Abstract
It is widely acknowledged that the presence of extracellular matrix components as substrates can drastically modulate the phenotype and gene expression of cultured cells, including tumor cells. A number of published reports indicated that substrates made from two peculiar collagen species, i.e. type V and OF/LB, which are abnormally deposited in the stroma of primary ductal infiltrating carcinoma (d.i.c.) of the breast “in vivo,” were able to exert marked and opposite effects on “in vitro” viability, growth and invasiveness of the 8701-BC cell line, isolated from d.i.c.-affected breast epithelium. To complement such functional data on the effect of cell-collagen interactions with information at molecular level, we have utilized a combination of differential display- and semi-quantitative multiplex-PCR techniques with the aim of detecting variations in the expression levels of selected genes by cells maintained in either culture condition. Here we report some prototypical data on the identification and semi-quantitation of three of the differentially-amplified PCR products found, i.e. HSP2A and MSF-B which are up-regulated in cells grown onto OF/LB collagen substrate, and SRCAP which is prominently down-regulated in the presence of type V collagen substrate. This protocol represents a powerful tool for evaluating changes in the levels and patterns of gene expression which can be theoretically adapted to any experimental model system.
Keywords: Polymerase Chain Reaction; Gene Expression; Collagen; Tumor Cells, Cultured
Introduction
The identification of differentially expressed genes to investigate a specific experimental system or the genetic changes associated with a given pathology is one of the most commonly pursued tasks in molecular and cellular biology. Although the use and application of DNA microarrays is in continuous expansion, the high specialization and cost of required equipment is so far a limiting parameter for most labs. Nonetheless, diverse other techniques allow the study of the modulation of gene expression including differential display (DD)-PCR, which permits the isolation of cDNA reverse transcribed from mRNA, allowing comparison of gene expression of differentially treated samples (1). The cDNA fragments are cloned and identified by sequencing and similarity searches. The differential expression of the cDNA can then be confirmed using more stringent techniques for the evaluation of steady-state mRNA such as semi-quantitative PCR.
It is generally acknowledged that the extracellular matrix does not function as a mere passive scaffold for connective tissue within organ architecture, but that it plays an “informational” role through a network of interactions between cells and signal molecules which is of primary importance in the control of cellular proliferation, in cell differentiation and motility, during histogenesis, for the maintenance of tissue homeostasis, and in cancer development. It has been previously reported that the phenotype of 8701-BC breast cancer cells is drastically modulated by the presence of different collagen matrices. In particular, when OF/LB and type V collagen, which have been proven to accumulate in breast stroma in cases of primary ductal infiltrating carcinoma (d.i.c.) (2, 3), are used as culture substrates, opposite cell behaviors can be observed. OF/LB collagen promotes cell growth, motility and invasion (4-6) whilst type V collagen prominently impairs cell survival, slowing-down cell proliferation rate and blocking cell invasive activity (5-8). Such cumulative results prompted a more detailed study about the effect of 8701-BC cell adhesion onto either OF/LB or type V collagen substrates on gene expression re-programming. Type IV collagen, being a “natural” support for cells of epithelial origin, was used as a control substrate. Here we report the successful adjustment of the protocol published by Sokolov and Prockop (9), which provides fragments from the internal sequences of the transcripts, to generate mRNA-DD electrophoretograms in polyacrylamide gels. This method provides the high resolution necessary for identifying genes that are selectively expressed under particular experimental conditions. Coupling of this technique with the multiplex relative RT-PCR protocol published by Spencer and Christensen (10) for verification of actual differences in transcript abundance allowed a fast, simple and radioisotope-free determination of some changes in the levels and patterns of gene expression following cell adhesion to collagen substrates, which can be theoretically adapted to any experimental condition.
Materials and Methods
Cell cultures and substrates
8701-BC cell line, derived from a d.i.c. biopsy fragment (11) was grown in 10% fetal calf serum (FCS)-containing RPMI 1640 medium (Gibco, Paisley/UK) plus antibiotics (100 U penicillin and 100 µg streptomycin/ml). For the production of bi-dimensional collagen substrates, type IV and type V collagens, purchased from Sigma (St.Louis, MO/USA), and OF/LB collagen (courtesy of Prof. I. Pucci-Minafra, Università di Palermo), were dissolved at a concentration of 100 μg/ml of 0.5 M acetic acid. Sterilization of the collagen preparations was obtained by dialysis versus chloroform-containing 0.5 M acetic acid (2 ml/l) for 2 h in the cold, followed by exhaustive dialysis versus chloroform-free 0.5 M acetic acid, as reported in Luparello et al. (7). The collagen solution was subsequently plated under sterile conditions in tissue culture containers at the concentration of 10 μg/cm2 and incubated for 48 h at 37°C, after which the excess liquid was discarded and the substrate exhaustively neutralized with sterile PBS just before use.
Messenger RNA extraction and reverse transcription
8701-BC cells were seeded onto collagen substrates at different concentrations (6 x 105cells/cm2 on type IV and OF/LB collagen, and 1.2 x 106 cells/cm2 on type V collagen) due to the diverse growth rate following cell attachment as reported by Schillaci et al. (4) and Luparello et al. (7), and grown for 48 h, which represented the minimum time lapse allowing stable adhesion onto type V collagen of the subpopulation of 8701-BC cells able to attach onto it (7). Isolation of polyA+ mRNA from cells, once detached with cell scrapers and collected in PBS by centrifugation, was performed using the mRNA Isolation kit (Roche, Mannheim/D) according to manufacturer’s instructions. cDNA synthesis was performed according to the protocol provided with M-MLV Reverse Transcriptase, RNase H Minus, Point Mutant (Promega, Madison,WI/USA) using 200 units of enzyme, 50ng of random hexamers (Promega) as primers, and 1µg of polyA+ mRNA.
DD-PCR
Before DD-PCR experiments, the quality of the cDNA preparations obtained from reverse transcription was checked by amplification in the presence of primers specific for human β-actin (see Table 1 for primer sequences).
Table 1.
Primers used for conventional and SM-PCR experiments
| Transcript detected | Oligonucleotides (5’→3’) | Product size |
| β actin (22) | GTGGGGCGCCCCAGGCACCA CTCCTTAATGTCACGCACGATTTC |
548 |
| HSP2A | AAGGCAGTGGAGTCCTTACGG CTTGTCAGCCACCATGTAGCG |
479 |
| MSF-B | GTCCAGACTCCCCTACTCCGA CTGACTCCTCAATATCGTGCG |
342 |
| KIAA0309/SRCAP | TTAGCCTCAGCTTCACCGGTA CTAGGGCCAGTGTCTGCGTAG |
126 |
PCR was performed using 2.5 units of RedTaq DNA Polymerase (Sigma) and 100 pmol of each primer in a 50µl reaction mixture; the thermal cycling profile was as follows: 94°C for 2 minutes, then 45 cycles of 94°C for 30 seconds, 55°C for 30 seconds and 72°C for 45 seconds, followed by a final elongation at 72°C for 5 minutes.
For differential expression analysis, DD-PCR was performed using the arbitrary 10-mer amplimers (see Table 2) designed by Sokolov and Prockop (9), in the following pairs: BS52/BS54, BS52/BS55, BS54/BS55, BS57/BS58, BS70/BS73 and BS76/BS78. The PCR amplification was carried out in an UNO Thermoblock (Biometra, Göttingen/D) using 25 pmoles of each of two primers, 1-2 µl of the cDNA template obtained from mRNA and 3.6 U of AmpliTaq DNA Polymerase, Stoffel fragment (Perkin Elmer, USA), as recommended by Doss (12), in 50 µl final volume. The thermal cycle used was a denaturation step of 94.5°C for 3 minutes, followed by 45 cycles of 94.5°C for 1 minute, 34°C for 1 minute, 72°C for 1 minute and a final extension of the product for 10 minutes at 72°C.
Table 2.
Primers used for DD-PCR experiments as designed by Sokolov and Prockop (9)
| Name | Sequence |
| BS52 | CAAGCGAGGT |
| BS54 | AACGCGCAAC |
| BS55 | GTGGAAGCGT |
| BS57 | GGAAGCAGCT |
| BS58 | CAGTGAGCGT |
| BS70 | GAGCTATGGC |
| BS73 | AGCCTGTGTC |
| BS76 | CTGGTCACAC |
| BS78 | CACAGTGAGC |
Electrophoresis and silver stain
After PCR amplification, 8 µl of the amplification products were electrophoresized in a non-denaturing 6% polyacrylamide gel casted in a sequencing apparatus (cm 21 x cm 40 x mm 0.4; Sequi-Gen, Bio-Rad, Richmond/USA) at constant 55 W and the band pattern visualized by silver staining as reported in Promega’s manual Silver Sequence-DNA Sequencing System (13). Briefly, the gel was first fixed for 20 min. in 10% acetic acid, exhaustively washed with ultrapure water and stained for 30 min. in a solution prepared by dissolving 2g of AgNO3 and 3 ml of 37% formaldehyde in 2 l of ultrapure water. After brief dipping into ultrapure water, the drained gel was transferred into a chilled developing solution prepared by dissolving 60g of Na2CO3 in 2 l of ultrapure water and adding, just before use, 3 ml of 37% formaldehyde and 400 µl of sodium thiosulphate (10 mg/ml). Developing was stopped by adding 1 l of 10% acetic acid for 2-3 min. Image of the gel was digitally recorded using a sequencing gel-sized scanner, such as the HP ScanJet 4C/T (Hewlett-Packard, Palo Alto CA/USA). For re-amplification of differentially-displayed bands, the silver-stained gel was immediately and exhaustively washed with several changes of ultrapure water and the bands of interest carefully scratched from the gel with a sterile syringe needle and used as template for PCR amplifications performed as described before. Several cycles of amplification and agarose gel electrophoresis were repeated until a single pure band was visualized in the ethidium bromide-stained gel and eluted using Ultrafree DA filter columns (Millipore, Bedford MA/USA).
DNA cloning and similarity search
The purified PCR products were subsequently cloned using the pGEM-T Easy vector system (Promega) and JM109 competent cells, high efficiency (Promega), according to Sirchia et al. (14). The recombinant plasmids were isolated with the High Pure Plasmid Isolation kit (Roche) according to manufacturer’s instructions. The insert sequence was determined by MWG Biotech sequencing service. DNA sequence similarity was searched with the BLAST algorithm (15) available on-line at http://blast.genome.ad.jp/.
Semi-quantitative multiplex polymerase-chain reaction (SM-PCR)
For SM-PCR we followed the protocol of Spencer and Christensen (10) with minor modifications. Essentially, for each cDNA species of interest and for that of a “housekeeping” gene, such as β-actin in present experiments, we preliminarly determined the range of cycles over which the species should be examined, i.e. starting from the minimum number necessary to visualize the desired product by 2% agarose electrophoresis and ethidium bromide stain (see Table 1 for primer sequences). Cycle profile was a denaturation step of 94.5°C for 3 minutes, followed by varying numbers of cycles of 94.5°C for 30 seconds, 50°C for 1 minute, 72°C for 1 minute and a final extension of the product for 5 minutes at 72°C. Subsequently, the cDNA of interest were co-amplified with β-actin cDNA over a range of cycles starting from the minimum necessary for detection, maintaining the intensity of the band of β-actin product constant by adding the related primers at the appropriate number of cycles before the end of the cycling protocol for each sample. Two different RNA preparations from each experimental condition were pooled to make more pronounced any differences between the expression levels and the experiments were repeated at least twice. Digital images of the gels were acquired using Kodak 1D software, and the intensities of the bands of interest, evaluated with SigmaScan software (SPSS, USA), were normalized for those of β-actin, and plotted as a function of cycle number. Exponential regression equations fitted to the curves, obtained using Excel software, were used to calculate the number of cycles necessary to reach a normalized intensity threshold value = 1 for each sample. Determination of R2 values were utilized to assess that the range of cycles considered were actually in the exponential phase of amplification. The relative difference in abundance between two samples was taken as 2 n where n is the difference between the numbers of cycles required by the samples to reach the threshold. n = 1 was taken as the minimum value for biological significance of differences in gene expression.
Results and Discussion
To search for genes differentially-expressed in 8701-BC breast cancer cells cultured onto either type IV, type V or OF/LB collagen substrates, we amplified cDNA from polyA+ mRNA isolated from cell preparations by reverse transcription using degenerate hexanucleotides. Internal regions of the isolated cDNAs were then subjected to PCR amplification using combinations of two of the 10mer primers with arbitrary but defined sequences reported in Table 2. DD-PCR experiments were replicated at least twice with different samples to reduce the number of false positive bands prior to further testing. All the combinations of arbitrary primers generated electrophoretograms that showed differential gene expression (not shown), although BS57/BS58 and BS76/BS78 primer pairs yielded larger total numbers of DNA fragments (about ten-fold more). Different DD-bands were chosen for further analysis and submitted to re-amplification with the same primers. Each of the cDNA fragments present in these bands gave a robust yield of amplified product, which was then isolated, cloned and sequenced as described before. Using BLASTN 2.2.6., similarity was found between the sequences obtained and those deposited either in the non-redundant nucleic acid sequence (nr-nt) or in the non-redundant protein sequence database (nr-aa). In the present paper, we focus our attention on three bands (named OF-A, OF-B and OF-C) from BS76/BS78 primer-amplified cDNA, and one band (named OF-D) from BS57/BS58 primer-amplified cDNA, all prepared from cells cultured on OF/LB collagen. The selected bands are pointed by arrows in Figure 1.
Fig. 1. DD-PCR electrophoretograms of cDNA preparations from 8701-BC cells grown onto type IV, type V and OF/LB collagen substrates, amplified in the presence of BS76/BS78 and BS57/BS58 primer pairs.
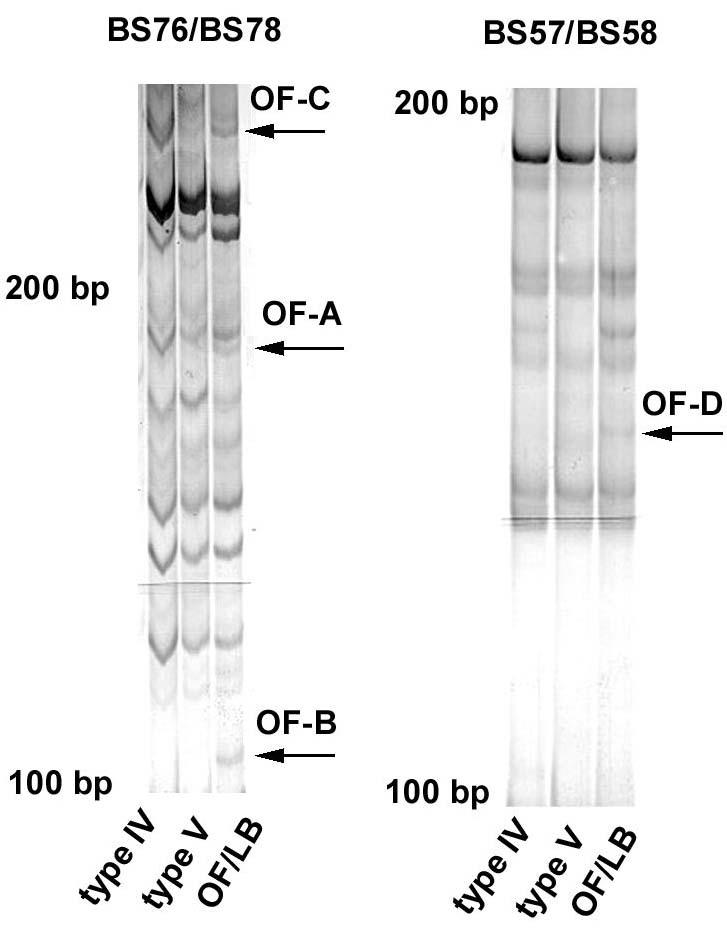
The arrows point to the differentially-displayed bands chosen for further analyses. Details of 6% sequencing PAGels, silver staining.
After sequencing and similarity search, as reported in Table 3, two bands (OF-C and -D) were found to contain a DNA fragment homologous to coding sequences of two known genes (MSF-B and HSP2A), whilst the translated sequence of the DNA fragment of OF-A band showed an elevated degree of similarity with a deposited protein sequence (KIAA0309/SRCAP protein). HSP2A (acc. number J02902) encodes the alpha-isotype of the regulatory subunit of human protein phosphatase 2A, MSF-B (acc. number AF189712) encodes an MLL septin-like fusion protein, whilst KIAA0309/SRCAP protein (acc. numbers AB002307_1 and AF143946_1) is a transcriptional activator (16). On the other hand, the DNA fragment contained in OF-B band showed similarity only with a so far non-characterized sequence from human chromosome 1q32.2-41. It was uncommon to obtain such result (about 30% of sequenced DD-PCR products, in our experience). No attempt was made to perform more detailed investigations on this unknown gene product.
Table 3.
Results obtained from DD-PCR experiments and band sequencing
| Band name | Alignment | Identities (%) | Positives (%) | Score | E value |
| OF-A | KIAA0309 gene product/SRCAP protein (protein alignment | 65 | 72 | 57 | 3.00E-008 |
| OF-B | Human DNA sequence from clone RP1 28O10 on chromosome 1q32.2-41 | 88 | / | 73.8 | 1.00E-010 |
| OF-C | Homo sapiens MLL septin-like fusion Protein MSF-B | 89.1 | / | 168.2 | 5.10E-040 |
| OF-D | Human protein phosphatase 2A regulatory Subunit, alpha-isotype | 94.6 | / | 119.1 | 1.40E-025 |
We then checked the differential expression of HSP2A, MSF-B and KIAA0309/SRCAP by PCR amplification in the presence of specific primers (see Table 1) designed using the Primer Selection software available on-line at http://alces.med.umn.edu/rawprimer.html. As shown in Figure 2, in a preliminary assay we found an amplification band of the expected size after conventional PCR of all samples tested, indicating that expression of genes of interest was switched-on in all the experimental conditions tested.
Fig. 2. Panel of PCR analyses showing the presence of amplification products for HSP2A, KIAA0309/SRCAP and MSF-B in cDNA preparations from 8701-BC cells grown onto type IV, type V and OF/LB collagen substrates.
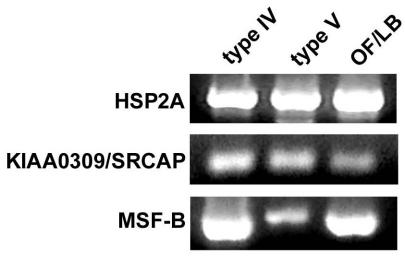
Ethidium bromide stain.
The identity of PCR products was further checked by sequencing (not shown). Subsequently, for semi-quantitative evaluation of expression levels, the cDNA preparations were submitted to SM-PCR as described. Figure 3 shows the plots of normalized data for HSP2A (Fig. 3A), MSF-B (Fig. 3B) and KIAA0309/SRCAP (Fig. 3C) versus cycle number, fit with exponential curves; from calculation of the relative difference in transcript abundance, growth of 8701-BC cells onto OF/LB collagen substrate resulted in the up-regulation of HSP2A and MSF-B of about 3.6- and 3.4-fold, respectively, if compared to the other substrates. No substantial difference was found between the expression levels of the two genes studied in cells cultured onto type IV and type V collagen substrates. On the other hand, growth of 8701-BC cells onto type V collagen substrate resulted in down-regulation of KIAA0309/SRCAP of about 17-fold, if compared to the other substrates. The difference between the expression levels of KIAA0309/SRCAP in cells seeded onto type IV and OF/LB collagen was not substantial. An explanation of the biological significance of the results obtained can be attempted if we consider that protein phosphatase 2A plays prominent roles in the control of cell growth and signalling (17), and, in particular, up-regulation of regulatory subunit A, α isotype, has been proven to drastically affect cell cycle and mitosis (18). In addition, septins, such as MSF-B, are thought to be prominently involved in cytokinesis and cell division (19). Therefore, up-regulation of both genes could be conceivably involved in the increase of proliferation rate observable when 8701-BC cells grow onto OF/LB collagen (4-6). Conversely, the pronounced down-regulation of the growth-promoting transcription factor KIAA0309/SRCAP (20) may be, at least in part, responsible of the restraining role played by type V collagen on the viability and growth of 8701-BC cells (4-6). Further study will be necessary to validate our postulates.
Fig. 3. SM-PCR for HSP2A (A), MSF-B (B) and KIAA0309/SRCAP (C).
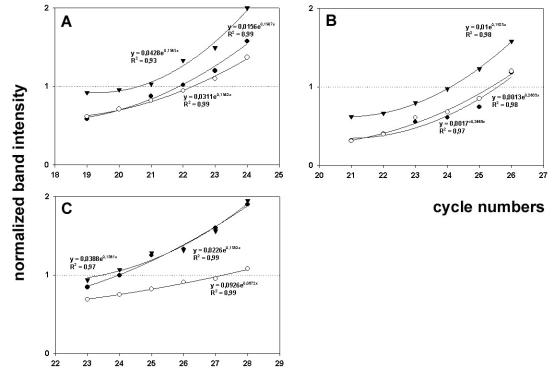
Representative plots of normalized data vs. cycle numbers fit with an exponential curve for 8701-BC cells cultured onto type IV (●), type V (○) and OF/LB (▼) collagen substrates.
In conclusion, our experiments highlight that a combination of DD-PCR and SM-PCR techniques can be a powerful tool for identifying genes that are selectively expressed by cells under particular experimental conditions, such as in the presence of different collagen matrices. The protocol we have reported does not exclude additional confirmation at mRNA level by non-PCR based assays (e.g. Northern blot and RNase protection) and at protein level. As an example, we have already utilized this technique in other investigations performed in our lab (21-23) and found that the up-regulation of hsp90 in breast cancer cells treated with PTHrP [67-86], demonstrated by the DD-PCR/SM-PCR protocol, could be supported by data obtained with Western blot (22). Thus, the agreement of the findings at both transcript and protein level further validates this experimental procedure as suitable for a detailed study of the molecular mechanisms of cell responses to micro-environmental stimuli.
Acknowledgments
This work was supported by funds form the University of Palermo (R.S. ex 60%).
References
- Liang P. A decade of differential display. BioTechniques. 2002;33:338–344. doi: 10.2144/02332rv01. [DOI] [PubMed] [Google Scholar]
- Pucci-Minafra I, Luparello C, Sciarrino S, Tomasino RM, Minafra S. Quantitative determination of collagen types present in the ductal infiltrating carcinoma of human mammary gland. Cell Biol Int Rep. 1985;9:291–296. doi: 10.1016/0309-1651(85)90047-5. [DOI] [PubMed] [Google Scholar]
- Luparello C, Rizzo CP, Schillaci R, Pucci-Minafra I. Fractionation of type V collagen from carcinomatous and dysplasic breast in the presence of alkaline potassium chloride. Anal Biochem. 1988;169:26–32. doi: 10.1016/0003-2697(88)90252-7. [DOI] [PubMed] [Google Scholar]
- Schillaci R, Luparello C, Minafra S. Type I and I-trimer collagens as substrates for breast carcinoma cells in culture. Effect on growth rate, morphological appearance and actin organization. Eur J Cell Biol. 1989;48:135–141. [PubMed] [Google Scholar]
- Luparello C, Sheterline P, Pucci-Minafra I, Minafra S. A comparison of spreading and motility behaviour of 8701-BC breast carcinoma cells on type I, I-trimer and type V collagen substrates. J Cell Sci. 1991;100:179–185. doi: 10.1242/jcs.100.1.179. [DOI] [PubMed] [Google Scholar]
- Pucci-Minafra I, Luparello C, Aquino A, Basiricò L, Minafra S, Franc S, Yakovlev L, Shoshan S. OF/LB collagen promotes chemoinvasion of breast cancer cells and directs epithelial cell migration into granulation tissue of experimental dermal wounds. Int J Oncol. 1995;6:1015–1020. doi: 10.3892/ijo.6.5.1015. [DOI] [PubMed] [Google Scholar]
- Luparello C, Schillaci R, Pucci-Minafra I, Minafra S. Adhesion, growth and cytoskeletal characteristics of 8701-BC breast carcinoma cells cultured in the presence of type V collagen. Eur J Cancer. 1990;26:231–240. doi: 10.1016/0277-5379(90)90217-h. [DOI] [PubMed] [Google Scholar]
- Luparello C. Adhesion to type V collagen and cloning efficiency in agar of 8701-BC breast cancer cells. Eur J Cancer. 1994;30A:1400–1401. doi: 10.1016/0959-8049(94)90196-1. [DOI] [PubMed] [Google Scholar]
- Sokolov BP, Prockop DJ. A rapid and simple PCR-based method for isolation of cDNAs from differentially expressed genes. Nucl Acid Res. 1994;22:4009–4015. doi: 10.1093/nar/22.19.4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer WE, Christensen MJ. Multiplex relative RT-PCR method for verification of differential gene expression. BioTechniques. 1999;27:1044–1052. doi: 10.2144/99275rr04. [DOI] [PubMed] [Google Scholar]
- Minafra S, Morello V, Glorioso F, La Fiura AM, Tomasino RM, Feo S, Mcintosh D, Woolley DE. A new cell line (8701-BC) from primary ductal infiltrating carcinoma of the human breast. Brit J Cancer. 1989;60:185–192. doi: 10.1038/bjc.1989.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doss RP. Differential display without radioactivity – a modified procedure. BioTechniques. 1996;21:408–409. doi: 10.2144/96213bm14. [DOI] [PubMed] [Google Scholar]
- Promega Technical Manual Silver Sequence DNA sequencing system. Promega Corporation 1995.
- Sirchia R, Ciacciofera V, Luparello C. Cloning differential display-PCR products with pGEM-T Easy vector system. Promega eNotes 2001. http://www.promega.com/enotes/applications/ap0025_tabs.htm.150
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl Acid Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston H, Kneer J, Chackalaparampil I, Yaciuk P, Chrivia J. Identification of a novel SNF2/SWI2 protein family member, SRCAP, which interacts with CREB-binding protein. J Biol Chem. 1999;274:16370–16376. doi: 10.1074/jbc.274.23.16370. [DOI] [PubMed] [Google Scholar]
- Janssens V, Goris J. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem J. 2001;353:417–439. doi: 10.1042/0264-6021:3530417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wera S, Fernandez A, Lamb NJC, Turowski P, Hemmings-Miezczack M, Mayer-Jaeckel RE, Hemmings BA. Deregulation of translational control of the 65-kDa regulatory subunit (PR65α) of protein phosphatase 2A leads to multinucleated cells. J Biol Chem. 1995;270:21374–21381. doi: 10.1074/jbc.270.47.28068. [DOI] [PubMed] [Google Scholar]
- Kalikin LM, Sims HL, Petty EM. Genomic and expression analyses of alternatively spliced transcripts of the MLL septin-like fusion gene (MSF) that map to a 17q25 region of loss in breast and ovarian tumors. Genomics. 2000;63:165–172. doi: 10.1006/geno.1999.6077. [DOI] [PubMed] [Google Scholar]
- Ghosh AK, Majumder M, Steele R, Yaciuk P, Chrivia J, Ray R, Ray RB. Hepatitis C virus NS5A protein modulates transcription through a novel cellular transcription factor SRCAP. J Biol Chem. 2000;275:7184–7188. doi: 10.1074/jbc.275.10.7184. [DOI] [PubMed] [Google Scholar]
- Pucci-Minafra I, Carella C, Cirincione R, Chimenti S, Minafra S, Luparello C. Type V collagen induces apoptosis of 8701-BC breast cancer cells and enhances m-calpain expression. Breast Cancer Res. 2000;2:E008–E008. doi: 10.1186/bcr60. [DOI] [Google Scholar]
- Luparello C, Sirchia R, Pupello D. PTHrP [67-86] regulates the expression of stress proteins in breast cancer cells inducing modifications in urokinase-plasminogen activator and MMP-1 expression. J Cell Sci. 2003;116:2421–2430. doi: 10.1242/jcs.00472. [DOI] [PubMed] [Google Scholar]
- Luparello C, David F, Campisi G, Sirchia R. T47-D cells and type V collagen: a model for the study of apoptotic gene expression by breast cancer cells. Biol Chem. 2003;384:965–975. doi: 10.1515/BC.2003.109. [DOI] [PubMed] [Google Scholar]


