Abstract
Previous studies have stipulated Hec1 as a conserved kinetochore component critical for mitotic control in part by directly binding to kinetochore fibers of the mitotic spindle and by recruiting spindle assembly checkpoint proteins Mad1 and Mad2. Hec1 has also been reported to localize to centrosomes, but its function there has yet to be elucidated. Here, we show that Hec1 specifically colocalizes with Hice1, a previously characterized centrosomal microtubule-binding protein, at the spindle pole region during mitosis. In addition, the C-terminal region of Hec1 directly binds to the coiled-coil domain 1 of Hice1. Depletion of Hice1 by small interfering RNA (siRNA) reduced levels of Hec1 in the cell, preferentially at centrosomes and spindle pole vicinity. Reduction of de novo microtubule nucleation from mitotic centrosomes can be observed in cells treated with Hec1 or Hice1 siRNA. Consistently, neutralization of Hec1 or Hice1 by specific antibodies impaired microtubule aster formation from purified mitotic centrosomes in vitro. Last, disruption of the Hec1/Hice1 interaction by overexpressing Hice1ΔCoil1, a mutant defective in Hec1 interaction, elicited abnormal spindle morphology often detected in Hec1 and Hice1 deficient cells. Together, the results suggest that Hec1, through cooperation with Hice1, contributes to centrosome-directed microtubule growth to facilitate establishing a proper mitotic spindle.
INTRODUCTION
In dividing eukaryotic cells, a dynamic bipolar spindle is pivotal for efficient and accurate chromosome congression and subsequent segregation into two progeny cells. Failure to do so often leads to segregation errors and aneuploidy, a hallmark feature of most cancers. The assembly of a proper mitotic spindle can be achieved by multiple cooperative mechanisms mediated by numerous motor and nonmotor proteins, including those with microtubule binding and modulating activities (Compton, 2000; Karsenti and Vernos, 2001; Kline-Smith and Walczak, 2004).
Hec1, also known as Ndc80, is an evolutionarily conserved coiled-coil protein critical for mitotic progression (Chen et al., 1997a,b; Wigge et al., 1998). It contains three leucine-heptad coiled-coil domains at the C-terminal region, which are thought to mediate multiple protein–protein interactions (Chen et al., 1997b; Kline-Smith et al., 2005; Ciferri et al., 2008). The N-terminal region of Hec1 is a microtubule binding domain structurally similar to the calponin-homology domain of the microtubule binding protein EB1 (Cheeseman et al., 2006; Wei et al., 2007; Ciferri et al., 2008). In cells, Hec1 is a major binding partner for another coiled-coil protein, Nuf2. The Hec1/Nuf2 dimer resides at the outer kinetochore layer and orients in such a way that their N-terminal microtubule binding module projects outward, whereas the C-terminal tail is anchored by the inner layer components Spc24/Spc25 (McCleland et al., 2003; Emanuele et al., 2005; Wilson-Kubalek et al., 2008). These four proteins can form a dumbbell-shaped heterotetramer in an equal molecular ratio in vitro (Ciferri et al., 2005; Wei et al., 2005). At kinetochores, the Hec1 complex associates with the KNL1/Spc105 and the Mtw1/Mis12 complexes, and together they form the KNL-1/Mis12 complex/Ndc80 complex network that can provide two microtubule binding interfaces for kinetochore fiber attachment (Cheeseman et al., 2006). The microtubule binding activity of Hec1 is inhibited upon phosphorylation of its N-terminal tail by kinetochore-associated kinase Aurora B. This serves as a critical mechanism for correcting improper microtubule attachment at the kinetochore (Cheeseman et al., 2006; DeLuca et al., 2006).
Importantly, Hec1 overexpression has been observed in a variety of human cancers and was found to associate with adverse clinical outcomes of primary breast cancers and cases with multiple cancers (Chen et al., 1997a; van't Veer et al., 2002; Glinsky et al., 2005). In an inducible mouse model, overexpression of Hec1 was shown to result in spindle checkpoint hyperactivation correlating with eventual significant tumor formation, mainly lung adenoma and hepatocellular adenoma (Diaz-Rodriguez et al., 2008). This animal phenotype is recapitulative of those observed in Mad2-overexpressing mice (Sotillo et al., 2007). Hec1 has now emerged as a novel therapeutic target for potential cancer intervention by using strategies of RNA interference (RNAi) or small molecular inhibitors in part because of its specific requirement in the mitotic process (Gurzov and Izquierdo, 2006; Li et al., 2007; Wu et al., 2008b). Therefore, the investigation of Hec1 function is becoming an increasingly important area of study.
The precise functions of Hec1 during mitosis are not fully understood at present, although previous studies have revealed important roles of Hec1 at the kinetochore. Interestingly, several reports have documented that a portion of cellular Hec1, as well its binding partners Nuf2 and Spc25, are located at the centrosome during interphase and at the spindle pole region during mitosis (Hori et al., 2003; Sauer et al., 2005; Lin et al., 2006; Goshima et al., 2007; Diaz-Rodriguez et al., 2008). Importantly, significant spindle abnormalities (e.g., multipolarity) were observed in Hec1-depleted cells (Martin-Lluesma et al., 2002; DeLuca et al., 2003; McCleland et al., 2003). However, it remains to be shown whether Hec1 is involved in regulating mitotic spindle assembly by interacting with a spindle-associated factor.
Previous yeast two-hybrid screens have identified multiple Hec1-interacting candidates known to be involved in centrosome or spindle regulation (Chen et al., 2002; Wong et al., 2007; Wu et al., 2008a), one of which is Hice1. Hice1 was shown to be a centrosome- and spindle-associated protein possessing a microtubule binding module at its N-terminal region (Wu et al., 2008a). Remarkably, Hice1 knockdown cells exhibited delayed mitotic progression in conjunction with evident spindle abnormalities, increased chromosome misalignment and segregation errors. This was later confirmed by other reports, which detected Hice1 as a subunit of the eight-member Augmin complex (Lawo et al., 2009; Uehara et al., 2009). Although accumulating evidence suggests the critical importance of Hice1 in maintaining proper spindle morphology, it has not been systematically tested whether Hice1 plays a regulatory role in spindle assembly, for example, microtubule nucleation initiated by the well-documented γ-tubulin ring complex (γ-TuRC) complex (Wiese and Zheng, 2006).
In this study, we showed that Hec1 colocalizes with Hice1 at the spindle pole region during mitosis and Hec1 interacts with the coiled-coil domain 1 of Hice1. Reproducibly, knockdown of Hice1 by small interfering RNA (siRNA) treatment resulted in reduction of Hec1 in cells, primarily from the centrosome and spindle pole vicinity. Antibody-mediated neutralization of Hec1 or Hice1 impaired microtubule aster formation from isolated mitotic centrosomes. Disruption of the Hec1/Hice1 interaction by overexpressing Hice1ΔCoil1, a mutant defective in Hec1 interaction, triggered spindle multipolarity characteristic of Hec1- and Hice1-deficient cells. These results suggest that the interaction of Hec1 with Hice1 is important for optimal centrosome-directed microtubule formation, so as to facilitate the establishment of a proper mitotic spindle.
MATERIALS AND METHODS
Interaction Assays
For the yeast two-hybrid assay, full-length (FL) Hice1 was tagged with the GAL4 transactivation domain. Hec1 deletion mutants were each tagged with the GAL4 DNA binding domain. The detailed procedure for yeast two-hybrid assay were described previously (Durfee et al., 1993; Chen et al., 1997a). For glutathione transferase (GST) pull-down assays, Hice1 and Hec1 deletion mutants were tagged with GST and prepared from Escherichia coli by using affinity binding with glutathione-Sepharose. The 35S-labeled Hice1 produced by in vitro translation was used for interacting with various GST-Hec1 fusions in the pull-down assay, and vice versa. Procedures for GST-pull-down assays and coimmunoprecipitation assays have been detailed previously (Xiao et al., 2001). For the Western blot analysis of the resultant coimmunoprecipitates, the Nuf2 blot was developed with a TrueBlot Ultra kit (eBioscience, San Diego, CA) to avoid cross-reactivity with the immunoglobulin (Ig)G heavy chain, because Nuf2 migrates closely to the IgG heavy chain and Hice1. The blot was then stripped with a stripping buffer (Millipore, Billerica, MA) to allow for the subsequent detection of Hice1.
For in vitro interacting assays with further purified proteins, Hec1/GST-Nuf2 dimer encoded by a bicistronic pGEX-6p-1 plasmid or GST-Nuf2 was expressed in BL21 bacteria and purified by affinity binding with glutathione-Sepharose followed by gel filtration chromatography (Superdex 200) under a high salt condition (Ciferri et al., 2005). Six his-tagged Hice1 (wild type) and Hice1 mutants lacking one of the two coiled-coiled regions were expressed in Rosetta bacterium strain and purified using immobilized metal affinity chromatography affinity resin according to manufacturer's instructions (Bio-Rad Laboratories, Hercules, CA). Hice1 proteins were also further purified by gel filtration chromatography (Superdex 200) using the AKATA FPLC system (Pharmacia/GE Healthcare, Chalfont St. Giles, Buckinghamshire, United Kingdom). For the in vitro interaction, equal amounts of each Hice1 versions were mixed with GST-Nuf2 or Hec1/GST-Nuf2 proteins for 30 min at room temperature (RT). Potential interacting proteins were pulled down by glutathione-Sepharose beads blocked with 5% bovine serum albumin in a binding buffer (30 mM Tris-HCl, pH 7.6, 500 mM NaCl, and 0.2% Triton X-100). The complexes were washed adequately and then lysed in the Lamelli buffer for SDS-polyacrylamide gel electrophoresis (PAGE) analysis followed by Western blot with various antibodies.
Cell Lines, Mutant Expression, and RNAi
Human cancer cell lines HeLa and U2OS were cultured in DMEM plus 10% fetal bovine serum (FBS). The KE37 cell line of T lymphoblastic origin (Mayer and Kinkel, 1982) were cultivated in RPMI medium plus 10% FBS. U2OS cells stably expressing Hice1-green fluorescent protein (GFP), Hec1-GFP, or Hice1ΔCoil1-GFP were established by infection of retrovirus produced in GP2 293 packaging cell line. Previously validated Hec1 and Hice1 targeting siRNA sequences were used (Martin-Lluesma et al., 2002; Lin et al., 2006; Wu et al., 2008a). siRNA was transfected into cells with Lipofectamine 2000 (Invitrogen, Carlsbad, CA).
Microscopy
Immunostaining procedure was adapted as described previously (Wu et al., 2000). In brief, cells grown on coverslips were gently washed with the PEMG buffer [80 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), pH 6.8, 5 mM EGTA, 1 mM MgCl2, and 4 M glycerol] or phosphate-buffered saline (PBS) before fixation with 100% methanol at –20°C or 4% paraformaldehyde in PEMG or PBS buffer. After permeabilization with 0.4% Triton-X 100, cells were blocked with 5% normal goat serum (NGS) in PBS and then incubated with primary antibodies in PBS with 5% NGS (1–2 h; RT). Secondary antibodies used were conjugated with Alexa 488 or 594 (Invitrogen, Carlsbad, CA). 4′,6-Diamidino-2-phenylindole (DAPI) staining was applied after secondary antibody incubation and cells were finally mounted on coverslides with Prolong gold anti-fade reagent (Invitrogen). Images were captured with an Axiovert 200M microscope (Carl Zeiss, Thornwood, NY) equipped with a charge-coupled device camera (Hamamatsu, Bridgewater, NJ) controlled by the Axiovision software. Further image analysis or quantification was performed with Image-Pro Plus (MediaCybernetics, Bethesda, MD) or Adobe Photoshop software (Adobe Systems, Mountain View, CA).
Centrosome Isolation and In Vitro Microtubule Nucleation Assay
Centrosomes were isolated from KE37 cells similarly to the previous report (Gosti-Testu et al., 1986). For mitotic centrosome isolation, HeLa or KE37 cells were first synchronized at the G1/S boundary via a double thymidine arrest procedure, released to progress through S phase, and finally arrested at prometaphase by adding nocodazole 8 h before harvest. The mitotic arrest efficiency was further confirmed by fluorescence-activated cell sorting analysis (>90% of cells with 4N DNA content). Centrosome fractions were collected and analyzed by SDS-PAGE and Western blotting. For centrosome-directed microtubule nucleation assay in vitro, reaction mixtures contained 1× BRB80 (80 mM PIPES, pH 6.9, 2 mM MgCl2, 0.5 mM EGTA, and 1 mM guanosine triphosphate). Before being used for neutralization, antibodies underwent buffer exchange in 1× BRB80 using Amicon centrifugal filter devices (Millipore). Antibodies were incubated with centrosomes for 30 min at 4°C before addition of microtubules containing 20% of rhodamine-labeled tubulins. Nucleation mixture was incubated for 37°C for 30 min and then applied on a coverslide and covered with a coverslip for subsequent microscopic imaging (Bornens et al., 1987).
Microtubule Regrowth Assays
Microtubule regrowth in interphase or mitotic cells was performed similarly to the previous report (Luders et al., 2006). In brief, cells were grown on coverslip and transfected with siRNA for 48 h. For mitotic microtubule growth, cells were treated with 400 ng/ml nocodazole at 37°C for 2 h to depolymerize microtubules, followed by drug washout and an additional incubation on ice for 30 min (cold shock). Note that for interphase cells only cold treatment is required. Cells were recovered in prewarmed growth medium at 37°C to allow microtubule regrowth and subsequently fixed at various time points for further immunostaining to reveal the tubulin structures. Microscopic z-sectioning of mitotic cells was performed to collect the images at varying z-axis focal planes. Images were deconvoluted, the microtubule aster intensity was quantified using ImageJ (National Institutes of Health, Bethesda, MD), and the maximal projection of the z-sections is presented.
Antibodies
Commercial antibodies used for immunostaining or Western blotting were as follows: mouse monoclonal anti-Hec1, rabbit anti-Aurora A, mouse anti-BubR1, mouse anti-Eg5, mouse anti-β-actin, rabbit anti-p150Glued, and rabbit anti-NUMA (GeneTex, San Antonio, TX); mouse anti-α-tubulin and rabbit anti-γ-tubulin (Sigma-Aldrich, St. Louis, MO); sheep anti-β-tubulin (Cytoskeleton, Denver, CO); mouse anti-Nuf2 antibodies (GeneTex and MBL international, Woburn, MA); and mouse anti-GFP monoclonal mixtures (Roche Diagnostics, Mannheim, Germany). Affinity-purified mouse polyclonal antibody for Hice1 was described previously (Wu et al., 2008a).
RESULTS
Hec1 Interacts with the Centrosome Component Hice1
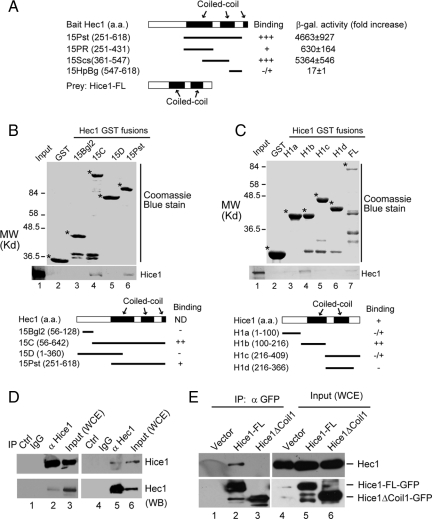
Hec1 was previously found to interact with Hice1 in a yeast two-hybrid screen by using the Hec1 C-terminal region as bait (Chen et al., 1997b). Hice1 has been demonstrated to be a centrosomal and microtubule binding protein (Wu et al., 2008a), and both proteins are shown to be localized at the centrosome and spindle. This potential interaction of Hec1 with Hice1 provides an opportunity to address the role of Hec1 at the centrosome. To do so, we first consolidated the interaction between Hec1 and Hice1. To refine the precise region of Hec1 responsible for Hice1 interaction, a yeast two-hybrid assay was used using different regions of Hec1 fused to the GAL4 DNA-binding domain, whereas the full-length Hice1 was tagged with the transactivation domain. A region covering part of the Hec1 C-terminal coiled-coil domains was sufficient for Hice1 interaction (Figure 1A). Next, a set of GST-tagged Hec1 or Hice1 deletion mutants were prepared from bacteria for GST-pull-down assays. The resultant GST preparations are mainly products of correct size, although some also contain small amount of degradation species (Figure 1, B and C). With these GST fusions, a preferential interaction of 35S-labeled Hice1 was detected for a Hec1 region spanning from coiled-coil 1–2 (Figure 1B). Inversely, the 35S-labeled Hec1 was found to primarily interact with a fragment encompassing the first coiled-coil domain of Hice1 (Figure 1C). Weak interactions with Hec1 were also observed for the other fragments of Hice1, probably due to nonoptimal binding condition for Hec1. Nonetheless, the data support that the Hec1/Hice1 interaction is primarily mediated through the coiled-coil 1 and 2 regions of Hec1 and the coiled-coil 1 domain of Hice1 in vitro.
Figure 1.
Mapping the interaction domains of Hec1 and Hice1. (A) Determination of the Hice1 binding region of Hec1 by yeast two-hybrid assay. The fold increase in β-galactosidase activity indicated relative interaction strength. Black boxes represent coiled-coil domains: Hec1 coiled-coil domain 1 (aa 261-403), domain 2 (aa 458-548), and domain 3 (aa 615–642); Hice1 coiled-coil domain 1 (aa 150-220) and domain 2 (aa 260-340). (B) Mapping the Hice1 binding region of Hec1 by in vitro GST pull-down assay. Comparable amount of GST (lane 2) or GST-tagged Hec1 fragments (lanes 3–6) were prepared from bacteria and used to interact with in vitro translated 35S-labeled Hice1. The autoradiograph after SDS-PAGE was shown to reveal Hice1 (bottom). Top panel of the gel shows the Coomassie Blue staining. Schematic depictions of the Hec1 fragments fused to the GST tag are also shown (ND, not determined). Bands marked by asterisks indicate correct GST-fusion proteins, whereas the unlabeled smaller bands are degradation products. (C) Determination of the Hec1 binding region of Hice1 by using similar in vitro GST pull-down assay as described in B. (D) Anti-Hice1 antibody immunoprecipitates Hec1 (lane 2) and anti-Hec1 reciprocally immunoprecipitates Hice1 (lane 5). Normal mouse IgG was used as a negative control (lanes 1 and 4). (E) GFP-fused Hice1-FL (full length) or Hice1ΔCoil1 (aa 150-228 deleted) was transiently expressed by retroviral infection into U2OS cells, which were then used for immunoprecipitation with a mixture of two monoclonal anti-GFP antibodies. Western blot result on the immunoprecipitates was shown.
To determine whether Hec1 and Hice1 associate with each other in vivo, a coimmunoprecipitation (co-IP) assay was performed using cell extracts prepared from U2OS cells. It was found that anti-Hice1 antibodies coimmunoprecipitated with Hec1 and reciprocally anti-Hec1 antibodies coimmunoprecipitated with Hice1 (Figure 1D), suggesting positive association in vivo. Next, co-IP experiments were carried out using U2OS cells expressing GFP-tagged Hice1-FL (full length), Hice1ΔCoil1 (coiled-coil domain 1 deleted), or GFP only, because the coiled-coil domain 1 of Hice1 is important for Hec1 interaction in vitro (Figure 1). Hice1-FL, but not Hice1ΔCoil1 or GFP alone, was capable of coimmunoprecipitating with endogenous Hec1 (Figure 1E), thereby confirming that the coiled-coil domain 1 of Hice1 is responsible for Hec1 interaction in cells.
Hice1 Interacts with Hec1 of the Hec1/Nuf2 Complex
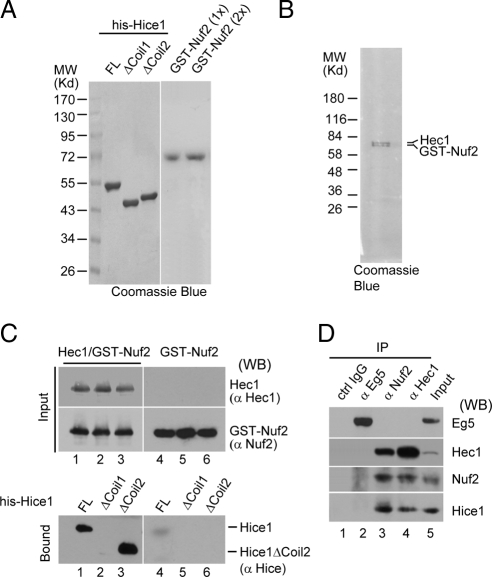
Nuf2 is thought to be a major binding partner of Hec1 and the Hec1/Nuf2 heterodimer may further complex with Spc24 and Spc25 to form a heterotetramer complex (McCleland et al., 2003; Ciferri et al., 2005; Emanuele et al., 2005; Wei et al., 2005). To test whether Hice1 may interact with the Hec1/Nuf2 dimer (presumably through Hec1), 6his-tagged Hice1-FL, Hice1ΔCoil1, and Hice1ΔCoil2 were individually expressed in bacteria and purified to near homogeneity by performing affinity binding followed by gel filtration chromatography (Figure 2A). GST-Nuf2 alone or the Hec1/GST–Nuf2 complex was also expressed and similarly purified to near homogeneity as described previously (Figure 2, A and B) (Ciferri et al., 2005). Purified Hec1/GST-Nuf2 dimer was able to interact with Hice1-FL and Hice1ΔCoil2, but not Hice1ΔCoil1 (Figure 2C, lanes 1–3). This indicates that Hice1 interacts with Hec1/Nuf2 and that the coiled-coil domain 1 of Hice1 is critically important for this binding event (Figure 1). Furthermore, little or no interaction was detected for GST-Nuf2 toward any version of the purified Hice1 proteins when Hec1 was absent (Figure 2C, lanes 4–6), suggesting that Hec1 but not Nuf2 plays a primary role in mediating the interaction between Hice1 and the Hec1/Nuf2 dimer. Consistently, both anti-Hec1 and anti-Nuf2 antibodies were capable of coimmunoprecipitating with endogenous Hice1 from cell extracts. In contrast, Eg5, an abundant spindle-associated kinesin, failed to associate with Hice1 in a similar assay (Figure 2D). Together, the results clearly demonstrate that Hice1 may use its coiled-coil domain 1 to specifically interact with Hec1 or with the Hec1/Nuf2 dimer via Hec1.
Figure 2.
Hice1 binds to the Hec1/Nuf2 complex. (A and B) Coomassie Blue staining of various purified His-tagged Hice1 versions and GST-Nuf2 (A), and Hec1/GST-Nuf2 dimer (B). Hice1ΔCoil1, amino acid 150-228 deleted; Hice1ΔCoil2, amino acids 263-329 deleted. (C) In vitro binding assay using the above purified proteins (detailed in Materials and Methods). Equal amounts of Hice1-FL, Hice1ΔCoil1, and Hice1ΔCoil2 were used to interact with Hec1/GST-Nuf2 dimer (lanes 1–3) or GST-Nuf2 (lanes 4–6). Interacting proteins were then pull downed by glutathione-Sepharose beads and subjected to SDS-PAGE followed by Western blot. (D) Hice1 coimmunoprecipitated with endogenous Nuf2 and Hec1. HeLa cell extract was subjected to coimmunoprecipitation assay with indicated antibodies. The image showed the Western blot developed with respective antibodies.
Hec1 and Hice1 Associates with the Centrosomal Structure
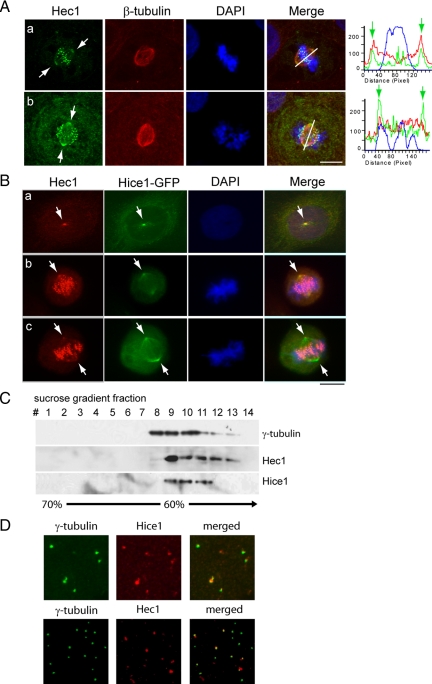
Both Hice1 and Hec1 are distributed to distinct subcellular locations during the cell cycle progression such as the spindle and/or kinetochore, the sites for their potential molecular interaction. We therefore examined where Hec1 and Hice1 may colocalize with each other in cells. Shown in Figure 3A, immunofluorescent staining revealed that Hec1, in addition to its discrete kinetochore-associated foci, colocalized with the spindle marker β-tubulin on the spindle pole and the vicinity. For a quantitative visualization, the centrosome and kinetochore positions were highlighted by fluorescence intensity peaks in a pole-to-pole line scanning graph (Figure 3A). Similarly, Hec1 was found to colocalize with Hice1-GFP on the centrosome during mitosis (Figure 3B). The localization of Hec1 and Hice1 at the centrosome was retained upon treatment with nocodazole (Supplemental Figure 1), indicating a microtubule independent mechanism of recruitment. To further confirm the centrosomal localization of these two proteins, a sucrose gradient centrifugation assay was used to isolate centrosomes from the human T lymphoblastic cell line KE37 (Bornens et al., 1987). Western blot analysis on the resultant centrifugational fractions indicated that Hec1 and Hice1 cofractionated with the centrosome marker γ-tubulin (Figure 3C). Last, immunofluorescent staining was performed using isolated centrosomes stuck on coverslips. As revealed by colocalization with γ-tubulin signals, the centrosomes were positive for Hice1 and Hec1 in 88 and 89% of the population, respectively (Figure 3D). Together, these results suggest that Hec1 and Hice1 may interact with each other at the centrosome.
Figure 3.
Hec1 and Hice1 are components of the centrosome. (A) Double immunostaining of Hec1 and β-tubulin in U2OS. Right, line scanning shows the relative fluorescence intensity along the line connecting the spindle poles. (B) Immunostaining for Hec1 in U2OS cells stably expressing Hice1-GFP. Arrows indicate centrosome/spindle poles (A and B). (C) Isolated KE37 centrosomes were prepared by discontinuous sucrose gradient centrifugation and serial fractions were separated by SDS-PAGE and subjected to Western blotting analysis with anti-γ-tubulin, Hec1, and Hice1 antibodies. (D) Double immunostaining of purified KE37 centrosomes with anti-γ-tubulin and Hec1 or Hice1 antibody. Bar, 10 μm. DAPI staining was used to reveal chromosome/nucleus in A and B.
Hice1 Is Required for Hec1 Stabilization and Proper Cellular Localization
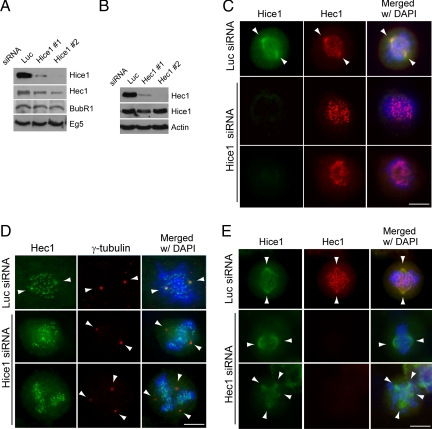
To determine the functional interrelationship between Hec1 and Hice1, we next examined whether Hice1 could regulate the stability and/or subcellular localization of Hec1, or vice versa. For this purpose, Hec1 or Hice1 was depleted in U2OS cells by siRNA transfection. Two sets of sequence-specific siRNAs for Hice1 or Hec1 were used to minimize potential RNAi off-target effects. Remarkably, Western blot results showed that the depletion of Hice1 resulted in a concomitant reduction of Hec1, but not the spindle molecule Eg5 or the kinetochore molecule BubR1 (Figure 4A). However, when Hec1 was depleted, the Hice1 level remained unchanged (Figure 4B), suggesting that Hice1 is important for regulating the cellular level of Hec1 directly or indirectly, and not vice versa.
Figure 4.
Depletion of Hice1 led to diminished Hec1 signal at the centrosome. (A) Western blot analysis of U2OS cells with indicated antibodies after depletion of Hice1 by using two different siRNAs (1 and 2). (B) Western blot analysis of U2OS cells after depletion of Hec1 using two different siRNAs (1 and 2). (C) Immunofluorescence staining for Hice1 and Hec1 in luciferase or Hice1 siRNA-treated mitotic U2OS cells. (D) Immunofluorescence staining for Hec1 and γ-tubulin in luciferase or Hice1 siRNA-treated mitotic U2OS cells. Note the spindle distribution of Hec1 (colocalization with Hice1 and γ-tubulin) and the numerous chromosome-associated Hec1 speckles corresponding to kinetochores. (E) Immunofluorescence staining for Hec1 and Hice1 in luciferase or Hec1 siRNA-treated mitotic U2OS cells. For C–E, arrowheads indicate the spindle pole position. Bars, 10 μm.
Because Hec1 localizes to both kinetochores and centrosomes, we next asked whether the Hec1 signals at these two subcellular locations were differentially affected by Hice1 depletion. Interestingly, Hice1 siRNA-treated U2OS cells were negative of Hec1 staining on the spindle pole and its vicinity, but they were still positive for the centrosomal marker γ-tubulin (Figure 4, C and D). In contrast, the kinetochore-associated Hec1 signal was not affected (Figure 4, C and D), suggesting that Hice1 depletion preferentially affects the centrosomal/spindle pool of Hec1. Fluorescent quantification of a 3-μm circular region around the centrosome revealed a 80–85% decrease of Hec1 signal in Hice1 siRNA-treated cells relative to control treated ones (n = 35 centrosomes for each treatment; p < 0.05). Inversely, knockdown of Hec1 by siRNA did not seem to affect the localization of Hice1 in cells displaying either a bipolar or multipolar spindle (Figure 4E). Importantly, the mitotic distribution of two other well documented spindle molecules, NUMA and p150Glued (a dynactin subunit), was not apparently affected by Hice1 depletion (Supplemental Figure 2), suggesting a specific effect on Hec1 subcellular localization. Together, these findings suggest that Hice1 is critically important for proper Hec1 localization at the centrosome and spindle.
Inactivation of Hec1 and Hice1 Resulted in Reduced Microtubule Regrowth from Centrosomes
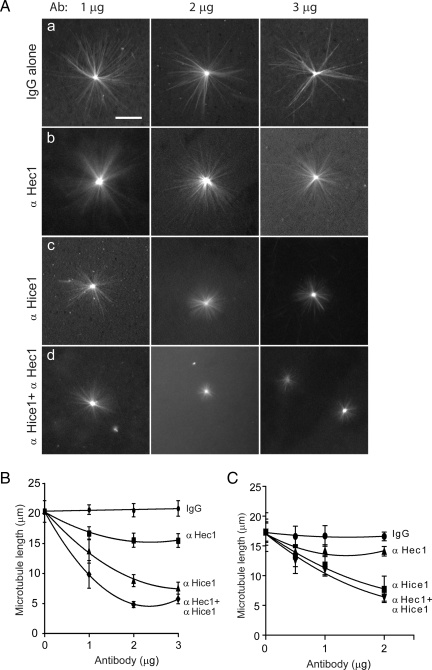
The centrosome is a major microtubule nucleation site for initiating spindle assembly. We were intrigued in testing the potential role of Hec1 and Hice1 in centrosome-directed microtubule formation. To test this possibility, a microtubule growth assay was undertaken with enriched centrosomes that were isolated as described in a previous experiment (Figure 3). Both Hec1 and Hice1 have a microtubule binding activity (Cheeseman et al., 2006; DeLuca et al., 2006; Wu et al., 2008a). To neutralize Hec1 and Hice1, specific antibodies were prepared and affinity purified. Isolated centrosomes were first neutralized with Hice1 or Hec1 antibodies and then incubated with rhodamine-labeled tubulin to allow microtubule outgrowth (Figure 5A). The addition of varying doses of normal mouse IgG did not alter the microtubule growth in comparison with the mock-treated reaction (Figure 5, Aa and B). Interestingly, adding Hec1 antibody resulted in shortened microtubule length (maximal reduction ∼25%) (Figure 5, Ab and B). In comparison, Hice1 antibody was able to inhibit microtubule growth more dramatically (maximal reduction ∼60%) (Figure 5, Ac and B). Notably, adding both Hice1 and Hec1 antibodies seemed to have an additive inhibitory effect on microtubule length (Figure 5B), but a low level of microtubule growth (5 μm) still occurred. Because centrosomes isolated from the above procedure represent a mixture of both interphase and mitotic centrosomes, we then proceeded to determine whether the phenotypes can be detected using a more pure mitotic population. To do so, mitotic centrosomes were isolated from KE37 or HeLa cells enriched at prometaphase by a double thymidine block followed by nocodazole arrest (>90% arrested at M phase; see Materials and Methods). Using the mitotic centrosomes, similar phenotypes were recapitulated in the microtubule nucleation assay (Figure 5C). Together, the results suggest that Hice1 and Hec1 are required for optimal microtubule aster formation in vitro, where Hice1 plays a more dominant role than Hec1. To further determine whether the reduction of microtubule length was due to Hec1's and Hice1's role in microtubule nucleation or stabilization of prenucleated microtubules, microtubule asters were first allowed to form from the centrosomes followed by adding polyclonal antibodies against Hice1 or Hec1. No reduction in final microtubule length was detected (data not shown), suggesting that Hice1 and Hec1 are unlikely to play a major role in maintaining the prenucleated microtubules.
Figure 5.
Microtubule nucleation assay after neutralization of Hec1 and Hice1 by antibodies. (A) Representative images of radial microtubule arrays emanating from purified KE37 centrosomes. Centrosomes were incubated with 1, 2, or 3 μg of antibody for 30 min at 4°C before addition of rhodamine-conjugated tubulins. Normal mouse IgG was used as a negative control. (B) Quantification of average microtubule length from centrosomes (n = 50 centrosomes each, mixtures of interphase and mitotic centrosomes). Error bars indicate 1 SD. Bar, 20 μm. The effect among groups is significantly different (p < 0.02, two-way ANOVA test). (C) Quantification of average microtubule length from mitotic centrosomes isolated from KE37 cells (>90% of cells enriched at prometaphase). Error bars indicate 1 SD. Bar, 20 μm. The effect among groups is significantly different (p < 0.033, two-way ANOVA test). Similar results were also obtained from HeLa cells (data not shown).
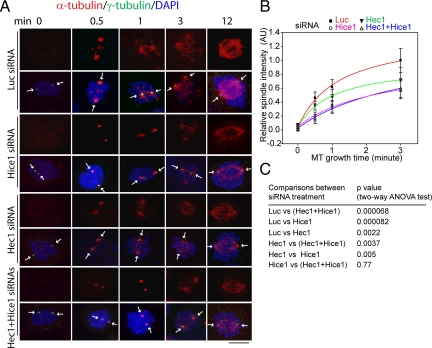
Next, we directly tested the role of Hec1 and Hice1 in centrosomal microtubule nucleation in vivo by examining the microtubule regrowth after cold-induced depolymerization in live cells. A time-dependent kinetic assay revealed no major microtubule growth defect in Hec1 or Hice1 siRNA-treated interphase cells, at least during the early time points (Supplemental Figure 3, A and B). Furthermore, the interphase centrosomes seemed to duplicate and mature normally in Hec1-depleted cells (Supplemental Figure 3, C and D) and Hice1-depleted cells, compared with control siRNA-treated cells (Wu et al., 2008a). Thus, the role of Hec1 and Hice1 in centrosome regulation seems to be subtle during interphase. Alternatively, the remaining residual Hec1 and Hice1 after siRNA treatment are sufficient for the interphase centrosome functionality. Therefore, a specific role of Hec1 and Hice1 in mitotic spindle assembly was then tested by adapting a mitotic microtubule regrowth assay in U2OS cells (Luders et al., 2006). In this assay, the microtubule aster formation was visualized and measured by quantitative confocal fluorescent microscopy. Because Hice1 or Hec1 depletion tends to trigger abnormal spindle pole configurations incompatible for faithful microtubule aster quantification, only those cells with two distinct spindle poles, reflecting bipolarity, were measured. Even under this stringent condition, it was found that, in comparison to the control siRNA-treated cells, the time-dependent microtubule regrowth from mitotic centrosomes was impaired significantly at early time points (≤3 min) in cells treated with Hec1 siRNA, Hice1 siRNA, or both (Figure 6, A and B). The relative reduction in microtubule aster intensity is significant (p < 0.0022, two-way analysis of variance [ANOVA] test) (Figure 6C). Consistent with the observation from the microtubule nucleation assay in vitro (Figure 5), Hec1 inactivation in cells seemed to have less effect on centrosomal microtubule growth than Hice1 inactivation. However, treating cells with Hec1 and Hice1 siRNAs together elicited a similar effect to Hice1 siRNA alone (p = 0.77; no significant difference) (Figure 6C). Together, these results suggest that Hec1 functions in microtubule aster formation from mitotic centrosomes, which is probably controlled by Hice1.
Figure 6.
Impaired microtubule regrowth from centrosomes in mitotic U2OS cells depleted of Hice1 and/or Hec1 by siRNA. (A) Immunostaining of α-tubulin and γ-tubulin in the mitotic U2OS cells subjected to microtubule regrowth assay. Cells were stained against α-tubulin and γ-tubulin. Confocal microscopy and image deconvolution were performed, and the maximal projections of z-axis series of images were shown. Luciferase siRNA-treated cells were analyzed as a control. Bar, 10 μm. (B) Time-dependent kinetics of centrosomal microtubule aster formation. Microtubule asters (0.5–3 min) were quantified by measuring the fluorescence intensity in a 4-μm circular area around each distinct centrosome. The mean value at each time point was derived from ≥30 randomly selected mitotic cells with a near bipolar spindle (three independent experiments) and used to fit a growth curve according to a multifactor exponential equation (SigmaPlot, Systat Software, San Jose, CA). The 12-min time point was not measured as the asters had become less distinct. AU, arbitrary units. (C) p values derived from a two-way ANOVA test (factor 1, time; factor 2, siRNAs) to show differences in the mean levels of microtubule aster intensity between various siRNA (factor 2) treatment groups. Differences among different time points (factor 1) are all significant (p < 0.002, data not shown).
It is noteworthy that spindle microtubules may also originate from within the spindle, which is a novel pathway shown to involve the eight-subunit Augmin complex (Goshima et al., 2008; Zhu et al., 2008). Importantly, FAM29A, an Augmin subunit, was found to associate with Hice1 but not Hec1 in a coimmunoprecipitation assay (Supplemental Figure 4), suggesting that the Hec1/Hice1 association is distinct to Hice1 itself but not necessarily to the integral Augmin complex. Thus, the role of Hec1/Hice1 in the centrosomal microtubule nucleation is probably specific to a subpopulation of Hice1 different from that in the Augmin complex.
The Interaction of Hec1 with Hice1 Is Required for Proper Mitotic Spindle Formation
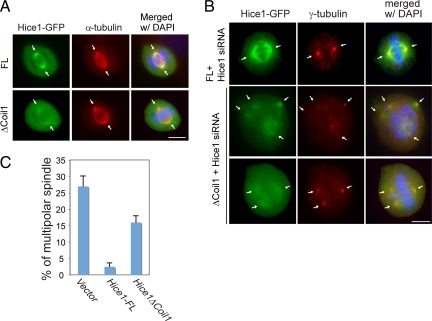
To further consolidate the importance of the Hec1/Hice1 interaction for spindle formation, we subsequently examined how the abrogation of Hec1/Hice1 interaction may affect the spindle configuration. To disrupt the Hec1/Hice1 interaction, the Hice1ΔCoil1 mutant defective in Hec1 binding (Figure 1E) was used. It was noted that GFP-tagged Hice1ΔCoil fails to localize to microtubule arrays during interphase (Wu et al., 2008a) but still associates with the mitotic spindle, albeit to a lesser extent than the full-length version (Figure 7A). This suggests a mitotic specific and Hec1-independent mechanism for Hice1 recruitment to spindles. Next, U2OS cells were transiently infected with Hice1-expressing retrovirus (RNAi-resistant Hice1-FL-GFP or Hice1ΔCoil1-GFP), or GFP-expressing control virus, followed by Hice1 siRNA transfection to deplete endogenous Hice1. Expectedly, depletion of Hice1 in GFP-only U2OS cells triggered significant formation of abnormal spindles with multiple poles (Figure 7B), similar to the previous report (Wu et al., 2008a). Expression of Hice1-FL in Hice1-siRNA treated cells restored the bipolar spindle formation to a normal rate (Figure 7, B and C). Remarkably, replacing endogenous Hice1 with the Hice1ΔCoil1 mutant led to a sixfold increase of multipolar spindle formation relative to the Hice1-FL rescued cells (Figure 7, B and C) in part due to defective Hec1 interaction, although non-Hec1 associated negative effects cannot be ruled out at present. The phenotypes are reminiscent of those observed in Hec1 or Hice1 depleted cells (Martin-Lluesma et al., 2002; DeLuca et al., 2003; Hori et al., 2003; McCleland et al., 2003; Lin et al., 2006; Wu et al., 2008a). Together, these results highlight the critical importance of the Hec1/Hice1 interaction for a proper mitotic spindle assembly in vivo, which requires the coiled-coil domain 1 of Hice1.
Figure 7.
Effects of expressing Hec1-binding deficient Hice1 mutant on spindle morphology. Cells were infected with retrovirus expressing Hice1-siRNA-resistant Hice1-FL-GFP, Hice1ΔCoil1-GFP. (A) In mitotic cells, Hice1-FL-GFP and Hice1ΔCoil1-GFP colocalize with the spindle maker β-tubulin as revealed by coimmunostaining. (B) The Hice1-FL-GFP or Hice1ΔCoil1-GFP cells were transfected with Hice1 siRNA for 2 d to deplete endogenous Hice1. Images showed the spindle configurations as revealed by costaining with γ-tubulin. (C) Bar graph was used to show the occurrence of spindles with multiple poles in each group. Bars (B and C), 10 μm.
DISCUSSION
In this communication, Hec1 is shown to directly interact with Hice1 and play a positive role in centrosome-directed microtubule spindle assembly. Hice1 and Hec1 are intimately connected, such that the depletion of Hice1 results in preferential loss of Hec1 from centrosomes and spindles. Depletion of Hec1 and Hice1 in cells by siRNA retards de novo mitotic microtubule regrowth from centrosomes and the subsequent spindle assembly. Consistently, the neutralization of Hec1 and Hice1 in purified mitotic centrosomes inhibits microtubule nucleation. These results suggest that Hec1 cooperates with Hice1 in the centrosome-directed microtubule growth to facilitate the establishment of a proper mitotic spindle.
Microtubule nucleation can be initiated from the centrosome, chromatid, or within the spindle. Hice1 was recently shown to be a subunit of the eight-member Augmin complex. RNAi experiments suggested that some Augmin subunits were involved in microtubule nucleation from within the spindle but not from the centrosome (Goshima et al., 2008; Zhu et al., 2008). Although it remains to be shown whether Augmin exclusively functions as an integral unit, it is likely that a pool of Hice1 may function in the intraspindle microtubule nucleation as an Augmin subunit, similar to FAM29A (Zhu et al., 2008). Consistent with this idea, we noticed that the spindle density in Hice1-depleted cells was often decreased (Wu et al., 2008). In contrast, our results suggest that Hice1 may additionally contribute to the centrosome-directed pathway. This latter activity of Hice1 is in part mediated through Hec1, which does not associate with FAM29A in a coimmunoprecipitation experiment (Supplemental Figure 4), although the potential association with the other six Augmin components remains to be tested. It is possible that the Hec1-associated pool of Hice1 might be distinct from that constituting the Augmin complex. Furthermore, Hice1 could be engaged in chromatid-directed microtubule nucleation pathway, possibly through associating with Aurora A and TPX2 (our unpublished data). Thus, it is very likely that Hice1 forms distinct complexes to regulate microtubule nucleation from various subcellular sites.
The activity of Hec1 seems to be subtle in the microtubule nucleation from mitotic centrosomes, but it can be detected reproducibly and with statistical significance. On the contrary, the activity of Hice1 is much more evident. Our preliminary study reveals that Hec1 and Hice1, when ectopically expressed in cells, can associate with the γ-TuRC via the GCP2 subunit. Furthermore, both Hice1 and Hec1 have microtubule binding and bundling activities in vitro (Cheeseman et al., 2006; Wu et al., 2008). It is possible that during microtubule nucleation by the γ-TuRC complex, Hec1 can assist Hice1 to hold or stabilize the microtubules and also help build bundled microtubule fibers important for establishing a bipolar spindle with sufficient microtubule density.
An obvious multipolar phenotype was observed in cells when endogenous Hice1 has been replaced by ectopically expressed Hice1ΔCoil1 (so as to disrupt the Hice1/Hec1 interaction) or in those cells depleted of Hice1 or Hec1. It is apparent that Hice1 and Hec1 play a positive role to maintain the spindle bipolarity. However, it remains to be elucidated with regards to the exact relationship between the microtubule nucleation activity of Hice1/Hec1 and the spindle polarity. It is possible that defects in microtubule nucleation may affect the ratio of free tubulin versus microtubules, which may then adversely affect the spindle microtubule density and consequently the bipolarity as a whole, as proposed in a previous study (Holmfeldt et al., 2003). Moreover, it cannot be ruled out that Hice1ΔCoil1 may also affect some other unknown aspects of Hice1 function, other than recruiting Hec1 to the spindle pole. For example, the coiled-coil domain 1 of Hice1 may be important for stabilizing the K-fibers to maintain the tension on the kinetochores and the spindle, as suggested for some Augmin subunits (Lawo et al., 2009). In this regard, however, the requirement for Hec1 in stabilizing K-fibers is well established (DeLuca, 2006). Whether Hec1 and Hice1 work together to regulate the K-fiber dynamics and stability at the kinetochore/K-fiber interface warrants further investigation.
It is known that overexpression of Hec1 associates with adverse clinical outcomes of human cancers. Consistently, we observed that both Hice1 and Hec1 are overexpressed with similar expression profiles in the NCI-60 panel of cancer lines and an additional collection of breast and ovarian cancer cell lines (total >70 cell lines; our unpublished data), underlying a common regulatory pathway for the expression of these two molecules, the significance of which in tumorigenesis is becoming increasingly clear and seems to implicate their roles in mitosis. The novel activity of Hec1 in mitotic centrosomes may help explain its role in tumorigenesis.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Drs. Claudio Ciferri and Andrea Mussachio (European Institute of Oncology) for the Hec1/Nuf2 expression plasmids, Dr. Yasushi Hiraoka for the Nuf2-GFP construct, Dr. Eric Karsenti (European Molecular Biology Laboratory) for providing the KE37 cell line, and Drs. Hui Zhu and Guowei Fang (Stanford University) for FAM29A reagents. We thank Connie Tsai and Angali Tapadia for part of the protein preparation work, and Dr. Yumay Chen for initiating the work. G. W. is supported by a postdoctoral fellowship from the Susan Komen Breast Cancer Foundation. R. W. is supported by a predoctoral fellowship from the DOD Congressionally Directed Medical Research Program in Breast Cancer and the National Institutes of Health Medical Scientist Training Program. This work was supported by National Institutes of Health grant R01 CA-107568) (to W.H.L.).
Abbreviations used:
- γ-TuRC
γ-tubulin ring complex
- DAPI
4′,6-diamidino-2-phenylindole
- GFP
green fluorescent protein
- GST
glutathione transferase.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-11-1123) on September 23, 2009.
REFERENCES
- Bornens M., Paintrand M., Berges J., Marty M. C., Karsenti E. Structural and chemical characterization of isolated centrosomes. Cell Motil. Cytoskeleton. 1987;8:238–249. doi: 10.1002/cm.970080305. [DOI] [PubMed] [Google Scholar]
- Cheeseman I. M., Chappie J. S., Wilson-Kubalek E. M., Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- Chen Y., Riley D. J., Chen P. L., Lee W. H. HEC, a novel nuclear protein rich in leucine heptad repeats specifically involved in mitosis. Mol. Cell. Biol. 1997a;17:6049–6056. doi: 10.1128/mcb.17.10.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Riley D. J., Zheng L., Chen P. L., Lee W. H. Phosphorylation of the mitotic regulator protein Hec1 by Nek2 kinase is essential for faithful chromosome segregation. J. Biol. Chem. 2002;277:49408–49416. doi: 10.1074/jbc.M207069200. [DOI] [PubMed] [Google Scholar]
- Chen Y., Sharp Z. D., Lee W. H. HEC binds to the seventh regulatory subunit of the 26 S proteasome and modulates the proteolysis of mitotic cyclins. J. Biol. Chem. 1997b;272:24081–24087. doi: 10.1074/jbc.272.38.24081. [DOI] [PubMed] [Google Scholar]
- Ciferri C., De Luca J., Monzani S., Ferrari K. J., Ristic D., Wyman C., Stark H., Kilmartin J., Salmon E. D., Musacchio A. Architecture of the human ndc80-hec1 complex, a critical constituent of the outer kinetochore. J. Biol. Chem. 2005;280:29088–29095. doi: 10.1074/jbc.M504070200. [DOI] [PubMed] [Google Scholar]
- Ciferri C., et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–439. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton D. A. Spindle assembly in animal cells. Annu. Rev. Biochem. 2000;69:95–114. doi: 10.1146/annurev.biochem.69.1.95. [DOI] [PubMed] [Google Scholar]
- DeLuca J. G., Gall W. E., Ciferri C., Cimini D., Musacchio A., Salmon E. D. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- DeLuca J. G., Howell B. J., Canman J. C., Hickey J. M., Fang G., Salmon E. D. Nuf2 and Hec1 are required for retention of the checkpoint proteins Mad1 and Mad2 to kinetochores. Curr. Biol. 2003;13:2103–2109. doi: 10.1016/j.cub.2003.10.056. [DOI] [PubMed] [Google Scholar]
- Diaz-Rodriguez E., Sotillo R., Schvartzman J. M., Benezra R. Hec1 overexpression hyperactivates the mitotic checkpoint and induces tumor formation in vivo. Proc. Natl. Acad. Sci. USA. 2008;105:16719–16724. doi: 10.1073/pnas.0803504105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durfee T., Becherer K., Chen P. L., Yeh S. H., Yang Y., Kilburn A. E., Lee W. H., Elledge S. J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- Emanuele M. J., McCleland M. L., Satinover D. L., Stukenberg P. T. Measuring the stoichiometry and physical interactions between components elucidates the architecture of the vertebrate kinetochore. Mol. Biol. Cell. 2005;16:4882–4892. doi: 10.1091/mbc.E05-03-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glinsky G. V., Berezovska O., Glinskii A. B. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J. Clin. Invest. 2005;115:1503–1521. doi: 10.1172/JCI23412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Mayer M., Zhang N., Stuurman N., Vale R. D. Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J. Cell Biol. 2008;181:421–429. doi: 10.1083/jcb.200711053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Wollman R., Goodwin S. S., Zhang N., Scholey J. M., Vale R. D., Stuurman N. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 2007;316:417–421. doi: 10.1126/science.1141314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosti-Testu F., Marty M. C., Berges J., Maunoury R., Bornens M. Identification of centrosomal proteins in a human lymphoblastic cell line. EMBO J. 1986;5:2545–2550. doi: 10.1002/j.1460-2075.1986.tb04533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurzov E. N., Izquierdo M. RNA interference against Hec1 inhibits tumor growth in vivo. Gene Ther. 2006;13:1–7. doi: 10.1038/sj.gt.3302595. [DOI] [PubMed] [Google Scholar]
- Holmfeldt P., Brattsand G., Gullberg M. Interphase and monoastral-mitotic phenotypes of overexpressed MAP4 are modulated by free tubulin concentrations. J. Cell Sci. 2003;116:3701–3711. doi: 10.1242/jcs.00685. [DOI] [PubMed] [Google Scholar]
- Hori T., Haraguchi T., Hiraoka Y., Kimura H., Fukagawa T. Dynamic behavior of Nuf2-Hec1 complex that localizes to the centrosome and centromere and is essential for mitotic progression in vertebrate cells. J. Cell Sci. 2003;116:3347–3362. doi: 10.1242/jcs.00645. [DOI] [PubMed] [Google Scholar]
- Karsenti E., Vernos I. The mitotic spindle: a self-made machine. Science. 2001;294:543–547. doi: 10.1126/science.1063488. [DOI] [PubMed] [Google Scholar]
- Kline-Smith S. L., Sandall S., Desai A. Kinetochore-spindle microtubule interactions during mitosis. Curr. Opin. Cell Biol. 2005;17:35–46. doi: 10.1016/j.ceb.2004.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline-Smith S. L., Walczak C. E. Mitotic spindle assembly and chromosome segregation: refocusing on microtubule dynamics. Mol. Cell. 2004;15:317–327. doi: 10.1016/j.molcel.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Lawo S., et al. HAUS, the 8-subunit human Augmin complex, regulates centrosome and spindle integrity. Curr. Biol. 2009;19:816–826. doi: 10.1016/j.cub.2009.04.033. [DOI] [PubMed] [Google Scholar]
- Li L., Yang L., Scudiero D. A., Miller S. A., Yu Z. X., Stukenberg P. T., Shoemaker R. H., Kotin R. M. Development of recombinant adeno-associated virus vectors carrying small interfering RNA (shHec1)-mediated depletion of kinetochore Hec1 protein in tumor cells. Gene Ther. 2007;14:814–827. doi: 10.1038/sj.gt.3302933. [DOI] [PubMed] [Google Scholar]
- Lin Y. T., Chen Y., Wu G., Lee W. H. Hec1 sequentially recruits Zwint-1 and ZW10 to kinetochores for faithful chromosome segregation and spindle checkpoint control. Oncogene. 2006;25:6901–6914. doi: 10.1038/sj.onc.1209687. [DOI] [PubMed] [Google Scholar]
- Luders J., Patel U. K., Stearns T. GCP-WD is a gamma-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat. Cell Biol. 2006;8:137–147. doi: 10.1038/ncb1349. [DOI] [PubMed] [Google Scholar]
- Martin-Lluesma S., Stucke V. M., Nigg E. A. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science. 2002;297:2267–2270. doi: 10.1126/science.1075596. [DOI] [PubMed] [Google Scholar]
- Mayer L., Fu S. M., Kunkel H. G. Human T cell hybridomas secreting factors for IgA-specific help, polyclonal B cell activation, and B cell proliferation. J. Exp. Med. 1982;1:1860–1865. doi: 10.1084/jem.156.6.1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleland M. L., Gardner R. D., Kallio M. J., Daum J. R., Gorbsky G. J., Burke D. J., Stukenberg P. T. The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev. 2003;17:101–114. doi: 10.1101/gad.1040903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer G., Korner R., Hanisch A., Ries A., Nigg E. A., Sillje H. H. Proteome analysis of the human mitotic spindle. Mol Cell Proteomics. 2005;4:35–43. doi: 10.1074/mcp.M400158-MCP200. [DOI] [PubMed] [Google Scholar]
- Sotillo R., Hernando E., Diaz-Rodriguez E., Teruya-Feldstein J., Cordon-Cardo C., Lowe S. W., Benezra R. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell. 2007;11:9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara R., Nozawa R. S., Tomioka A., Petry S., Vale R. D., Obuse C., Goshima G. The augmin complex plays a critical role in spindle microtubule generation for mitotic progression and cytokinesis in human cells. Proc. Natl. Acad. Sci. USA. 2009;106:6998–7003. doi: 10.1073/pnas.0901587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van 't Veer L. J., et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- Wei R. R., Al-Bassam J., Harrison S. C. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat. Struct. Mol. Biol. 2007;14:54–59. doi: 10.1038/nsmb1186. [DOI] [PubMed] [Google Scholar]
- Wei R. R., Sorger P. K., Harrison S. C. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc. Natl. Acad. Sci. USA. 2005;102:5363–5367. doi: 10.1073/pnas.0501168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese C., Zheng Y. Microtubule nucleation: gamma-tubulin and beyond. J. Cell Sci. 2006;119:4143–4153. doi: 10.1242/jcs.03226. [DOI] [PubMed] [Google Scholar]
- Wigge P. A., Jensen O. N., Holmes S., Soues S., Mann M., Kilmartin J. V. Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J. Cell Biol. 1998;141:967–977. doi: 10.1083/jcb.141.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson-Kubalek E. M., Cheeseman I. M., Yoshioka C., Desai A., Milligan R. A. Orientation and structure of the Ndc80 complex on the microtubule lattice. J. Cell Biol. 2008;182:1055–1061. doi: 10.1083/jcb.200804170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J., et al. A protein interaction map of the mitotic spindle. Mol. Biol. Cell. 2007;18:3800–3809. doi: 10.1091/mbc.E07-06-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Lee W. H., Chen P. L. NBS1 and TRF1 colocalize at promyelocytic leukemia bodies during late S/G2 phases in immortalized telomerase-negative cells. J. Biol. Chem. 2000;275:30618–30622. doi: 10.1074/jbc.C000390200. [DOI] [PubMed] [Google Scholar]
- Wu G., Lin Y. T., Wei R., Chen Y., Shan Z., Lee W. H. Hice1, a novel microtubule-associated protein required for maintenance of spindle integrity and chromosomal stability in human cells. Mol. Cell. Biol. 2008a;28:3652–3662. doi: 10.1128/MCB.01923-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Qiu X. L., Zhou L., Zhu J., Chamberlin R., Lau J., Chen P. L., Lee W. H. Small molecule targeting the Hec1/Nek2 mitotic pathway suppresses tumor cell growth in culture and in animal. Cancer Res. 2008b;68:8393–8399. doi: 10.1158/0008-5472.CAN-08-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J., Liu C. C., Chen P. L., Lee W. H. RINT-1, a novel Rad50-interacting protein, participates in radiation-induced G(2)/M checkpoint control. J. Biol. Chem. 2001;276:6105–6111. doi: 10.1074/jbc.M008893200. [DOI] [PubMed] [Google Scholar]
- Zhu H., Coppinger J. A., Jang C. Y., Yates J. R., 3rd, Fang G. FAM29A promotes microtubule amplification via recruitment of the NEDD1-gamma-tubulin complex to the mitotic spindle. J. Cell Biol. 2008;183:835–848. doi: 10.1083/jcb.200807046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.