 h. Each concentration and exposure period used in this study was non-cytotoxic, as measured by lactate dehydrogenase leakage using a commercially available kit (Promega, Madison, WI). Our criteria for selecting the high concentration of each phytochemical were (1) no adverse cytotoxic effects compared with the vehicle-only controls, as described in more detail previously (Gross-Steinmeyer et al., 2004), (2) maximizing induction effect on CYP1A1 or CYP1A2 as described previously (Gross-Steinmeyer et al., 2004), and (3) maximizing the detection of AFB-DNA adducts. One additional lower concentration was chosen for each compound to assess dose-response effects for the various end points (Gross-Steinmeyer et al., 2004)
Effects of CUR, DIM, IXN, and 8PN on AFB-mediated genotoxicity.
Changes in AFB-DNA adduct formation were determined in hepatocyte preparations obtained from six individual human livers. Hepatocytes were pretreated for 48 h with two non-cytotoxic concentrations of six phytochemicals, followed by 6 h coincubations of phytochemicals and 0.2–0.4μM 3H-AFB or vehicle as shown in Table 1. In all covalent binding experiments, concentrations of 3H-AFB ranged from 0.2 to 0.4μM, depending on the specific activity of the particular product lot, to provide sufficient sensitivity for adduct determination. In order to assess the relationship between the concentration of AFB used and AFB-DNA adducts formed, we conducted a dose-response assessment with a single preparation of human hepatocytes, using 0.1, 0.2, 0.3 and 0.4μM AFB, and the same experimental conditions as described above. A linear regression analysis showed an almost perfect linear correlation with an R2 of 0.998 (data not shown). The concentrations used in our study fell within the concentration range we had tested empirically. The linear dose-response for AFB-DNA adduct formation in human hepatocytes is consistent with data obtained previously with hepatocytes from rat and trout (reviewed by Eaton and Gallagher, 1994). DNA adducts were normalized for 0.4μM 3H-AFB in each hepatocyte preparation based on these data, demonstrating a highly linear dose-response relationship in the applied concentration range.
TABLE 1.
Experimental Design for Initial Genotoxicity Experiment
| Sample | 48 h pretreatment (0–48 h) | 6 h AFB treatment (48–54 h) |
| Non-modulated control | 0.1% DMSO | 0.2–0.4μM 3H-AFB |
| CUR (low) | 25μM CUR | 0.2–0.4μM 3H-AFB + 25μM CUR |
| CUR (high) | 50μM CUR | 0.2–0.4μM 3H-AFB + 50μM CUR |
| DIM (low) | 10μM DIM | 0.2–0.4μM 3H-AFB + 10μM DIM |
| DIM (high) | 50μM DIM | 0.2–0.4μM 3H-AFB + 50μM DIM |
| IXN (low) | 10μM IXN | 0.2–0.4μM 3H-AFB + 10μM IXN |
| IXN (high) | 25μM IXN | 0.2–0.4μM 3H-AFB + 25μM IXN |
| 8PN (low) | 10μM 8PN | 0.2–0.4μM 3H-AFB + 10μM 8PN |
| 8PN (high) | 25μM 8PN | 0.2–0.4μM 3H-AFB + 25μM 8PN |
Note. Different AFB concentrations were applied due to sensitivity reasons (see also the “Materials and Methods” section). DMSO: dimethyl sulfoxide.
Dimethyl sulfoxide concentration was 0.1% in vehicle controls and samples during 48 h pretreatments, and 0.2% during subsequent 6 h coincubations. Cell harvesting and DNA isolation of 3H-AFB-treated cells were performed using the Qigaen Genomic DNA Purification kit (Qiagen, Valencia, CA) according to the manufacturer's recommendation. DNA concentrations were determined by fluorescence using Hoechst dye 33258 and known amounts of calf thymus DNA as a standard. Known DNA amounts were subsequently subjected to liquid scintillation counting for quantification of covalently bound 3H-AFB on DNA. A subset of cells, heated to 95°C for 5 min to inactivate biotransformation enzymes prior to AFB exposure, was treated identically to correct for non-covalent binding of 3H-AFB and/or remaining 3H-AFB in DNA isolates. The AFB-DNA adduct levels were corrected by subtracting the radioactivity obtained from this nonspecific binding control to represent the extent of metabolically bioactivated AFB that bound covalently to DNA. Additionally, a non-treated cell control was subjected to DNA isolation in the same manner as the radiolabeled treatments to account for any background radioactivity in the final DNA samples. Final 3H-AFB-DNA adduct levels were calculated in units of (fmol adduct/100 × μmol DNA) which is equal to adducts per 107 nucleotides.
mRNA expression analyses.
Changes in mRNA expression were determined in hepatocyte preparations obtained from four to eight individual human livers. Hepatocytes were treated for 48 h with DIM or 8PN at the same concentrations as used during the pretreatment period in the genotoxicity study (see also Table 1). mRNA expression levels of human CYP1A1, CYP1A2, CYP3A4, CYP3A5, mEH, GSTM1, and GSTT1 were determined by “real-time” reverse transcriptase (RT)-PCR. At 0 and 48 h after exposure, the hepatocytes were lysed on the plates, and total RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA) as recommended by the supplier. The quality of the RNA preparations was assessed electrophoretically via 18S and 28S band intensities. Reverse transcription of 0.1- to 2 μg total RNA using oligod(T)15 primer and Superscript II RNaseH (Gibco, Grand Island, NY) was performed according to the manufacturer's instructions. Fluorogenic 5′-nuclease assays (TaqMan) were carried out using an ABI Prism 7700 Sequence detection system (Perkin-Elmer Applied Biosystems, Foster City, CA). The thermal cycling conditions comprised an initial denaturation step at 95°C for 10 min, followed by 40 cycles at 95°C for 20 s and 62°C for 60 s. The gene-specific sequences of primer pairs and probes used in the TaqMan assays are as follows: CYP3A5 (NM_000777): forward primer, acagatccccttgaaattagacacg; reverse primer, cttagggttccatctcttgaatcca; probe, aaggacttcttcaaccagaaaaacccattgttcta. mEH (NM_000120): forward primer, catctcctcccagcgcttctac; reverse primer, cagagaagccagtgggcacat; probe, ttcatccgctcatgcttctgggtcatc. All other primer pairs and probes for TaqMan assays have been listed previously (Gross-Steinmeyer et al., 2004, 2005).
GSTM1 and GSTT1 genotyping was performed as described by Chen et al. (1996).
Human CYP1A1, CYP1A2, and CYP3A4 inhibition assays.
Methoxyresorufin-O-demethylase (MROD), ethoxyresorufin-O-deethylase (EROD), and benzyloxyresorufin-O-debenzylase (BROD) assays were conducted with microsomes from recombinant yeast strains expressing human CYP1A2 or CYP1A1 and Supersomes expressing human CYP3A4 (Gentest, Woburn, MA). The recombinant yeast strains were derived from the parental Saccharomyces cerevisiae strain yHE2, and yeast microsomes were prepared as previously described (Eugster and Sengstag, 1993). Standard enzyme activity assay protocols using the yeast microsomes as well as Supersomes were performed as described previously (Gross-Steinmeyer et al., 2004). A concentration range of 0.15–50μM phytochemical was assessed for inhibitory effects and compared to the noninhibited (solvent) control, as well as to corresponding negative controls. Two independent experiments were performed with each phytochemical, and six measurements were taken per condition within each experiment.
Effects of DIM on AFB-mediated genotoxicity.
Hepatocytes obtained from three individual human livers were treated with the following combinations of treatment conditions to discriminate between treatment effects on transcription and direct effects on enzyme activity: (1) 48 h pretreatment with DIM, followed by 6 h treatment with 0.4μM 3H-AFB, (2) 48 h pretreatment with DIM, followed by 6 h treatment with 0.4μM 3H-AFB and DIM, and (3) 48 h pretreatment with solvent, followed by 6 h treatment with 0.4μM 3H-AFB and DIM. DIM was applied at 10μM each, and treatment conditions were as described above. The temporal relationship between all incubations was kept consistent, and vehicle solvents were used to make all treatment conditions identical. AFB-DNA adduct levels were determined as described above.
Statistical analyses.
Statistical significances of fold alteration values over controls (e.g., modulated DNA adduct levels, modulated transcriptional expression) were determined by one-way ANOVA using Dunnett's post test. IC50 values were calculated using BioDataFit 1.02 software (Chang Bioscience, Inc., Castro valley, CA).
RESULTS
Effects of Phytochemicals on AFB-DNA Adduct Formation
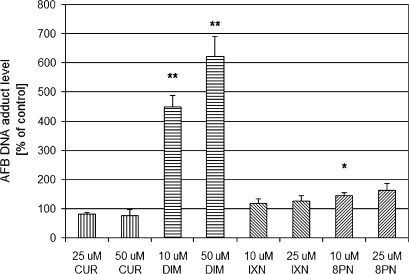
To determine the effects of phytochemicals on AFB genotoxicity, primary cultures of isolated human hepatocytes were treated with CUR, DIM, IXN, or 8PN for 48 h and subsequently coincubated for 6 h with 3H-AFB and phytochemicals (Table 1). The untreated (control) AFB-DNA adduct level averaged 5.4 adducts per 107 nucleotides (six individual hepatocyte preparations), with a range from 1.7 to 9.9 adducts per 107 nucleotides (Fig. 1).
FIG. 1.
Modulation of AFB-DNA adduct formation by phytochemicals in cultured human primary hepatocytes. Hepatocytes were treated with two doses of each phytochemical for 48 h and subsequently coincubated with 0.4μM 3H-AFB and phytochemical. Control represents non-modulated AFB-DNA adduct level and was treated with solvent instead of phytochemical. AFB-DNA adduct levels are expressed as percentages over non-modulated control and represent means and SEs from six independent experiments. The control AFB-DNA adduct level averaged 5.4 adducts per 107 nucleotides (range 1.7–9.9 adducts per 107 nucleotides). *Denotes 0.01 < p < 0.05, **denotes p < 0.01.
Relative to their respective control values, DIM (10 and 50μM) and 8PN (10μM) significantly increased DNA adduct formation (Fig. 1). CUR (10 and 50μM) and IXN (10 and 25μM) did not alter AFB-DNA adduct levels. DIM increased AFB-related DNA damage by 4.5- and 6.2-fold at 10 and 50μM, respectively. DNA damage was increased more than eightfold at 50μM DIM in a single hepatocyte preparation (data not shown). The hops-derived flavanoid, 8PN, produced a modest increase in AFB-mediated genotoxicity, reaching 1.5- and 1.6-fold increase in AFB-DNA adduct formation at 10 and 25μM, respectively, whereas IXN (another hops-derived flavanoid) and CUR had no significant effect on AFB-DNA adduct formation.
Effects of DIM and 8PN on Expression of Genes Involved in Biotransformation of AFB
To evaluate whether DIM and/or 8PN modulate (up- or downregulate) the expression of AFB biotransformation pathways, we determined mRNA expression of genes involved in biotransformation of AFB following treatments with DIM (10 and 50μM) and 8PN (10 and 25μM) for 48 h, using the same incubation conditions used for AFB-DNA adduct determination. CUR and IXN were excluded from these studies as these compounds had no significant effect on AFB-DNA adduct formation. Effects on mRNA expression of CYP1A1, CYP1A2, CYP3A4, CYP3A5, GSTM1, GSTT1, and mEH were determined in hepatocyte preparations derived from four to eight different donors, depending on the genotype. The results of the quantitative real-time RT-PCR analyses (Table 2) demonstrated that DIM caused a dramatic increase in CYP1A1 and CYP1A2 mRNA in a dose-related manner that was very consistent within all hepatocyte preparations. The average increase in CYP1A1 and CYP1A2 mRNA by the highest concentration of DIM (50μM) was 625-fold and 90-fold, respectively.
TABLE 2.
Modulation of Transcriptional Gene Expression by Phytochemicals in Cultured Human Primary Hepatocytes
| Treatment | CYP1A1 | CYP1A2 | CYP3A4 | CYP3A5 | mEH | GSTM1a | GSTT1a |
| Control | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| DIM, 10μM | 113 ± 16 (8)** | 24.9 ± 3.1 (8)** | 1.1 ± 0.2 (8) | 1.1 ± 0.2 (8) | 1.4 ± 0.3 (8) | 0.79 ± 0.36 (4) | 1.2 ± 0.4 (6) |
| DIM, 50μM | 625 ± 156 (8)** | 90.2 ± 20.1 (8)** | 1.2 ± 0.2 (8) | 1.1 ± 0.1 (8) | 2.0 ± 0.9 (8) | 0.56 ± 0.09 (4)* | 2.0 ± 1.2 (6) |
| 8PN, 10μM | 1.6 ± 0.2 (8) | 2.0 ± 0.3 (8)* | 1.2 ± 0.3 (8) | 0.8 ± 0.2 (8) | 4.1 ± 1.6 (8) | 1.4 ± 0.3 (4) | 0.9 ± 0.2 (6) |
| 8PN, 25μM | 1.7 ± 0.4 (8) | 2.3 ± 0.4 (8)* | 0.8 ± 0.1 (8) | 0.7 ± 0.2 (8) | 2.2 ± 0.5 (8) | 1.1 ± 0.2 (4) | 0.9 ± 0.1 (6) |
Note. Hepatocytes were treated with two doses of DIM or 8PN for 48 h, and mRNA levels of genes listed were determined by quantitative RT-PCR analysis as described in the “Materials and Methods” section. Control represents non-modulated transcription level following solvent control treatment for 48 h. Values are expressed as fold alteration relative to solvent control (with the control equal to 1) and represent means of fold alteration values and SEs from hepatocytes from N individual preparations (N is displayed in parentheses).
includes only samples that had a positive genotype (e.g., were not homozygous null for the gene deletion).
*Denotes 0.01 < p < 0.05, **denotes 0.001 < p < 0.01, ***denotes p < 0.001.
Further, DIM caused a significant, dose-dependent downregulation of GSTM1 expression in those hepatocyte preparations that were GSTM1 positive. 8PN had a modest, but significant effect on CYP1A2 expression and increased mRNA levels by 2.0- and 2.3-fold at 10 and 25μM 8PN, respectively.
Direct Effects of Phytochemicals on AFB Biotransformation Enzyme Catalytic Activity
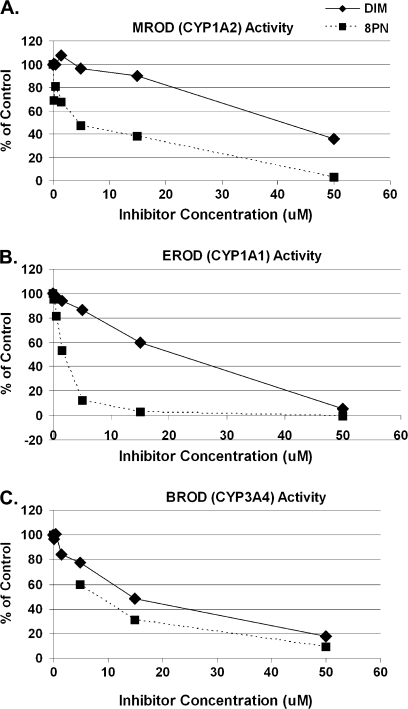
We further explored the ability of these phytochemicals to inhibit catalytic activities of key enzymes involved in the oxidation of AFB: CYP1A2 (AFB-epoxide formation [activation] and aflatoxin M1 formation [detoxification]), CYP1A1 (aflatoxin M1 formation [detoxification]), and CYP3A4 (aflatoxin Q1 [AFQ] formation [detoxification] and AFB-epoxide formation [bioactivation]), using recombinant yeast strains expressing human CYP1A1 and CYP1A2 and commercially available Supersomes (Gentest) expressing human CYP3A4. Both DIM and 8PN inhibited human CYP1A2-mediated MROD, CYP1A1-mediated EROD, and CYP3A4-mediated BROD activities in a dose-dependent manner (Fig. 2). The potency of enzyme inhibition occurred in the order 8PN > DIM for CYP1A1, and 8PN >> DIM for CYP1A2, and 8PN > DIM for CYP3A4, as judged by the IC50 values. 8PN showed the most potent inhibitory effects of all phytochemicals tested, with IC50 values of approximately 4.5, 1.7, and 8.4μM for CYP1A2, CYP1A1, and CYP3A4, respectively. DIM had higher IC50 values of 40.9, 21.4, and 14.5μM for CYP1A2, CYP1A1, and CYP3A4, respectively.
FIG. 2.
Modulation of catalytic activity of CYP1A1, CYP1A2, and CYP3A4 by DIM and 8PN in vitro. EROD (B), MROD (A), and BROD (C) activities were determined using microsomes from recombinant yeast strains expressing human CYP1A1, CYP1A2, and Supersomes (Gentest) expressing human CYP3A4, respectively, after adding increasing concentrations of DIM or 8PN. Values are expressed as % inhibition relative to solvent controls. Means and corresponding SDs are derived from two independent inhibition experiments (with six measurements per experiment). All inhibition mean values at phytochemical concentrations at 1.5μM and higher were statistically significant (p < 0.01), as well as 8PN at 0.15 and 0.5μM in MROD and DIM and 8PN both at 0.5μM in EROD. IC50 values for 8PN were approximately 4.5, 1.7, and 8.4μM for CYP1A2, CYP1A1, and CYP3A4, respectively. IC50 values for DIM were 40.9, 21.4, and 14.5μM for CYP1A2, CYP1A1, and CYP3A4, respectively.
Modulation of AFB-DNA Adduct Formation by DIM under Different Treatment Conditions
DIM significantly increased the expression of CYP1A1 and CYP1A2 at the mRNA and protein levels (Table 2; Gross-Steinmeyer et al., 2004). DIM also directly inhibited catalytic activity of CYP1A1 and CYP3A4 and to a lesser extent CYP1A2 (Fig. 2).
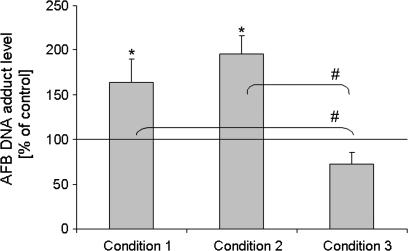
In order to determine to what extent the transcriptional (i.e., induction) versus the enzyme inhibitory effects contribute to DIM's modulation of AFB-DNA adduct formation, we measured adducts under three conditions. Under condition 1, cells were pretreated with DIM for 48 h, after which time the media was removed, cells were rinsed with PBS, and incubated for 6 h in media containing 3H-AFB and no DIM (this was the same protocol that was used to generate the data displayed in Fig. 1). Condition 2 was as condition 1, with the exception that cells were coexposed for the last 6 h with both 3H-AFB and DIM. Condition 3 was as condition 2, except cells were not treated with DIM for the first 48 h but with vehicle instead. Condition 1 was designed to detect mainly transcriptional effects resulting in induction of CYP1A1 and CYP1A2, whereas condition 3 was chosen to assess effects that were due to direct interaction (inhibition) of DIM with CYP enzymes. Effects measured under condition 2 detected transcriptional as well as direct enzyme interaction effects.
The results depicted in Figure 3 show that the increase in AFB-mediated genotoxicity by DIM was qualitatively similar (and not significantly different) under condition 1 (48 h DIM pretreatment and 6 h 3H-AFB) and condition 2 (48 h DIM pretreatment and 6 h DIM/3H-AFB coincubation). DIM protected somewhat from AFB damage under condition 3 (no DIM pretreatment, 6 h DIM/3H-AFB coincubation). However, the inhibitory effect of coincubation was counterbalanced when coincubation was preceded by pretreatment with DIM (condition 2). These findings suggest that the induction of CYP1A2 gene expression by DIM outweighs its inhibitory effect on CYP1A2 and/or 3A4 catalytic activity responsible for AFB activation to the DNA-binding metabolite AFB-epoxide, at least at the DIM concentration used.
FIG. 3.
Modulation of AFB-DNA adduct formation by DIM in cultured human primary hepatocytes under different treatment conditions. To assess contribution of transcriptional versus enzyme inhibition effects, we measured AFB-DNA adduct levels under three different conditions: (1) following a 48 h pretreatment with DIM, media was removed, cells were rinsed with PBS, and incubated for 6 h in media containing tritiated AFB but no DIM; (2) following a 48 h pretreatment with DIM, media was removed, cells were rinsed with PBS, and incubated for an additional 6 h in media containing tritiated AFB and DIM; (3) following a 48 h treatment with vehicle only (no DIM), media was removed, cells were rinsed with PBS, and incubated for an additional 6 h in media containing tritiated AFB and DIM. DIM was applied at 10μM. Condition 1 is designed to detect transcriptional effects, condition 3 detects potential enzyme inhibition effects, and condition 2 detects the overall effect of both. Conditions 1, 2, and 3 were compared to an analogous treatment without DIM application, which served as control (equal to 100%), for example, following a 48 h treatment with vehicle only (no DIM), media was removed, cells were rinsed with PBS, and incubated for an additional 6 h in media containing tritiated AFB only (no DIM). AFB-DNA adduct levels are expressed as percentages of control and represent means and SDs from three independent experiments (e.g., hepatocytes from three individual preparations). Conditions 1 and 2 were not significantly different from each other, but each was different from control (p ≤ 0.05), marked as “*.” Condition 3 was significantly different from conditions 1 and 2 (p ≤ 0.05), but not from the control, marked as “#.”
DISCUSSION
Increased AFB-Mediated Genotoxicity and Significance for Humans
DIM and 8PN increased AFB-DNA adduct formation in primary human hepatocytes from six individual preparations, all in a dose-dependent manner. No significant effects were observed following CUR and IXN treatments. DIM had the most substantial effect with a more than eightfold increase of AFB adduct levels relative to control in single hepatocyte preparations.
As CYP1A2 is a major bioactivating enzyme for AFB-epoxide formation at low concentrations of AFB (Gallagher et al., 1996), the increased AFB-DNA damage may be explained by the potent upregulation of CYP1A2 expression by DIM in cultured human hepatocytes, as observed in this study as well as in a previous report of our laboratory (Gross-Steinmeyer et al., 2004). A similarly potent upregulation of CYP1A2 was shown by Horn et al. (2002) who reported induction of both CYP1A1 and CYP1A2 by I3C (the precursor of DIM) in rat. Other studies have shown that both I3C and DIM are ligands of the aryl hydrocarbon receptor (AhR). Once a ligand binds to AhR, it can activate transcription of genes which have a xenobiotic response element in their promoters.
I3C may be responsible for the in vivo induction of CYP1A2 enzyme activity in humans following consumption of controlled diets consisting of cruciferous vegetables (Kall and Clausen, 1995; Lampe et al., 2000; Shertzer and Senft, 2000; Walters et al., 2004). However, it is still uncertain what concentrations of DIM are achieved in the human liver following a typical serving of the corresponding vegetable. Another significant route of exposure to DIM is through “nutraceuticals.” Recent clinical studies examined the pharmacokinetics aspects of DIM after either consumption of purified I3C (Reed et al., 2006) or DIM (Reed et al., 2008) in humans, revealing DIM plasma levels in the lower micromolar range (∼0.1–1.0μM). Consequently, the potent induction effects at concentrations as low as 0.1μM (Gross-Steinmeyer et al., 2004) may have further implications for other genotoxic carcinogens that are formed through hepatic CYP1A2 activity in humans (e.g., several nitrosamines, food-related heterocyclic aromatic amines). Repeated intake of DIM may be associated with an increased risk due to procarcinogens that are bioactivated via human hepatic CYP1A2 activity. For example, dietary supplements containing I3C and/or DIM are used for their putative anticancer effects of estrogen-dependent cancers (for review, see Bradlow, 2008).
Potential Mechanisms through which DIM Modulates AFB-Mediated Genotoxicity
The two primary mechanisms proposed in the literature for chemoprotective action against AFB-induced carcinogenesis are induction of GSTs that detoxify AFBO and competitive inhibition of CYP-mediated activation of AFB to AFBO, based on a presumed common mechanism of action with the dithiolthione drug, oltipraz (Kwak et al., 2001).
Our data revealed that CYP1A2 was strongly upregulated by DIM and to a much lower extent by 8PN. However, both compounds were shown to be effective inhibitors of CYP1A2 as well. We performed a series of systematic incubations which allowed for discrimination between effects on transcription and direct effects on enzyme activity (see Fig. 3). The focus was on DIM in these experiments as it showed by far the strongest modulation of AFB-mediated DNA adduct formation compared to the other phytochemicals tested (Fig. 1). The results of combination treatments (Fig. 3) strongly suggest that the induction of gene expression by DIM (Table 2) rather than its inhibitory effect on catalytic activity (Fig. 2) gives rise to the elevation of AFB-mediated genotoxicity in intact human hepatocytes. Thus, we conclude that the increase in AFB-DNA adducts by DIM may be explained through its substantial induction of CYP1A2 (bioactivation) and/or its downregulation of GSTM1 (detoxification).
Interestingly, Dashwood et al. (1991) also reported a dual action of I3C (parent compound of DIM) in trout; I3C acted as an AFB tumor inhibitor when given prior to or during AFB initiation but was a tumor promoter when fed after AFB initiation. I3C's potential tumor promoting activity was approximately as great as its tumor inhibitory activity.
Previous kinetic data from our laboratory suggested that, at the relatively low hepatic concentrations of AFB that might result from human dietary exposures (< 1μM), human CYP1A2 is very effective at bioactivating AFB and, thus, was predicted to be the dominant CYP involved in activation of AFB in vivo (Gallagher et al., 1996). At these low substrate concentrations, CYP1A2 catalyzes the formation of the reactive intermediate, AFBO, and CYP3A4 metabolizes AFB preferentially to the hydroxylated metabolite AFQ. Due to a well-described allosteric mechanism, the formation rate of AFBO by CYP3A4 increases with higher concentrations of AFB (> 100μM), at least in vitro (Gallagher et al., 1996).
Recently, Kamdem et al. (2006) assessed the contribution of CYP1A2, CYP3A4, CYP3A5, and CYP3A7 to overall AFBO production in a panel of human microsomes (n = 13). Their analysis used a hepatic abundance model which took both the expression levels of CYPs and their kinetic parameters into account. While the kinetic parameters reported in our previous study (Gallagher et al., 1996) and Kamdem et al. were very similar, they concluded that CYP3A4 was more important than CYP1A2 in the overall activation of AFB to AFBO, mainly due to its significantly higher expression in most livers compared to CYP1A2.
CYP1A1 predominantly forms aflatoxin M1 (AFM) (Kelly et al., 2002). As AFM has significantly lower toxic and carcinogenic potential than AFB, its formation is considered to be a detoxification pathway. The present study shows that DIM dramatically induced CYP1A1 but also was an effective inhibitor of the CYP1A1 enzyme activity.
CYP3A5 has qualitatively similar activity toward AFB as CYP3A4 and is capable of forming AFBO (Yamazaki et al., 1995; Kamdem et al., 2006). However, there are several common polymorphisms in CYP3A5 that result in little or no expression of functional protein, and thus, most people do not express a functional phenotype of CYP3A5 (Thompson et al., 2006). Neither compound in this study affected the expression of CYP3A5 (Table 2).
In rodents, certain alpha-class GSTs have high catalytic activity toward AFBO and, thus, are important determinants of susceptibility to AFB-induced liver cancer (Eaton et al., 2001; Hayes et al., 1998; Kwak et al., 2001), although the significance of human GSTs in detoxifying AFBO remains uncertain. Human and nonhuman primate liver alpha-class GSTs were found not to have any activity toward AFBO (Eaton et al., 2001; Wang et al., 2002). However, both in vitro studies and human epidemiological data suggest that human GSTM1 has some conjugation activity toward AFBO. Another relevant route of detoxication in humans may be the enzymatic hydrolysis of AFBO to the less toxic AFB-dihydrodiol catalyzed by mEH (Kelly et al., 2002; Kirk et al., 2005; McGlynn et al., 1995). Neither DIM nor 8PN had any significant inductive effects on GSTM1 or mEH in the present study. In fact, DIM caused some modest downregulation of GSTM1 expression at the higher concentration.
Our findings further support two hypotheses: (1) In isolated human hepatocytes with basal (noninduced) CYP450 expression, CYP3A4 appears to be the major bioactivating enzyme of AFB. This is due to its higher expression relative to CYP1A2, following hepatocyte isolation procedures, even though we had found that in vitro (human liver microsomes), CYP1A2 may be the major enzyme to bioactivate AFB at concentrations comparable to those used in the present study (Gallagher et al., 1996). However, it should be noted that the kinetic studies of CYP1A2 and CYP3A4 toward AFB utilized a cell-free system in which precise concentrations of AFB substrate could be determined. The AFB concentration at the smooth endoplasmic reticulum, the intracellular site of CYP activity, in cultured human hepatocytes remains unknown but may potentially be different from the extracellularly applied concentration of 0.4μM (maximum) due to lipid partitioning of lipophilic substrates such as AFB and/or due to other factors present in intact cells but absent in microsomal preparations; these could potentially lead to different kinetic profiles in intact cells from that observed in microsomal preparations. (2) Alternatively, in hepatocytes with induced CYP1A2, such as that occurs following treatment with DIM (which did not affect CYP3A4 expression), CYP1A2 expression is increased substantially to similar or even higher transcription levels compared to CYP3A4 as observed in the present study (based on comparison of quantitative RT-PCR cycle threshold number), and thus, it may contribute significantly more to bioactivation of AFB than constitutive levels of CYP3A4.
In conclusion, the substantial increase of AFB-DNA damage by DIM may be explained through its substantial induction of CYP1A2, perhaps, in combination with the observed downregulation of GSTM1. The increase in DNA damage by DIM raises potential safety risks for dietary supplements of DIM and/or its precursor I3C. The lack of induction of phase II biotransformation by all tested phytochemicals in this study, unlike numerous induction effects observed in animal-derived models, emphasizes once more the importance of utilizing primary human-derived test systems in the evaluation of potential chemoprotective mechanisms of relevance to humans.
FUNDING
National Institutes of Environmental Health Sciences (R01ES05780 and P30ES07033); Cancer Research and Prevention Foundation (to K.G.S.). Human hepatocyte preparation was supported in part by National Institute of Diabetes and Digestive and Kidney Diseases (DK-7-0004 to S.C.S.).
References
- Bradlow HL. Review. Indole-3-carbinol as a chemoprotective agent in breast and prostate cancer. In Vivo. 2008;22:441–445. [PubMed] [Google Scholar]
- Buetler TM, Slone D, Eaton DL. Comparison of the aflatoxin B1-8,9-epoxide conjugating activities of two bacterially expressed alpha class glutathione S-transferase isozymes from mouse and rat. Biochem. Biophys. Res. Commun. 1992;188:597–603. doi: 10.1016/0006-291x(92)91098-b. [DOI] [PubMed] [Google Scholar]
- Chen CL, Liu Q, Relling MV. Simultaneous characterization of glutathione S-transferase M1 and T1 polymorphisms by polymerase chain reaction in American whites and blacks. Pharmacogenetics. 1996;6:187–191. doi: 10.1097/00008571-199604000-00005. [DOI] [PubMed] [Google Scholar]
- Ciska E, Martyniak-Przybyszewska B, Kozlowska H. Content of glucosinolates in cruciferous vegetables grown at the same site for two years under different climatic conditions. J. Agric. Food Chem. 2000;48:2862–2867. doi: 10.1021/jf981373a. [DOI] [PubMed] [Google Scholar]
- Dashwood RH, Fong AT, Williams DE, Hendricks JD, Bailey GS. Promotion of aflatoxin B1 carcinogenesis by the natural tumor modulator indole-3-carbinol: influence of dose, duration, and intermittent exposure on indole-3-carbinol promotional potency. Cancer Res. 1991;51:2362–2365. [PubMed] [Google Scholar]
- Deng ZL, Wei YP, Ma Y. Polymorphism of glutathione S-transferase mu 1 and theta 1 genes and hepatocellular carcinoma in southern Guangxi, China. World J. Gastroenterol. 2005;11:272–274. doi: 10.3748/wjg.v11.i2.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton DL, Bammler TK, Kelly EJ. Interindividual differences in response to chemoprotection against aflatoxin-induced hepatocarcinogenesis: Implications for human biotransformation enzyme polymorphisms. Adv. Exp. Med. Biol. 2001;500:559–576. doi: 10.1007/978-1-4615-0667-6_85. [DOI] [PubMed] [Google Scholar]
- Eaton DL, Gallagher EP. Mechanisms of aflatoxin carcinogenesis. Annu. Rev. Pharmacol. Toxicol. 1994;34:135–172. doi: 10.1146/annurev.pa.34.040194.001031. [DOI] [PubMed] [Google Scholar]
- Eugster HP, Sengstag C. Saccharomyces cerevisiae: An alternative source for human microsomal liver enzymes and its use in drug interaction studies. Toxicology. 1993;82:61–73. doi: 10.1016/0300-483x(93)90060-6. [DOI] [PubMed] [Google Scholar]
- Gallagher EP, Kunze KL, Stapleton PL, Eaton DL. The kinetics of aflatoxin B1 oxidation by human cDNA-expressed and human liver microsomal cytochromes P450 1A2 and 3A4. Toxicol. Appl. Pharmacol. 1996;141:595–606. doi: 10.1006/taap.1996.0326. [DOI] [PubMed] [Google Scholar]
- Gross-Steinmeyer K, Stapleton PL, Liu F, Tracy JH, Bammler TK, Quigley SD, Farin FM, Buhler DR, Safe SH, Strom SC, et al. Phytochemical-induced changes in gene expression of carcinogen-metabolizing enzymes in cultured human primary hepatocytes. Xenobiotica. 2004;34:619–632. doi: 10.1080/00498250412331285481. [DOI] [PubMed] [Google Scholar]
- Gross-Steinmeyer K, Stapleton PL, Tracy JH, Bammler TK, Lehman T, Strom SC, Eaton DL. Influence of Matrigel-overlay on constitutive and inducible expression of nine genes encoding drug-metabolizing enzymes in primary human hepatocytes. Xenobiotica. 2005;35:419–438. doi: 10.1080/00498250500137427. [DOI] [PubMed] [Google Scholar]
- Hayes JD, Pulford DJ, Ellis EM, McLeod R, James RF, Seidegard J, Mosialou E, Jernstrom B, Neal GE. Regulation of rat glutathione S-transferase A5 by cancer chemopreventive agents: Mechanisms of inducible resistance to aflatoxin B1. Chem. Biol. Interact. 1998;111–112:51–67. doi: 10.1016/s0009-2797(97)00151-8. [DOI] [PubMed] [Google Scholar]
- Henderson MC, Miranda CL, Stevens JF, Deinzer ML, Buhler DR. In vitro inhibition of human P450 enzymes by prenylated flavonoids from hops, Humulus lupulus. Xenobiotica. 2000;30:235–251. doi: 10.1080/004982500237631. [DOI] [PubMed] [Google Scholar]
- Higdon JV, Delage B, Williams DE, Dashwood RH. Cruciferous vegetables and human cancer risk: Epidemiologic evidence and mechanistic basis. Pharmacol. Res. 2007;55:224–236. doi: 10.1016/j.phrs.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn TL, Reichert MA, Bliss RL, Malejka-Giganti D. Modulations of P450 mRNA in liver and mammary gland and P450 activities and metabolism of estrogen in liver by treatment of rats with indole-3-carbinol. Biochem. Pharmacol. 2002;64:393–404. doi: 10.1016/s0006-2952(02)01190-5. [DOI] [PubMed] [Google Scholar]
- Kall MA, Clausen J. Dietary effect on mixed function P450 1A2 activity assayed by estimation of caffeine metabolism in man. Hum. Exp. Toxicol. 1995;14:801–807. doi: 10.1177/096032719501401004. [DOI] [PubMed] [Google Scholar]
- Kamdem LK, Meineke I, Gödtel-Armbrust U, Brockmöller J, Wojnowski L. Dominant contribution of P450 3A4 to the hepatic carcinogenic activation of aflatoxin B1. Chem Res. Toxicol. 2006;19:577–586. doi: 10.1021/tx050358e. [DOI] [PubMed] [Google Scholar]
- Kelloff GJ, Crowell JA, Steele VE, Lubet RA, Malone WA, Boone CW, Kopelovich L, Hawk ET, Lieberman R, Lawrence JA, et al. Progress in cancer chemoprevention: Development of diet-derived chemopreventive agents. J. Nutr. 2000;130:467S–471S. doi: 10.1093/jn/130.2.467S. [DOI] [PubMed] [Google Scholar]
- Kelly EJ, Erickson KE, Sengstag C, Eaton DL. Expression of human microsomal epoxide hydrolase in Saccharomyces cerevisiae reveals a functional role in aflatoxin B1 detoxification. Toxicol. Sci. 2002;65:35–42. doi: 10.1093/toxsci/65.1.35. [DOI] [PubMed] [Google Scholar]
- Kensler TW, Curphey TJ, Maxiutenko Y, Roebuck BD. Chemoprotection by organosulfur inducers of phase 2 enzymes: Dithiolethiones and dithiins. Drug Metabol. Drug Interact. 2000;17:3–22. doi: 10.1515/dmdi.2000.17.1-4.3. [DOI] [PubMed] [Google Scholar]
- Keum YS, Jeong WS, Kong AN. Chemopreventive functions of isothiocyanates. Drug News Perspect. 2005;18:445–451. doi: 10.1358/dnp.2005.18.7.939350. [DOI] [PubMed] [Google Scholar]
- Kirk GD, Turner PC, Gong Y, Lesi OA, Mendy M, Goedert JJ, Hall AJ, Whittle H, Hainaut P, Montesano R, et al. Hepatocellular carcinoma and polymorphisms in carcinogen-metabolizing and DNA repair enzymes in a population with aflatoxin exposure and hepatitis B virus endemicity. Cancer Epidemiol. Biomarkers Prev. 2005;14:373–379. doi: 10.1158/1055-9965.EPI-04-0161. [DOI] [PubMed] [Google Scholar]
- Kong AN, Yu R, Hebbar V, Chen C, Owuor E, Hu R, Ee R, Mandlekar S. Signal transduction events elicited by cancer prevention compounds. Mutat. Res. 2001;480–481:231–241. doi: 10.1016/s0027-5107(01)00182-8. [DOI] [PubMed] [Google Scholar]
- Kwak MK, Egner PA, Dolan PM, Ramos-Gomez M, Groopman JD, Itoh K, Yamamoto M, Kensler TW. Role of phase 2 enzyme induction in chemoprotection by dithiolethiones. Mutat. Res. 2001;480–481:305–315. doi: 10.1016/s0027-5107(01)00190-7. [DOI] [PubMed] [Google Scholar]
- Lampe JW, King IB, Li S, Grate MT, Barale KV, Chen C, Feng Z, Potter JD. Brassica vegetables increase and apiaceous vegetables decrease cytochrome P450 1A2 activity in humans: Changes in caffeine metabolite ratios in response to controlled vegetable diets. Carcinogenesis. 2000;21:1157–1162. [PubMed] [Google Scholar]
- Long XD, Ma Y, Wei YP, Deng ZL. The polymorphisms of GSTM1, GSTT1, HYL1*2, and XRCC1, and aflatoxin B1-related hepatocellular carcinoma in Guangxi population, China. Hepatol. Res. 2006;36:48–55. doi: 10.1016/j.hepres.2006.06.004. [DOI] [PubMed] [Google Scholar]
- McGlynn KA, Rosvold EA, Lustbader ED, Hu Y, Clapper ML, Zhou T, Wild CP, Xia XL, Baffoe-Bonnie A, Ofori-Adjei D, et al. Susceptibility to hepatocellular carcinoma is associated with genetic variation in the enzymatic detoxification of aflatoxin B1. Proc. Natl. Acad. Sci. U.S.A. 1995;92:2384–2387. doi: 10.1073/pnas.92.6.2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda CL, Stevens JF, Ivanov V, McCall M, Frei B, Deinzer ML, Buhler DR. Antioxidant and prooxidant actions of prenylated and nonprenylated chalcones and flavanones in vitro. J. Agric. Food Chem. 2000;48:3876–3884. doi: 10.1021/jf0002995. [DOI] [PubMed] [Google Scholar]
- Reed GA, Arneson DW, Putnam WC, Smith HJ, Gray JC, Sullivan DK, Mayo MS, Crowell JA, Hurwitz A. Single-dose and multiple-dose administration of indole-3-carbinol to women: Pharmacokinetics based on 3,3′-diindolylmethane. Cancer Epidemiol. Biomarkers Prev. 2006;15:2477–2481. doi: 10.1158/1055-9965.EPI-06-0396. [DOI] [PubMed] [Google Scholar]
- Reed GA, Sunega JM, Sullivan DK, Gray JC, Mayo MS, Crowell JA, Hurwitz A. Single-dose pharmacokinetics and tolerability of absorption-enhanced 3,3′-diindolylmethane in healthy subjects. Cancer Epidemiol. Biomarkers Prev. 2008;17:2619–2624. doi: 10.1158/1055-9965.EPI-08-0520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogan EG. The natural chemopreventive compound indole-3-carbinol: State of the science. In Vivo. 2006;20:221–228. [PubMed] [Google Scholar]
- Shertzer HG, Senft AP. The micronutrient indole-3-carbinol: Implications for disease and chemoprevention. Drug Metabol. Drug Interact. 2000;17:159–188. doi: 10.1515/dmdi.2000.17.1-4.159. [DOI] [PubMed] [Google Scholar]
- Singh S, Khar A. Biological effects of curcumin and its role in cancer chemoprevention and therapy. Anticancer Agents Med. Chem. 2006;6:259–270. doi: 10.2174/187152006776930918. [DOI] [PubMed] [Google Scholar]
- Stevens JF, Taylor AW, Clawson JE, Deinzer ML. Fate of xanthohumol and related prenylflavonoids from hops to beer. J. Agric. Food Chem. 1999a;47:2421–2428. doi: 10.1021/jf990101k. [DOI] [PubMed] [Google Scholar]
- Stevens JF, Taylor AW, Deinzer ML. Quantitative analysis of xanthohumol and related prenylflavonoids in hops and beer by liquid chromatography-tandem mass spectrometry. J. Chromatogr. A. 1999b;832:97–107. doi: 10.1016/s0021-9673(98)01001-2. [DOI] [PubMed] [Google Scholar]
- Strom SC, Pisarov LA, Dorko K, Thompson MT, Schuetz JD, Schuetz EG. Use of human hepatocytes to study P450 gene induction. Meth. Enzymol. 1996;272:388–401. doi: 10.1016/s0076-6879(96)72044-x. [DOI] [PubMed] [Google Scholar]
- Thompson EE, Kuttab-Boulos H, Yang L, Roe BA, Di Rienzo A. Sequence diversity and haplotype structure at the human CYP3A cluster. Pharmacogenomics J. 2006;6:105–114. doi: 10.1038/sj.tpj.6500347. [DOI] [PubMed] [Google Scholar]
- Walters DG, Young PJ, Agus C, Knize MG, Boobis AR, Gooderham NJ, Lake BG. Cruciferous vegetable consumption alters the metabolism of the dietary carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) in humans. Carcinogenesis. 2004;25:1659–1669. doi: 10.1093/carcin/bgh164. [DOI] [PubMed] [Google Scholar]
- Wang C, Bammler TK, Eaton DL. Complementary DNA cloning, protein expression, and characterization of alpha-class GSTs from Macaca fascicularis liver. Toxicol. Sci. 2002;70:20–26. doi: 10.1093/toxsci/70.1.20. [DOI] [PubMed] [Google Scholar]
- Yamazaki H, Inui Y, Wrighton SA, Guengerich FP, Shimada T. Procarcinogen activation by cytochrome P450 3A4 and 3A5 expressed in Escherichia coli and by human liver microsomes. Carcinogenesis. 1995;16:2167–2170. doi: 10.1093/carcin/16.9.2167. [DOI] [PubMed] [Google Scholar]