Abstract
BACKGROUND:
The genetics of intestinal lactase divide the world’s population into two phenotypes: the ability (a dominant trait) or inability (a recessive trait) to digest lactose. A prebiotic effect of lactose may impact the colonic flora of these phenotypes differently.
OBJECTIVE:
To detect and evaluate the effects of lactose on subjects divided according to their ability to digest lactose.
METHODS:
A total of 57 healthy maldigesters (n=30) and digesters (n=27) completed diet questionnaires, genetic and breath hydrogen testing, and quantitative stool analysis for species of bacteria. Log10 transformation of bacterial counts was compared with lactose intake in both groups using multiple regression analysis.
RESULTS:
There was a significant relationship between genetic and breath hydrogen tests. Daily lactose intake was marginally lower in lactose maldigesters (median [interquartile range] 12.2 g [31 g] versus 15 g [29.6 g], respectively). There was no relationship between lactose intake and breath hydrogen tests in either group. There were no differences in bacterial counts between the two groups, nor was there a relationship between bacterial counts and lactose intake in either group.
CONCLUSION:
The differential bacterial effects of lactose were not quantitatively detected in stool samples taken in the present study.
Keywords: Bacteria, Colonic, Lactose
Abstract
HISTORIQUE :
La génétique de la lactase intestinale divise la population mondiale en deux phénotypes : la capacité (un trait dominant) ou l’incapacité (un trait récessif) de digérer le lactose. Un effet prébiotique du lactose pourrait avoir des conséquences différentes sur la flore du côlon de ces phénotypes.
OBJECTIF :
Déceler et évaluer les effets du lactose sur des sujets divisés selon leur capacité de digérer le lactose.
MÉTHODOLOGIE :
Au total, 57 personnes en santé incapables (n=30) et capables (n=27) de digérer le lactose ont rempli des questionnaires sur leur régime, effectué le test de l’hydrogène expiré et le test génétique et subi une analyse quantitative de leurs selles pour déterminer les espèces de bactéries en cause. Les auteurs ont comparé la transformation log10 des numérations bactériennes dans les deux groupes lors de la consommation de lactose au moyen de l’analyse à régression multiple.
RÉSULTATS :
Les auteurs ont constaté un lien significatif entre le test de l’hydrogène expiré et le test génétique. La consommation quotidienne de lactose était légèrement plus faible chez les personnes incapables de digérer le lactose (médiane [plage interquartile] de 12,2 g [31 g] par rapport à 15 g [29,6 g], respectivement). Ils n’ont constaté aucun lien entre la consommation de lactose et les tests de l’hydrogène expiré dans les deux groupes. Ils n’ont remarqué ni différence de numérations bactériennes dans les groupes ni de lien entre les numérations bactériennes et la consommation de lactose dans l’un ou l’autre groupe.
CONCLUSION :
Les auteurs n’ont pas décelé d’effets bactériens différentiels quantitatifs du lactose dans les coprocultures prélevées dans le cadre de la présente étude.
Increasing levels of research in the past two decades have focused on the relationships between intestinal commensal (nonpathogenic) microflora and host interactions. These concepts are important for both pathogenesis and therapeutic intervention. Contextually, the concepts of prebiotics (mainly undigested carbohydrates that promote host beneficial bacteria in the lower intestine [1]) and probiotics (live microorganisms, derived mainly from human lower intestine, that deliver health benefits to the host [2]) are relevant. In gastroenterology, these putative relationships specifically apply to colorectal cancer and inflammatory bowel disease (IBD) (3). The possibility that lactose, a natural disaccharide present in most dairy foods (DF), may confer prebiotic effects, has not been extensively investigated.
The genetics of intestinal lactase divide the world’s population into two phenotypes: the ability (a dominant trait) or inability (a recessive trait) to digest lactose (4,5). In general, northern European populations and their descendants are lactose digesters (lactase persistent [LP]), more equatorial populations and their descendants are lactose maldigesters (lactase nonpersistent [LNP]). There are exceptions in Africa and the Middle East, with spotty distribution of LP populations amid predominantly LNP populations (6,7). Most Asians are LNP, with some LP Asians originating from Northern China and India (8).
The differential ability to digest lactose impacts DF consumption, generally with LNP consuming less DFs than LP populations (9). The reason for this relates, in part, to the development of symptoms in many LNP subjects when lactose is consumed above a threshold of 6 g to 10 g in a single dose (10,11). This occurs because lactose exerts an osmotic effect in the colon that increases water volume and colonic motility, thereby causing bloating, gas, cramps and diarrhea.
The consumption of DFs is also influenced by cultural habits. For example, in peri-Mediterranean cultures DFs are consumed more regularly than in most Asian cultures, even though LNP subjects represent the majority of the total population in the two regions. Cultural modification of regular DF consumption is facilitated by colonic adaptation (12–17), which putatively develops through enhanced bacterial metabolism and/or expansion of desired species such as Bifidobacteria and Lactobacilli (18–20).
As such, lactose may exert prebiotic effects at lower doses and may be more pronounced in LNP populations because much larger doses are required in LP individuals to spill into the colon (21). A differential impact of lactose on the two phenotypic populations could exert modifications of risk for the diseases mentioned above. Recently, we reported (9) that for five common cancers and IBD, quantifiable risk changes can be related to the national proportion of LP or LNP individuals. While meta-analyses (9) of patient-level data examining the effects of DFs on prostate, ovarian and breast cancer showed inverse effects with LNP subjects that approximated our epidemiological findings, opposite effects were found in both LP- and LNP-dominant populations for colorectal cancer. A protective effect was noted, raising the possibility that in LNP individuals, a lactose prebiotic effect reduces risk.
If lactose exerts such a differential bacterial effect on LNP and LP individuals, there may be bacterial floral differences between the two populations. As part of a study investigating the adaptive abilities of LNP and LP subjects to a daily lactose consumption regimen, we obtained baseline cross-sectional data that could test this hypothesis.
METHODS
Participants
The Sir Mortimer B Davis Jewish General Hospital in Montreal, Quebec, is a university-affiliated community-based hospital. It serves a large catchment area that includes both an indigenous working to middle class and a large, multiethnic, first- and second-generation immigrant population. All procedures were performed without knowledge of the genetic status of the study participants.
Healthy men and women were recruited via advertising within the hospital and the creation of a Web site outlining the study. Men and women of different ethnic and racial groups who were between 18 and 49 years of age were included. Participants did not have a priori knowledge of the purpose of any aspect of the study. However, subject recruitment targeted subjects who believed that they were lactose intolerant. Participants with stable chronic conditions such as dyslipidemia and thyroid disorders requiring treatment were included. Subjects who were pregnant, used antibiotics within the previous month, had acute or chronic digestive diseases including irritable bowel syndrome, chronic use of probiotics, nonsteroidal anti-inflammatory agents, narcotics or prokinetic drugs were excluded. Three included LNP participants were tested for celiac disease because their ethnic group is associated with a lower probability for lactose maldigestion; however, none were positive for antitissue transglutaminase antibodies. The study was approved by the Research and Ethics Committee of the Sir Mortimer B Davis Jewish General Hospital, and was registered through the Protocol Registration System, www.ClinicalTrials.gov, ID NCT00599859.
Qualified participants were asked to fast overnight (water allowed) after a suggested low-carbohydrate and low-DF supper, and presented to the gastroenterology laboratory between 08:00 and 10:00. Smoking was prohibited. Four variables were evaluated. Participants completed a three-day recall questionnaire. The use of lactose digestive aids (enzymes or lactose-free products) was noted; no subject admitted to using these agents. Blood was drawn for genetic testing of lactase. Participants were asked to provide approximately 10 g of stool (produced during the visit) in a sterile container. Every subject underwent a 4.5 h breath hydrogen test immediately following a 50 g lactose challenge, in which symptoms were also recorded (see below for details).
Diet questionnaire
Participants completed a three-day diet recall questionnaire that included 27 items deemed to contain lactose. A dietitian (LFC) analyzed the questionnaires and a daily mean intake of lactose, based on existing food tables (22,23), was calculated using a previously published validated model (24).
Genetic lactase tests
Genetic classification in adults may reflect obligatory lactase/lactose interactions more effectively than indirect tests because most adult LNP subjects develop a reduced ability to digest lactose by 20 years of age (25). Also, any factors that could affect the results of indirect tests should be controlled for (26). Blood was analyzed for the predominant genetic polymorphism in the lactase gene promoter region. The polymorphism C/T-13910, associates completely with the promoter region of LP and LNP status in the majority of European and Asian populations (27, 28). DNA was prepared using a DNA isolation kit (Gentra Systems, USA). A real-time polymerase chain reaction (PCR) assay based on fluorescence resonance energy transfer (29,30) was used with the LightCycler DNA Master hybridization probes kit (Roche Diagnostics, Germany) for analysis of the C/T genetic polymorphism. The forward primer: 5′–GCTTGGTAAGCATTTGAGT–3′ and the reverse primer: 5′–GTTGAAGCGAAGATGGG–3′, were used for PCR amplification. The sensor probe: 5′–ATGTAGCCCCTGGCCT–3′ was labelled with LC-640 at the 5′ end, and the anchor probe: 5′–CCTCTGCGCTGGCAATACAGATAAGA–3′ was labelled with fluorescein at the 3′ end. The mutation was detected by melting curve analysis. In this polymorphism, the C/C genotype is fully LNP, the T/T genotype is LP, the heterozygote C/T is also considered LP, but with a reduced measured level of intestinal lactase (27). The cohort was thereby classified into LP (T/T and C/T) and LNP (C/C) phenotypes.
Breath hydrogen test
The breath hydrogen test is the most effective indirect test to physiologically assess bacterial action and response to dietary manipulation (26,31). Participants refrained from smoking or eating (water was allowed) during the test, and generally remained calm throughout. After an overnight fast and a baseline breath hydrogen recording, 50 g of lactose mixed in water (200 mL to 250 mL) was ingested over several minutes. Breath hydrogen was measured (in parts per million [ppm]) using a validated handheld hydrogen chemical sensor (EC60 gastrolyzer, Bedfont Scientific Ltd, United Kingdom). This instrument has an analytical range of 0 ppm to 2000 ppm (32,33). Breath hydrogen was measured initially at 15 min intervals for the first 90 min and every 30 min thereafter to the completion at 4.5 h. After establishing a baseline, it was subtracted from values recorded at each subsequent time interval up to 4.5 h. In general, an acceptable baseline value is 20 ppm or less. A definite positive value is defined as 20 ppm above baseline (26,31). Hydrogen readings at each 30 min interval (after subtracting the baseline score) were summed to obtain a value for ‘total breath hydrogen’ (TBH2).
Symptom scores
Symptoms were recorded at the same time intervals as TBH2 on a Likert scale measuring from 0 to 3. Each symptom associated with sugar malabsorption (bloating, gas and cramps) were assigned a score of 0 for no symptom, 1 for mild, 2 for moderate and 3 for severe symptoms. Diarrhea was scored as 0 (none) or 1 (present). The total symptom score (TSS) for each participant was the sum of the scores for the same time points as TBH2, and has a range from 0 to 90 (ie, 9×3×3+9×1).
Stool analysis
In general, participants were successful in providing stool samples during each visit. However, in two cases, stool samples were obtained after consuming coffee or lactulose 150 min into the breath test. Both of these participants were LP. Samples were placed on ice and were shipped to the microbiology laboratory (at McGill University, Montreal, Quebec) within 2 h. The samples were analyzed on the same day. No samples were sent for analysis on weekends.
A modification from a previous report on fecal bacteria was used for quantification of total anaerobes, total enterobactericiae, total bifidobacteria and total lactobacilli (34,35). Fresh fecal samples (corrected to 100 g) were introduced into an anaerobic jar containing a gas mixture (GasPak, BBL, USA). A sample of 1.0 g of feces was removed and homogenized in 4 mL of prepared brain heart infusion broth supplemented with yeast extract and 5-cysteine hydrochloride. A serial dilution was made and plated on bacteroides (bile esculin), Enterobacteriaceae (MacConkey), Bifidobacterium (Beerens) and Lactobacillus (Rogosa) agar. The plates were incubated anaerobically for two to three days at 37°C with the exception of the MacConkey plates, which were incubated aerobically overnight at 37°C.
Stool enzyme analyses
Both beta (β)-D-galactosidase (β-gal, units/g stool) and N-acetyl-β-D-glucosaminidase (NAG, mL/min) were measured. Both of these enzymes are universal bacterial markers. β-gal, which is found in multiple species of bacteria, is considered part of the clinical definition of colonic adaptation (17) and may precede bacterial colony forming units (CFU) expansion in response to prebiotics (36) and, because of this, elevated β-gal in LNP individuals would support colonic adaptation. Measurement of fecal β-gal was performed by the O-nitrophenyl-β-D-galactopyranoside (ONPG) method. Briefly, 20 µL of stool in buffer was added to 480 μg of ONPG in sodium phosphate buffer (pH 7.0). The reaction was allowed to proceed for 10 min at 45°C. Sodium carbonate 1 M was added to stop the reaction and optical density at 420 nm (37) was subsequently read.
NAG was measured because it is also a ubiquitous bacterial enzyme involved in mucus digestion and may exert bacteriocidal effects (38,39); evaluating this enzyme may suggest whether it is also affected by colonic adaptation. NAG was analyzed using a buffered sample of 0.1 mL of stool sample added to 1 mL of substrate. After incubation at 37°C for 30 min with 0.5 mL of sodium carbonate buffer to stop the reaction, the optical density was read at 405 nm (37).
Statistical analysis
Age (years, normally distributed) was reported as mean ± SD. The remaining continuous variables (skewed distributions) were reported as medians with interquartile range (IQR). Categorical variables were reported as proportions ± SE. The bacterial counts and enzyme activity values were close to normally distributed after log10 transformation and, therefore, analyzed as such.
The demographic variables TBH2, TSS, daily lactose intake, sex and race for each group were reported separately. The primary outcome of interest (stool counts for Bifidobacteria and Lactobacilli species) of LP and LNP subjects were compared using the unpaired t test. The unpaired t test was also used to compare LP and LNP subjects for secondary outcomes of total anaerobic counts, total enterobacteriaceae counts, β-gal and NAG. As an exploratory analysis, the ratio of bifidobacteria to total anaerobes and lactobacilli to total anaerobes between both populations were also compared using the unpaired t test. This analysis examined the relationship between the ratio of bacterial counts and lactose intake in LNP and LP subjects, and used multiple regression analysis to account for any possible interaction. For all analyses, P<0.05 was considered to be statistically significant.
The power calculation was based on the results of the only study to date evaluating microfloral response to DFs in humans (20). Setting alpha at 0.05 and power at 0.8, 45 subjects per group were required to detect a twofold increase in bifidobacteria or lactobacilli counts.
RESULTS
Fifty-seven multiethnic, healthy participants (23 men and 34 women) were recruited between September 2006 and March 2008. Demographic information for LNP and LP subjects is presented in Table 1. The median daily intake of lactose was slightly less in LNP subjects than in LP subjects. The proportion of subjects who consumed 20 g of lactose or more per day was 40.0% (12 of 30) in the LNP group and 40.7% (11 of 27) in LP subjects. As expected, both TBH2 and TSS were higher with lactose challenge in LNP subjects than in LP subjects.
TABLE 1.
Demographic variables
| Demographic |
Genetic variant at position 13910 of the lactase structural gene |
|
|---|---|---|
| Lactase nonpersistent (C/C) (n=30) | Lactase persistent (C/T or T/T) (n=27) | |
| Categorical variables | ||
| Women | 63 (9) | 56 (10) |
| Race | ||
| African | 27 (8) | 7 (5) |
| Asian | 30 (8) | 0 (0) |
| Caucasian | 43 (9) | 93 (5) |
| Positive hydrogen breath test | 97 (3) | 7 (5) |
| C/T genetic status | NA | 59 (10) |
| Continuous variables | ||
| Age, years | 32.1±8.7 | 26.3±6.5 |
| Lactose intake, mL | 12.2 (31) | 15 (29.6) |
| Lactose consumption ≤10 g/day, % | 47 (9) | 44 (10) |
| Total breath hydrogen, ppm | 424.5±377.5 | 36±79 |
| Total symptom score | 10±15 | 0±5.5 |
Categorical variables are presented as % (SE). Continuous variables are presented as mean ± SD if normally distributed and as median (interquartile range) if there is a skewed distribution. Total breath hydrogen was calculated by subtracting the baseline breath hydrogen from each of the breath hydrogen scores at 30 min, 60 min, 90 min, 120 min, 150 min, 180 min, 240 min and 270 min, and then adding them together. Total symptom score was calculated by summing the symptom score for each time point at 30 min, 60 min, 90 min, 120 min, 150 min, 180 min, 240 min and 270 min. For symptoms of bloating, gas and cramps, the following scores were used: 0 = absent, 1 = mild, 2 = moderate, 3 = severe. For diarrhea, a score of 0 = none and 1 = present. The theoretical maximum total symptom score is 90. C Cytosine; NA Not available; ppm Parts per million; T Thymine
Defining genetic homozygous mutants C/C as LNP, the breath hydrogen data demonstrated a high sensitivity (96.7%) and specificity (92.6%) for the genetic categorization. Within the LP population, 59.3% were C/T and 40.7% were homozygous T/T.
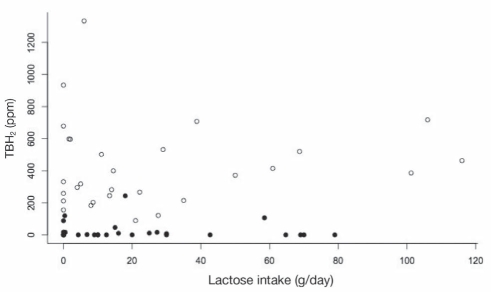
When TBH2 was plotted against average daily lactose consumption, there was a poor relationship in both LP subjects (R2=0.01; P=0.68) and LNP subjects (R2=0.01; P=0.60) (Figure 1). When we explored the relationship only in LNP subjects who consumed 20 g or more of lactose per day (n=12), there was still no relationship (slope=3.0, R2=0.26, P=0.09).
Figure 1).
The total breath hydrogen (TBH2) during the lactose challenge (sum of the increase in breath hydrogen over baseline at each 30 min interval from 30 min to 270 min) is plotted against the daily dairy food intake for lactase persistent (LP) subjects (solid circles) and lactase nonpersisitent (LNP) subjects (open circles). Regression lines are not plotted (R2=0.01; P>0.6 for both LP and LNP subjects). Note that some points overlap and, therefore, not all points are clearly visible. ppm Parts per million
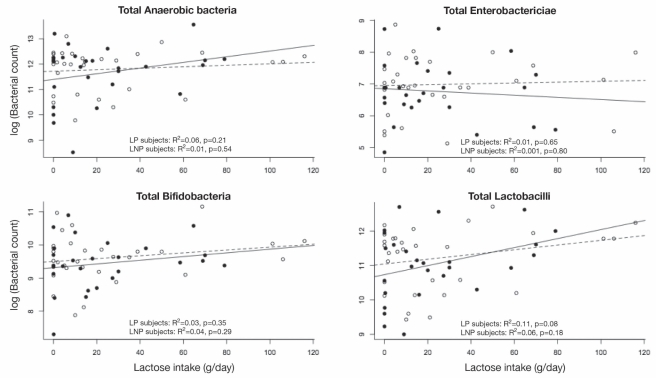
Comparison of colony counts of four different bacterial species (log10 CFU/g stool [IQR]) showed no statistical differences for LNP versus LP subjects: total anaerobes 11.8 (0.8) versus 11.6 (1.1), P=0.58; total enterobacteriaceae 7.0 (0.9) versus 6.8 (0.9), P=0.39; total bifidobacteria species 9.6 (0.7) versus 9.4 (0.8), P=0.39; and total lactobacilli species 11.2 (0.9) versus 11.0 (1.0), P=0.45.
Figure 2 shows scatter plots of the relationships for each of the four types of bacteria, with the associated regression lines (calculated for LNP and LP groups separately) superimposed. There was no difference in the relationship between LNP and LP subjects for lactose intake and any of the bacterial species counts (P>0.35 for all interactions, P>0.34 for all LNP and LP comparisons). There was no relationship between any of the bacterial counts and lactose intake among either LNP or LP subjects.
Figure 2).
The log10 of bacterial counts are plotted against the daily lactose intake for each of the different types of bacteria. The results for lactase persistent (LP) subjects are plotted as solid circles, and the regression line for LP subjects is plotted as a solid line. The results for lactase nonpersistent (LNP) subjects are shown as open circles and a dashed line. Multiple regression analyses for the relationship between each of the bacterial counts and lactose intake failed to show an effect of dairy food, genetic status or an interaction between lactose intake and genetic status
To compensate for possible variability in analyzing stool samples on different days, the log10 of the ratio of total bifidobacteria to total anaerobes, and lactobacilli to total anaerobes were also compared between LNP and LP subjects. There were no statistically significant differences observed in ratios for log10(bifidobacteria/total anaerobes) (−2.2 LNP, −2.2 LP; P=0.92) or for log10(lactobacilli/total anaerobes) (−0.57 LNP, −0.62 LP; P=0.76). On a numerical scale, the corresponding ratios are 0.6% for both LNP and LP subjects for bifidobacteria to total anaerobes, and 26.9% for LNP and 24.0% for LP subjects for lactobacilli to total anaerobes, respectively.
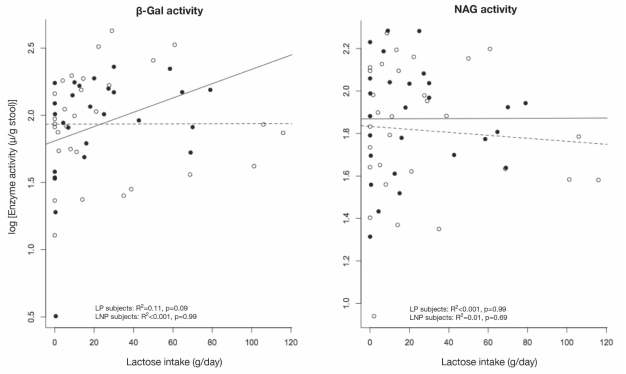
Enzyme activities between LNP and LP subjects also did not reveal any significant differences. The activity for log10(β-gal) in CFU/g stool (IQR) was 1.93 (0.38) for LNP and 1.93 (0.40) for LP (P=0.96). The activity for log10(NAG) was 1.82 (0.31) for LNP and 1.87 (0.26) for LP (P=0.47). There was no relationship between log10(β-gal) to increasing daily lactose intake (P=0.99), genetic status (P=0.36) or any interaction (P=0.16; R2 adjusted = 0.002 for the model). Similar results were obtained for log10(NAG) (increased lactose P=0.67; genetic status P=0.73; interaction P=0.79). The observed data are plotted in Figure 3, with the superimposed regression lines calculated for each group separately.
Figure 3).
The log of enzyme activities are plotted against the daily lactose intake for beta (β)-D-galactosidase (B-gal, left panel) and N-acetyl-β-D-glucosaminidase (NAG, right panel). The results for lactase persistent (LP) subjects are plotted as solid circles, and the regression line for LP subjects is plotted as a solid line. The results for lactase nonpersistent (LNP) subjects are shown as open circles and a dashed line. Multiple regression analyses for the relationship between each of the enzyme activities and dairy food intake failed to show an effect of dairy, genetic status or an interaction between lactose and genetic status
DISCUSSION
To our knowledge, the present study is the first clinical attempt to examine the relationship between fecal bacterial counts and lactose intake in LNP and LP subjects. We confirm that the lactose breath test accurately reflects genetic findings in a mixed indigenous (largely European ancestral) and immigrant population, and is similar to other reports (40–42). Average lactose intake, however, by both groups was similar and there was no relationship between lactose intake and TBH2.
Quantitative results of four bacterial groups, including two that contained subspecies with potential health benefits (bifidobacteria and lactobacilli), did not show any clinically relevant differences between LNP and LP participants. Similarly, there were no clinically relevant differences in bacterial lactase (β-gal) or NAG activity.
The intake of similar quantities of lactose (representing the equivalent of approximately one to two cups of milk per day) by both groups is somewhat unexpected because there is a clear, inverse relationship with the national prevalence of LNP status in the population (9). Furthermore, we previously demonstrated lower lactose intake in LNP subjects of Asian descent compared with LP subjects (43). As such, our population of LP subjects may have been somewhat atypical in that they may have restricted lactose intake believing that they were lactose intolerant.
We also failed to observe an expected inverse relationship between TBH2 and increased lactose intake (44). The putative reason for the inverse relationship is that regular larger threshold values might lead to colonic adaptation (17). Therefore, our LNP population was either not adapted or too few subjects consumed high-enough amounts of lactose. While only 12 of 30 LNP participants (40%) consumed 20 g of lactose or more (a daily intake determined to clearly separate hydrogen response from those taking less than this amount), there was no trend toward reduced hydrogen and, in fact, the slope of the relationship was in the opposite direction. The lack of a relationship may have occurred by chance, given our sample size or due to different study methods; we do not have a clear explanation for this finding.
Some studies evaluating the effects of different diets on fecal microflora have published qualitative differences (44), and ingestion of prebiotics like inulin, oligofructose and even lactulose (derived from lactose [45]) cause measurable changes in Bifidobacteria species (46,47). Contrary to the predictions based on the above results, we found almost identical quantitative distribution of bacteria (and enzyme activity) between LP and LNP populations. Ranges of CFUs in the different groups are comparable with ranges previously published using different methods (48–50). However, our results are also consistent with other studies. There were no differences in microbial counts between LP participants with or without milk sensitivity (51). As well, there were no differences in β-gal activity among LNP subjects who had or did not have symptoms of lactose intolerance (52). Therefore, lactose intake at doses described in the present study does not appear to exert distal bacterial effects in the two populations.
Therefore, can we conclude that lactose has no prebiotic potential based on our findings? While this remains a possibility, a number of issues need to be addressed. First, despite the expectation that the three-day diet recall reflects actual diet intake, this may be inaccurate. Furthermore, the random errors inherent in diet recall methods have been well delineated (53). It is also important, however, to note that the median intake of lactose for LNP individuals was quite close to the maximum limit of lactose digestibility (10,11). Very little lactose would then reach the colon, reducing the likelihood that these participants were adapted. Second, there is a large variation between individuals in bacterial counts and activity, which is one reason why bacterial counts are traditionally reported on a log scale. Therefore, a doubling of bacterial counts amounts to only a 0.3 log unit difference – this may be missed in an observational study such as the one we performed. Therefore, future longitudinal studies or large population-based studies may still reveal differences. Possible qualitative differences in species of bacteria may also exist and these could be explored in future studies.
SUMMARY
We report a cross-sectional analysis of colonic microbial flora in LP and LNP subjects. No meaningful differences in bacterial counts or enzyme activity between the groups were observed. However, our LP population may have been somewhat atypical in their lactose intake, which would restrict the generalizability of the results.
Footnotes
FUNDING AND DISCLOSURE: This study was supported by a Grant in Aid from the Danone Institute of Canada, Quebec Branch. None of the authors have any conflicts of interest to disclose.
REFERENCES
- 1.Gibson GR, Probert HM, Van Loo J, et al. Dietary modulation of the human colonic microbiota: Updating the concept of prebiotics. Nutr Res Rev. 2004;17:257. doi: 10.1079/NRR200479. [DOI] [PubMed] [Google Scholar]
- 2.Fuller R. Probiotics in man and animals. J Appl Bacteriol. 1989;66:365–78. [PubMed] [Google Scholar]
- 3.Szilagyi A. Risk modification of inflammatory bowel diseases by vitamin D, calcium and lactase/lactose interactions: Arguments based on relationships between IBD and colorectal cancer. In: Urster JP, editor. Research Trends in Nutrition for the Middle Aged and Elderly. New York: Nova Science Publishers; 2008. pp. 7–51. [Google Scholar]
- 4.Gilat T. Lactase deficiency: The world pattern today. Isr J Med Sci. 1979;15:369–73. [PubMed] [Google Scholar]
- 5.Sahi T, Isokoski M, Jussila J, et al. Recessive inheritance of adult-type lactose malabsorption. Lancet. 1973;2:823–6. doi: 10.1016/s0140-6736(73)90862-3. [DOI] [PubMed] [Google Scholar]
- 6.Ingram CJ, Elamin MF, Mulcare CA, et al. A novel polymorphism associated with lactose tolerance in Africa: Multiple causes for lactase persistence? Hum Genet. 2007;120:779–88. doi: 10.1007/s00439-006-0291-1. [DOI] [PubMed] [Google Scholar]
- 7.Scrimshaw NS, Murray EB. The acceptability of milk and milk products in populations with high prevalence of lactose intolerance. Am J Clin Nutr. 1988;48S:1079. doi: 10.1093/ajcn/48.4.1142. [DOI] [PubMed] [Google Scholar]
- 8.Sun HM, Qiao YD, Chen F, et al. The lactase gene-13910T allele can not predict the lactase-persistence phenotype in north China, Asia. Pac J Clin Nutr. 2007;16:598. [PubMed] [Google Scholar]
- 9.Shrier I, Szilagyi A, Correa JA. Impact of lactose containing foods and the genetics of lactase on diseases: An analytical review of population data. Nutr Cancer. 2008;60:292–300. doi: 10.1080/01635580701745301. [DOI] [PubMed] [Google Scholar]
- 10.Hertzler SR, Huynh BC, Savaiano DA. How much lactose is low lactose? Am J Diet Assoc. 1996;96:243–6. doi: 10.1016/S0002-8223(96)00074-0. [DOI] [PubMed] [Google Scholar]
- 11.Oku T, Nakamura S, Ichinose M. Maximum permissive dose of lactose and lactitol for transitory diarrhea, and utilizable capacity for lactose in Japanese female adults. J Nutr Sci Vitaminol. 2005;51:51. doi: 10.3177/jnsv.51.51. [DOI] [PubMed] [Google Scholar]
- 12.Habte D, Sterky G, Hjalmarsson B. Lactose malabsorption in Ethiopian children. Acta Paediatr Scand. 1973;62:649. doi: 10.1111/j.1651-2227.1973.tb17080.x. [DOI] [PubMed] [Google Scholar]
- 13.Sadre M, Karbasi K. Lactose intolerance in Iran. Am J Clin Nutr. 1979;32:1948. doi: 10.1093/ajcn/32.9.1948. [DOI] [PubMed] [Google Scholar]
- 14.Villar J, Kestler E, Castillo P, et al. Improved lactose digestion during pregnancy: A case of physiologic adaptation? Obstet Gynecol. 1988;71:697. [PubMed] [Google Scholar]
- 15.Szilagyi A, Salomon R, Martin M, et al. Lactose handling by women with lactose malabsorption is improved during pregnancy. Clin Invest Med. 1996;19:416. [PubMed] [Google Scholar]
- 16.Pribila BA, Hertzler SR, Martin BR, et al. Improved lactose digestion and intolerance among African-American adolescent girls fed a dairy-rich diet. J Am Diet Assoc. 2000;100:524. doi: 10.1016/S0002-8223(00)00162-0. [DOI] [PubMed] [Google Scholar]
- 17.Hertzler SR, Savaiano DA. Colonic adaptation to daily lactose feeding in lactose maldigesters reduces lactose intolerance. Am J Clin Nutr. 1996;64:232. doi: 10.1093/ajcn/64.2.232. [DOI] [PubMed] [Google Scholar]
- 18.Jiang T, Savaiano DA. In vitro lactose fermentation by human colonic bacteria is modified by Lactobacillus acidophilus supplementation. J Nutr. 1997;127:1489–95. doi: 10.1093/jn/127.8.1489. [DOI] [PubMed] [Google Scholar]
- 19.Jiang T, Savaiano DA. Modification of colonic fermentation by bifidobacteria and pH in vitro. Impact on lactose metabolism, shortchain fatty acid, and lactate production. Dig Dis Sci. 1997;42:2370–77. doi: 10.1023/a:1018895524114. [DOI] [PubMed] [Google Scholar]
- 20.Ito M, Kimura M. Influence of lactose on faecal microflora in lactose maldigesters. Microb Ecol Health Dis. 1993:73–76. [Google Scholar]
- 21.Szilagyi A. Review article: Lactose – a potential prebiotic. Aliment Pharmacol Ther. 2002;16:1591. doi: 10.1046/j.1365-2036.2002.01321.x. [DOI] [PubMed] [Google Scholar]
- 22.Canadian Nutrient File. 2007b. < http://www.hc-sc.gc.ca/fn-an/nutrition/fiche-nutri-data/index_e.html> (Version current at March 3, 2009).
- 23.USDA National Nutrient Database for Standard Reference, Release 19. < http://www.ars.usda.gov/main/site_main.htm?modecode=12-35-45-00> (Version current at March 3, 2009).
- 24.Cooper GS, Busby MG, Fairchild AP. Measurement of lactose consumption reliability and comparison of two methods. Ann Epidemiol. 1995;5:473–7. doi: 10.1016/1047-2797(95)00063-1. [DOI] [PubMed] [Google Scholar]
- 25.Seppo L, Tuure T, Korpela R, et al. Can primary hypolactasia manifest itself after the age of 20 years? A two-decade follow-up study. Scand J Gastroenterol. 2008;43:1082–7. doi: 10.1080/00365520802095485. [DOI] [PubMed] [Google Scholar]
- 26.Romagnuolo J, Schiller D, Bailey RJ. Using breath tests wisely in a gastroenterology practice: An evidence-based review of indications and pitfalls in interpretation. Am J Gastroenterol. 2002;97:1113–26. doi: 10.1111/j.1572-0241.2002.05664.x. [DOI] [PubMed] [Google Scholar]
- 27.Enattah NS, Kuokkanen M, Forsblom C, et al. Correlation of intestinal disaccharidase activities with the C/T(13910) variant and age. World J Gastroenterol. 2007;13:3508–12. doi: 10.3748/wjg.v13.i25.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuokkanen M, Enattah NS, Oksanen A, et al. Transcriptional regulation of the lactase-phlorizin hydrolase gene by polymorphisms associated with adult-type hypolactasia. Gut. 2003;52:647–52. doi: 10.1136/gut.52.5.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nilsson TK, Johansson CA. A novel method for diagnosis of adult hypolactasia by genotyping of the 13910 C/T polymorphism with pyrosequencing technology. Scand J Gastroenterol. 2004;39:287–90. doi: 10.1080/00365520310008304. [DOI] [PubMed] [Google Scholar]
- 30.Chao CK, Sibley E. PCR-RFLP genotyping assay for a lactase persistence polymorphism upstream of the lactase-phlorizin hydrolase gene. Genet Test. 2004;8:190–3. doi: 10.1089/gte.2004.8.190. [DOI] [PubMed] [Google Scholar]
- 31.Newcomer AD, McGill DB, Thomas PJ, et al. Prospective comparison of indirect methods for detecting lactase deficiency. N Engl J Med. 1075;293:1232–6. doi: 10.1056/NEJM197512112932405. [DOI] [PubMed] [Google Scholar]
- 32.Metz G, Gassull MA, Leeds AR, et al. A simple method of measuring breath hydrogen in carbohydrate malabsorption by end-expiratory sampling. Clin Sci Mol Med. 1976;50:237–40. doi: 10.1042/cs0500237. [DOI] [PubMed] [Google Scholar]
- 33.Van der Klei-van Moorsel JM, Douwes AC, van Oeveren JP. New principle for estimation of hydrogen in expired air. Eur J Pediatr. 1984;141:221–4. doi: 10.1007/BF00572764. [DOI] [PubMed] [Google Scholar]
- 34.Jung SJ, Houde R, Baurhoo B, Zhao X, Lee BH. Effects of galacto-oligosaccharides and a Bifidobacteria lactis-based probiotic strain on the growth performance and fecal microflora of broiler chickens. Poultry Sci. 2008;87:1694–9. doi: 10.3382/ps.2007-00489. [DOI] [PubMed] [Google Scholar]
- 35.Kim GB, Miyamoto CM, Meighen EA, et al. Cloning and characterization of the bile salt hydrolase genes (bsh) from Bifidobacterium bifidum strains. Appl Environ Microbiol. 2004;70:5603–12. doi: 10.1128/AEM.70.9.5603-5612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tannock GW, Munro K, Bibiloni R, et al. Impact of consumption of oligosaccharide-containing biscuits on the fecal microbiota of humans. Appl Environ Microbiol. 2004;70:2129–36. doi: 10.1128/AEM.70.4.2129-2136.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuen CT, Price RG, Chattagoon L, et al. Colorimetric assays for N-acetyl b-D glucosaminidase and b-D-galactosidase in human urine using newly-developed-nitrosyl substrates. Clin Chim Acta. 1982;124:195–204. doi: 10.1016/0009-8981(82)90387-4. [DOI] [PubMed] [Google Scholar]
- 38.Mayer C, Vocadio DJ, Mah M, et al. Characterization of a N-acetylhexosaminidase/-glucosidase from Cellulomonas fimi. FEBS. 2006;273:2929–41. doi: 10.1111/j.1742-4658.2006.05308.x. [DOI] [PubMed] [Google Scholar]
- 39.Langley DB, Harty DW, Jacques NA, et al. Structure of N-acetyl-beta-D-glucosaminidase (GcnA) from the endocarditis pathogen Streptococcus gordonii and its complex with the mechanism-based inhibitor NAG-thiazoline. J Mol Biol. 2008;377:104–16. doi: 10.1016/j.jmb.2007.09.028. [DOI] [PubMed] [Google Scholar]
- 40.Hogenauer C, Hammer HF, Melltzer K, et al. Evaluation of a new DNA test compared with the lactose hydrogen breath test for the diagnosis of lactase non-persistence. Eur J Gastroenterol Hepatol. 2005;17:371–6. doi: 10.1097/00042737-200503000-00018. [DOI] [PubMed] [Google Scholar]
- 41.Szilagyi A, Malolepszy E, Hamard X, et al. Comparison of a real-time PCR assay for lactase genetic polymorphism with standard indirect tests for lactose maldigestion. Clin Gastroenterol and Hepatol. 2007;5:192–6. doi: 10.1016/j.cgh.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 42.Waud JP, Mathews SB, Campbell AK. Measurement of breath hydrogen and methane, together with lactase genotype, defines the current best practice for investigations of lactose sensitivity. Ann Clin Biochem. 2008;45:50–8. doi: 10.1258/acb.2007.007147. [DOI] [PubMed] [Google Scholar]
- 43.Noack-Loebel C, Kuster E, Rusch V, et al. Influence of different dietary regimens upon the composition of the human fecal flora. Prog Food Nutr Sci. 1983;7:127–31. [PubMed] [Google Scholar]
- 44.Szilagyi A, Malolepszy P, Yesovitch S, et al. Inverse dose effect of pretest dietary lactose intake on breath hydrogen results and symptoms in lactase nonpersistent subjects. Dig Dis Sci. 2005;50:2178–82. doi: 10.1007/s10620-005-3028-4. [DOI] [PubMed] [Google Scholar]
- 45.Montgomery EM, Hudson CS. Relationship between rotary power and structure in the sugar group XXVII. Synthesis of a new saccharide ketose (lactulose) from lactose. J Am Chem Soc. 1930;52:2101. [Google Scholar]
- 46.Kruse H-P, Kleesen B, Blaut M. Effects of inulin on faecal bifidobacteria in human subjects. Br J Nutr. 1999;82:375–82. doi: 10.1017/s0007114599001622. [DOI] [PubMed] [Google Scholar]
- 47.Bouhnik Y, Attar A, Joly FA, et al. Lactulose ingestion increases faecal bifidobacterial counts: A randomised double-blind study in healthy humans. Eur J Clin Nutr. 2004;58:462–6. doi: 10.1038/sj.ejcn.1601829. [DOI] [PubMed] [Google Scholar]
- 48.McLean Baird I, Walters RL, Davies PS, et al. The effects of two dietary fiber supplements on gastrointestinal transit, stool weight and frequency, and bacterial flora, and fecal bile acids in normal subjects. Metabolism. 1977;26:117–28. doi: 10.1016/0026-0495(77)90047-6. [DOI] [PubMed] [Google Scholar]
- 49.Franks AH, Harmsen HJM, Raangs GC, et al. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl Environ Microbiol. 1998;64:3336–45. doi: 10.1128/aem.64.9.3336-3345.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swidsinski A, Loening-Baucke V, Vaneechoutte M, et al. Active Crohn’s disease and ulcerative colitis can be specifically diagnosed and monitored based on the biostructure of the fecal flora. Inflamm Bowel Dis. 2008;14:147–61. doi: 10.1002/ibd.20330. [DOI] [PubMed] [Google Scholar]
- 51.Apostolou E, Pelto L, Kirjavainen PV, et al. Differences in the gut bacteria flora in flora of healthy and milk-sensitive adults, as measured by fluorescence in situ hybridization. FEMS Immunol Med Microbiol. 2001;30:217–21. doi: 10.1111/j.1574-695X.2001.tb01573.x. [DOI] [PubMed] [Google Scholar]
- 52.He T, Priebe MG, Vonk RJ, et al. Identification of bacteria with galactosidase activity in faeces from lactase non-persistent subjects. FEMS Micrbiol Ecol. 2005;54:463–9. doi: 10.1016/j.femsec.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 53.Bingham SA. Limitations of the various methods for collecting dietary intake data. Ann Nutr Metab. 1991;35:117–27. doi: 10.1159/000177635. [DOI] [PubMed] [Google Scholar]