Abstract
Chromosome movements are linked to the active depolymerization of spindle microtubule (MT) ends. Here we identify the kinesin-13 family member, KLP59D, as a novel and uniquely important regulator of spindle MT dynamics and chromosome motility in Drosophila somatic cells. During prometaphase and metaphase, depletion of KLP59D, which targets to centrosomes and outer kinetochores, suppresses the depolymerization of spindle pole–associated MT minus ends, thereby inhibiting poleward tubulin Flux. Subsequently, during anaphase, loss of KLP59D strongly attenuates chromatid-to-pole motion by suppressing the depolymerization of both minus and plus ends of kinetochore-associated MTs. The mechanism of KLP59D's impact on spindle MT plus and minus ends appears to differ. Our data support a model in which KLP59D directly depolymerizes kinetochore-associated plus ends during anaphase, but influences minus ends indirectly by localizing the pole-associated MT depolymerase KLP10A. Finally, electron microscopy indicates that, unlike the other Drosophila kinesin-13s, KLP59D is largely incapable of oligomerizing into MT-associated rings in vitro, suggesting that such structures are not a requisite feature of kinetochore-based MT disassembly and chromosome movements.
INTRODUCTION
The position and movement of chromosomes on the mitotic spindle is linked to the tightly regulated polymerization dynamics of kinetochore-associated microtubule (kMT) ends. During metaphase, when chromosomes persist at the spindle equator, kMT minus-end depolymerization at poles is balanced by plus-end polymerization at kinetochores, allowing kMTs to maintain a constant length but inducing the poleward flux of tubulin subunits through the MT polymer lattice (Flux). Subsequently at the onset of anaphase, kMT plus-end polymerization ceases, allowing unbalanced minus-end depolymerization and Flux to exert poleward pulling forces on chromosomes. At the same time, factors within kinetochores depolymerize kMT plus ends exerting an additional force for moving chromatids to poles termed “Pacman” (Mitchison and Salmon, 2001).
MT destabilizing kinesin-13s have been found to be important effectors of spindle MT dynamics, generally, and “Pacman-Flux”–based chromatid-to-pole motion in anaphase (anaphase A; Wordeman, 2005). In Drosophila early embryos, two kinesin-13s, KLP10A and KLP59C, cooperate in anaphase A by stimulating the depolymerization of opposite kMT ends. KLP10A targets to spindle poles and is required for Flux, whereas KLP59C targets to centromeres and is involved in Pacman-based chromosome motility (Rogers et al., 2004). Electron microscopy has also shown that these proteins oligomerize into ring or spiral structures around MTs, providing a potential mechanism for coupling chromosome movement to kinesin-13 stimulated MT depolymerization (Moores et al., 2006; Tan et al., 2006). Surprisingly, recent work has shown that KLP59C does not directly influence anaphase A in Drosophila S2 cells (Goshima and Vale, 2005; Buster et al., 2007), indicating that kinesin-13s are utilized differently in different cell types.
This study focuses on the third Drosophila kinesin-13 family member, KLP59D, whose function has remained elusive. Our initial goal was to test the hypothesis that KLP59D replaces KLP59C as the Pacman depolymerase in S2 cells. Instead, we found KLP59D to be a uniquely important regulator of spindle dynamics and chromosome motility. Most significantly, KLP59D is utilized to stimulate the depolymerization of both kMT plus and minus ends during anaphase, making it the only known regulator of MT dynamics that simultaneously drives Pacman- and Flux-based chromosomal motility. Our data support a model in which KLP59D directly depolymerizes kMT plus ends, but acts on minus ends indirectly by targeting KLP10A.
MATERIALS AND METHODS
S2 Cell Culture and Double-stranded RNA Interference
Enhanced green fluorescent protein (EGFP)-α-tubulin– expressing S2 cells (a gift from R.Vale, UCSF) were cultured at 23°C in Schneider's medium supplemented with 10% heat inactivated fetal bovine serum (HI-FBS, Invitrogen, Carlsbad, CA) and penicillin/streptomycin (Invitrogen) (Buster et al., 2007). For live cell analysis and immunostaining, S2 cells were plated onto Concanavalin A–coated 35-mm glass-bottomed dishes (MatTek, Ashland, MA) and coverslips, respectively, for 2 h.
Double-stranded RNA (dsRNA) oligonucleotides for RNA interference (RNAi) were generated as described in Rogers et al. (2004). PCR primers were composed of the T7 polymerase promoter sequence (5′-TAATACGACTCACTATAGGG-3′) appended to the 5′ end each of the following gene-specific sequences: 1) KLP59D coding sequence (the first 910 bp): 5′-GGATCGCATCAAAATTGG-3′; 5′-CGTAGACCAGCGCATTG-3′; 2) KLP59D 5′UTR (99 bp): 5′-CGCTTGTGAACTGGAAATCA-3′; 5′-TGGTGGCAACGGATTATACA-3′; 3) KLP59D 3′UTR (85 bp): 5′-TTCGCCCACCACCTACTAAT-3′; 5′-GGGGTCTTCCAAACGCTATT-3′; 4) KLP10A coding sequence (the first 894 bp): 5′-ATGATTACGGTGGGGCA-3′; 5′-GACATCGATCTCCTTGCG-3′; 5) KLP59C coding sequence (the first 689 bp): 5′-ATGGATAAGTTGTCGATCG-3′; 5′-ACCAGGTTCACATGCTTGCG-3′; 6) CNN coding sequence (309 bp; targets an exon shared by all isoforms): 5′-GCGCTGCCTCGATATTACTC-3′; 5′-TGCACGGCATCTAGTTTGAG-3′; and 7) Control template (900 bp of noncoding sequence from pBluescript SK):
5′-TAAATTGTAAGCGTTAATATTTTG-3′; 5′-AATTCGATATCAAGCTTATCGAT-3′.
Immunofluorescence
RNAi-treated S2 cells were plated on concanavalin-A–coated coverslips and fixed in 100% methanol at −20°C. Rehydration was done in phosphate-buffered saline with 0.04% Tween-20 (PBST) followed by blocking in PBST containing 5% normal goat serum. The following primary antibodies were used: anti-KLP59D, anti-KLP10A (Rogers et al., 2004), anti-CID (a gift from G. Karpen lab, Lawrence Berkeley National Laboratory), anti-Asp (a gift from D. Glover lab), anti-α-tubulin antibody (DM1A, Sigma-Aldrich, St. Louis, MO) and anti-γ-tubulin antibody (GTU-88, Sigma-Aldrich). After treatment with fluorescent 2° antibodies (from Jackson ImmunoResearch, West Grove, PA), immunostained samples were mounted in anti-fade solution (1% N-propyl-gallate, 100 mM Tris, pH 8, and 50% glycerol). Imaging was done using a spinning-disk confocal system (Ultraview, Perkin Elmer-Cetus, Waltham, MA) mounted on a Nikon TE300 inverted microscope with a 100×, 1.4 NA objective (Melville, NY). Images were captured with an Orca ER digital camera (Hamamatsu, Hamamatsu City, Japan) as z-stacks and are presented as maximum intensity projections.
To induce C-mitosis, S2 cells were treated with 25 μM colchicine for 16 h and then fixed in 100% methanol at −20°C. Rehydration and immunostaining were done as described above.
Measurement of Anaphase A Chromatid-to-Pole Rates
After RNAi, S2 cells constitutively expressing eGFP-α-tubulin were plated on concanavalin-A–treated MatTek dishes and imaged as z-stacks at 10–15-s intervals using a spinning disk confocal system (Ultraview, Perkin Elmer-Cetus) as described above. Negatively stained profiles of anaphase chromatids were tracked at the ends of well-resolved kinetochore fibers (k-fibers). Shortening of these kMTs toward the spindle pole was measured over time manually using ImageJ (NIH; http://rsb.info.nih.gov/ij/). The rate of chromatid-to-pole motion was calculated as change in length of kMTs over time. Alternatively, the chromosomes were vital stained with Hoechst 33258 (300 ng/ml final concentration in the medium) just before visualizing the cells, and chromatid-to-pole rates were measured from the poleward translocation of the fluorescent anaphase chromatids through time. For each half-spindle, visually distinct kMTs or chromosomes were chosen for measurement. Both methods yielded similar results for similarly RNAi-treated S2 cells, demonstrating that brief exposure to Hoechst 33258 does not alter anaphase chromatid-to-pole movement. All measurements for each spindle were averaged, and the average used as a single data point. Therefore, the N values presented in the figures represent the number of “spindle averages” acquired for a particular treatment.
Quantification of Spindle Length and Axial Ratio
To quantitate the spindle length, pole-to-pole distance in each spindle was measured in RNAi-treated S2 cells after immunostaining with anti-α-tubulin using the calipers tools of MetaMorph software (Molecular Devices, Sunnyvale, CA). Axial ratio was determined by calculating the ratio between the long axis (spindle length) and the spindle width (metaphase plate).
Fluorescence Recovery after Photobleaching
Preanaphase S2 cells stably expressing eGFP-α-tubulin were photobleached (two bleaching pulses of 1.2 s each) at the spindle equator (assumed to consist of primarily MT plus ends) and a pole (assumed to consist primarily of MT minus ends). The two bleach zones were created in opposing half-spindles to prevent cross-interference during recovery. After photobleaching, time-lapse videos were captured at 3.6-s time intervals in a single z-plane using a TCS SP2 confocal system (Leica, Heidelberg, Germany) on a DMIRE2-Leica inverted microscope (Plan Apo 63× objective, 1.4 NA) for 2–3 min. MetaMorph software (Molecular Devices) was used to quantitate the recovery of the fluorescence intensities in the photobleached regions. Same-sized and - shaped regions-of-interest within the photobleached zones were used for the analyses. For each time point after photobleaching, the gradual decrease of fluorescence intensity resulting from image capture was compensated for by calculating the decrease in whole-cell fluorescence as a percentage of the whole-cell fluorescence measured for the prebleach image. This value was used to adjust the measured fluorescence recovery within the region-of-interest accordingly. The adjusted fluorescence recovery measurements of the bleached region were curve-fitted to a one-phase exponential equation: y(t) = A(1 − e(−τ · t)), where y is the measured fluorescence within the region-of-interest at time point t, A is the end point of fluorescence recovery, and τ is the fitted parameter, using nonlinear regression (GraphPad Prism4, San Diego, CA). The half-time of recovery (T1/2) was calculated as follows: T1/2 = (ln0.5)/τ. Collapsed spindles and spindles showing less that 30% recovery were not used in the calculations of T1/2.
To gain insight into the fluorescence recovery process in photobleached areas, kymographs were used to measure the rate of fluorescence encroachment into each photobleached area. Kymographs were generated from narrow, linear regions-of-interest that were drawn to overlay individual bleached k-fibers (whose position within the bleached area could be determined from later postbleach images in which fluorescence recovery had begun) and to span the bleached area. In photobleached areas of typical control RNAi spindles, fluorescence gradually extends into the area from the plateward side (i.e., from the side oriented toward the spindle equator), presumably as a result of flux of the k-fiber. Specifically, the rate of this poleward elongation of fluorescence was measured. Kymographs were generated with MetaMorph software. Then, the rate of poleward elongation was calculated manually from the slope (measured with ImageJ) of the track of the advancing fluorescence.
Measurement of Flux and Pacman Rates
S2 cells stably expressing eGFP-α-tubulin were treated by RNAi to knock down specific target proteins. In the case of KLP59D, three different nonoverlapping dsRNA oligomers were used. dsRNA derived from the 5′ UTR, 3′ UTR, or KLP59D coding sequence all produced similar effects on Flux and Pacman rates, indicating that the observed phenotypes result from a specific knock down of KLP59D. Because the 5′ and 3′ UTR dsRNAs were not used for all assays; however, only data for coding sequence dsRNA is presented.
In mitotic RNAi-treated S2 cells (expressing eGFP-α-tubulin), narrow rectangular bars were photobleached across each half-spindle, perpendicular to the spindle axis and approximately midway on each half-spindle. Just before imaging, chromosomes were vital stained by addition of Hoechst 33258 (300 ng/ml final) to the medium. Time-lapse recordings were generated at 3.26-s time intervals from a single z-plane using the Leica TCS system described above. ImageJ was used to manually measure the movement (change in distance) of the bleached bar toward the spindle pole as a function of time to calculate the flux rate. Anaphase Pacman rates were determined by measuring chromatid movement relative to the bleached bar over time. (Depending on the relative positions of a particular chromatid and the bleach mark on a half-spindle, the segregating chromatid could either be moving toward —or away from—the bleach mark.) For each half-spindle, visually distinct k-fibers or chromosomes were chosen for measurement. For each spindle, all of the measurements (either flux or Pacman rates) were averaged, and this “spindle average” was used as a single data point. Therefore, the values for N represent the number of spindle averages acquired for a particular treatment.
Protein Expression, Purification, and Interactions
Proteins used for electron microscopy (EM) and MT depolymerization assays, KLP59D full-length (KLP59D FL) and the neck-linker/motor domain (KLP59D NMD, amino acid residues 159-570), were expressed using the Bac-to-Bac baculovirus protein expression system in SF9 cells. Coding sequences were subcloned into the pFASTBAC HTA vector containing a 6x-His tag. Expressed proteins were purified using Ni-NTA beads (Qiagen, Valencia, CA) and buffer exchanged into BRB80 (80 mM PIPES, pH 6.8, 2 mM MgCl2, and 1 mM EGTA) containing 50 μM ATP. Immunoprecipitation assays were performed with protein G/A–coupled antibodies anti-KLP59D, anti-KLP10A, and anti-GFP (ab290, Abcam, Cambridge, MA) and lysates of mitotic S2 cells stably expressing KLP59D-GFP as described in Rath et al. (2004). Mitotic lysates were prepared from S2 cells treated with 25 μM colchicine for 16 h. Before the end of colchicine treatment, the cells were treated with 0.5 mM CuSO4 for 3–4 h to induce KLP59D-eGFP or KLP10A-eGFP expression. KLP59D antibody was preadsorbed with purified GST-tagged KLP10A NT to eliminate cross reactivity. KLP10A antibody was preadsorbed with GST-KLP59D NT. GST-KLP59D NT (N-terminus, aa 1-205) and GST-KLP10A NT (aa 1-229) fusion proteins were bacterially expressed and purified with glutathione-agarose resin (Sigma-Aldrich) according to manufacturer's instructions.
Quantification of KLP10A and Asp Immunofluorescence
After RNAi, S2 cells were fixed in methanol at −20°C and immunostained with anti-KLP10A (Rogers et al., 2004) or anti-Asp and anti-α-tubulin antibody (DM1A, Sigma-Aldrich) as described above. Immunostained samples were imaged using the spinning-disk confocal system (Ultraview, Perkin Elmer-Cetus) under identical conditions. Maximum intensity projections images were analyzed with MetaMorph, and the intensities of KLP10A and ASP immunofluorescence were measured at centrosomes, kinetochores, and the spindle poles. Fluorescence intensities were adjusted by subtraction of background fluorescence.
Quantification of MT Density and Tubulin-KLP10A Ratios
To test the possibility that KLP59D RNAi decreases KLP10A immunostaining by altering the amount of MTs at poles, two measurements were made using the same images prepared in the previous section. First, the near-pole MT density was measured and compared for control and KLP59D RNAi-treated cells. For this, each spindle was quartered into four regions-of-interest along its pole-to-pole length, and the α-tubulin immunofluorescence intensities of the two regions containing the poles were measured. After correcting the measurements by subtracting backgrounds, MT density was calculated by dividing the fluorescence intensities by the areas of the measured regions. The average values were not significantly different for the two treatments.
Second, the ratio of α-tubulin to KLP10A immunofluorescence intensities was calculated and compared for control and KLP59D RNAi cells. If the amount of KLP10A immunofluorescence at poles were strictly and solely a function of the amount of MTs at poles, then the ratio of tubulin to KLP10A should remain constant for all treatments. To test this prediction, both α-tubulin and KLP10A immunofluorescence intensities were measured from the same region-of-interest at each spindle pole, appropriate background subtractions were made, and the ratio at each pole was calculated.
MT Depolymerization Assay
Taxol-stabilized MTs used for the depolymerization assay were generated by incubating purified bovine brain tubulin in BRB80 buffer containing 2 mM GTP and 20 μM taxol at 37°C for 30 min and then isolating the polymer by centrifugation at 217,000 × g for 5 min at 28°C. MT depolymerization was measured with sedimentation and turbidity (light scattering) assays. For the sedimentation assay, taxol-stabilized MTs (1 μM final) were incubated with varying concentrations of purified KLP59D-NMD (from 0 to 0.1 μM) in BRB80 containing 1 mM ATP for 20 min at 23°C. Then the reactions were ultracentrifuged at 217,000 × g for 10 min at 28°C to separate soluble tubulin subunits (in the supernatant) from MTs (in the pellet). Pellets were resuspended in a volume of BRB80 equal to the original reaction volume. The relative tubulin content of the supernatant and pellet fractions was measured by densitometry of samples resolved on the same Coomassie-stained 10% SDS-PAGE gel.
For the MT turbidity (light-scattering) assay, taxol-stabilized MTs (1 μM) were mixed with varying concentrations of purified 6xHis-tagged full-length KLP59D in BRB80 buffer containing 1 mM ATP. Immediately after mixing, the optical density of each sample was measured through time at 23°C using a Lambda25 UV/VIS spectrometer (Perkin Elmer-Cetus). Turbidity change was measured by the change in 350-nm light scattering. A decrease in turbidity (A350) indicates depolymerization of MTs into tubulin subunits.
Electron Microscopy
Samples for negative stain electron microscope experiments were prepared by mixing 5 μM of MTs (prepared as described above) with either 1 or 2.5 μM KLP59D NMD or KLP10A NMD proteins in BRB80 buffer supplemented with 2 mM Mg2+/AMP-PNP and incubating for 20 min at 23°C. Five microliters of this mixture was then loaded onto freshly glow-discharged carbon supported grids (Electron Microscopy Sciences, Hatfield, PA). The grid was washed and stained with 1% uranyl acetate, followed by observation in a Tecnai F20 microscope (FEI, Eindhoven, The Netherlands) operating at 120 kV with a nominal magnification of ×50,000. Electron micrographs were recorded with a F224HD CCD camera (Tietz Video and Image Processing Systems, Gauting, Germany).
Statistical Data Analysis
The statistical differences between treatments were analyzed by either a one-way ANOVA on ranks (Kruskal-Wallis) for multiple groups or a nonparametric t test (Mann-Whitney) for two-group comparisons (SigmaStat, Systat Software; GraphPad Prism4, GraphPadA). Dunn's posttest was used after a nonparametric ANOVA to compare pairs of group medians. Medians are assumed to differ significantly when p ≤ 0.05.
The need to use nonparametric tests was determined by preliminary tests for normality and variance. When data sets were found to have nominally normal distributions and equal variance, then the appropriate parametric one-way ANOVA or t test was used.
RESULTS
KLP59D Targets to Kinetochores and Centrosomes
We performed immunofluorescence microscopy using affinity-purified anti-KLP59D antibodies to label mitotic S2 cells (antibodies characterized in Rogers et al., 2004). This revealed KLP59D to be a spindle component (Figure 1A) that associates with kinetochores, as indicated by colocalization with the inner kinetochore protein Cid (Figure 1B), and centrosomes, as indicated by colocalization with γ-tubulin (Figure 1C). KLP59D also localizes to the spindle midbody late in telophase (Supplemental Figure S1A). An identical localization was observed in S2 cells transfected with full-length eGFP-KLP59D (Supplemental Figure S1B).
Figure 1.
KLP59D localizes to kinetochores and centrosomes. (A) Immunofluorescence of an anaphase S2 cell double-labeled for KLP59D and α-tubulin. Centrosomes denoted by yellow arrowheads. (B) High-magnification image showing the relative localizations of KLP59D, Cid, and α-tubulin in early anaphase. (C) Top, immunofluorescence image of a metaphase spindle double-labeled for KLP59D and γ-tubulin. Bottom panels, magnified images of one centrosome (inset in the top panel).
KLP59D's localization to kinetochores initiates at prometaphase and persists through telophase (Supplemental Figure S1A). Some nuclear staining was also apparent during prophase (before nuclear envelope breakdown) but not specifically associated with kinetochores. Within kinetochores, KLP59D extends poleward and away from the interior base of the structure defined by the position of Cid (Figure 1B). This suggests that KLP59D is a component of outer kinetochores, where the majority of kMT plus ends terminate (Maiato et al., 2006). This contrasts the other two Drosophila kinesin-13s, KLP10A and KLP59C, which are generally confined to the inner kinetochore/centromere in this cell type (Rogers et al., 2004). Finally, no substantial redistribution of kinetochore KLP59D was apparent when MTs were depolymerized by colchicine (not shown), suggesting that this kinesin-13 is not associated with the fibrous corona, a region of outer kinetochores that extends out along the MT lattice and expands in the absence of MTs (Maney et al., 2000).
The centrosomal targeting of KLP59D becomes apparent somewhat earlier in the cell cycle, initiating during prophase, and continues throughout mitosis (Supplemental Figure S1A). KLP59D's association with centrosomes is MT-independent as it persists when MTs are depolymerized by colchicine (data not shown). It should be noted that centrosomes generally do not appear as organized structures during interphase in S2 cells (Rogers et al., 2008). Though KLP59D binds to the centrosome proper, as indicated by colabeling with γ-tubulin, it does not extend onto the focus of MT minus ends, which define the spindle pole. The majority of MT minus ends in S2 cell spindles are not directly attached to centrosomes, making the pole and centrosome two distinct structures (Rogers et al., 2005). This is notably different from the Flux-promoting kinesin-13 KLP10A, which labels both centrosomes and poles (Supplemental Figure S1C), or KLP59C, which does not associate with either structure (Rogers et al., 2004).
KLP59D Stimulates Anaphase A
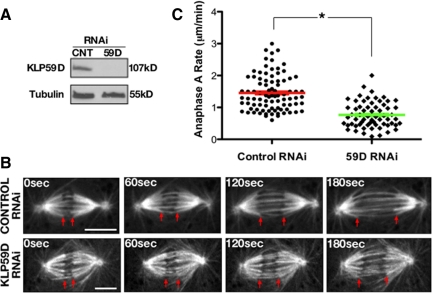
Earlier work suggested that KLP59D is not required for timely progression through mitosis because its depletion does not induce a significant elevation of mitotic index (% of mitotic cells; Rogers et al., 2004). Because such analyses provide limited information regarding KLP59D's impact on spindle and chromosome dynamics, we reassessed KLP59D's mitotic function using four-dimensional (4D) spinning disk confocal microscopy on live control and KLP59D RNAi-treated S2 cells expressing eGFP-α-tubulin. Observations were made after 7 d of RNAi treatment when KLP59D levels were reduced by ∼90% (Figure 2A). KLP59D-depleted cells generally assembled bipolar metaphase spindles, which proceeded into anaphase. However, significant defects in poleward chromosome motility were apparent upon entry into anaphase (Figure 2, B and C; Supplemental Movies S1 and S2). Specifically, KLP59D depletion decreased the rate of anaphase A nearly twofold, from an average control velocity of 1.34 ± 0.06 to 0.76 ± 0.05 μm/min in KLP59D RNAi-treated cells (p < 0.0001; values are means ± SEM throughout the text). Identical results were obtained using two other nonoverlapping KLP59D dsRNA constructs. By comparison, RNAi depletion of KLP10A induced a similar reduction of anaphase A rates to 0.86 ± 0.06 μm/min. Depletion of KLP59C had no significant impact on anaphase A in this cell type (1.2 ± 0.07 μm/min; Supplemental Table S1), consistent with earlier work.
Figure 2.
KLP59D drives anaphase A. (A) Western blots of lysates from S2 cell cultures treated with control (CNT) or KLP59D RNAi and probed with antibodies against KLP59D or α-tubulin (loading control). (B) Time-lapse images of anaphase A in control and KLP59D RNAi-treated S2 cells stably expressing eGFP-α-tubulin. Red arrows track the segregating chromatids, which appear as dark, negatively stained objects at the ends of k-fibers. Scale bars, 5 μm. (C) Anaphase A rates from each condition. Each symbol (●, ♦) represents the averaged anaphase A rate from a single spindle, and the red and green bars mark the overall average from each condition. Error bars, SEM. Significant difference between the two treatments; *p < 0.001.
Although KLP59D RNAi treatment had no obvious impact on spindle assembly in our live analyses, reexamination of spindle morphologies in fixed cells (allowing for a larger data set) did reveal a statistically significant (about twofold) increase of monopolar spindles and a modest increase in multinucleated cells (Supplemental Table S2). A similar KLP59D RNAi-induced increase in monopolar spindles was also reported in Rogers et al. (2004) but not identified as statistically significant. Bipolar spindles in KLP59D RNAi-treated cells were also slightly but significantly shorter than in controls (9.0 ± 0.36 and 10.4 ± 0.42 μm in KLP59D RNAi- and control-treated cells, respectively; p = 0.014), with a more rounded morphology as indicated by a decrease in the average axial ratio (spindle length-width at metaphase plate; 1.66 ± 0.07 in KLP59D RNAi vs. 2.05 ± 0.067; p = 0.0005). Therefore, although our data suggest that KLP59D has its most prominent impact on anaphase A, its activity influences other phases of mitosis, as well.
KLP59D Promotes Metaphase Flux
KLP59D's localization and impact on spindle morphology and chromosome motility suggests a role in the regulation of kMT dynamics. As mentioned above, a manifestation of kMT dynamics is poleward tubulin Flux which, in the steady-state metaphase spindle, emerges from a balance of MT plus-end polymerization (at the kinetochore) and minus-end depolymerization (at poles). During anaphase, Flux produces poleward pulling forces on chromosomes. To quantify KLP59D's impact on Flux in metaphase spindles, reference lines were photobleached across the half-spindles of control and KLP59D RNAi-treated eGFP-α-tubulin–expressing S2 cells, and the rates at which bleached subunits moved poleward were measured. The Flux rate of spindles in cells lacking KLP59D (0.4 ± 0.07 μm/min) decreases about threefold relative to controls (1.09 ± 0.08 μm/min; p < 0.001; Figure 3, A–C).
Figure 3.
KLP59D regulates metaphase Flux. (A) Time-lapse images from eGFP- α-tubulin–labeled metaphase spindles photobleached within each half-spindle; Pre, prebleach image; Post, postbleach images. Yellow and red arrowheads mark the starting and final positions, respectively, of a bleachmark. Scale bars, 2 μm. (B) Kymographs generated from the spindles shown in A. t and d, the time and distance axes, respectively. (C) Metaphase Flux rates measured from control and KLP59D RNAi-treated cells, plotted as in Figure 2. Significant difference between the two treatments; *p < 0.001.
KLP59D could plausibly induce Flux by 1) contributing to MT minus-end depolymerization at spindle poles, 2) promoting plus-end polymerization near the equator, or 3) generally promoting MT dynamics and turnover throughout the spindle. To assess this in further detail, we utilized fluorescence recovery after photobleaching (FRAP) to measure the relative turnover rate of fluorescent tubulin from MT ends at metaphase spindle poles and equators (Figure 4A). KLP59D RNAi strongly suppressed the turnover of fluorescent tubulin by ∼50% at poles (fluorescence recovery half-time, T1/2, increases about twofold relative to controls; KLP59D RNAi, 42.2 ± 6.8 s; control RNAi, 23.3 ± 2.1 s). In contrast, no significant effect was observed at the spindle equator (Figure 4B, Supplemental Figure S2). Thus, even though KLP59D is positioned at kinetochores and centrosomes during metaphase, it does not globally influence spindle MT dynamics but instead exerts its strongest influence at poles, like KLP10A (Buster et al., 2007).
Figure 4.
KLP59D stimulates depolymerization of MT minus ends. (A) Time-lapse images of a control, eGFP-α-tubulin–labeled metaphase spindle photobleached near the spindle equator and poles. Numbers denote time, in seconds, after bleaching. Dotted lines and arrowheads indicate the bleached regions at the equator and pole. (B) Average T1/2 of fluorescence recovery measured at spindle poles and equators. (C and D) Fluorescence reentry into bleached regions was analyzed with kymographs generated from prominent MT bundles (probably k-fibers; yellow lines, C, top panel). Track angles (C, bottom; red lines) of encroaching fluorescence moving poleward (orange arrows) were used to calculate the rates of movement shown in D. Poleward tubulin flow rate was significantly slower at the minus but not plus ends after KLP59D RNAi. Statistical significance; *p < 0.01. N indicated in each bar; error bars, SEM.
Does KLP59D specifically impact tubulin turnover at poles by stimulating kMT minus-end depolymerization? This cannot be addressed directly by our T1/2 measurements, because recovery is likely influenced by several additional factors, including new MT nucleation and dynamic instability of kMT ends. However, if KLP59D affects tubulin turnover because it regulates Flux, then any changes in tubulin turnover at different regions of an RNAi-treated spindle should be accompanied by similar changes in Flux rate. We reanalyzed our FRAP data to measure fluorescence encroachment into the bleached sections of individual k-fibers. Kymographs were generated from k-fibers spanning the bleached regions in order to measure rates of fluorescence movement into these regions (Figure 4C). We found that a change of tubulin turnover after KLP59D RNAi was mirrored by a change in the rate that poleward-directed fluorescent tubulin extended into the bleached section. At poles of KLP59D RNAi-treated cells, this poleward movement was reduced, and it matched the Flux velocity measured midway along the half-spindle (Figure 4D), consistent with a suppression of minus-end depolymerization.
On the other hand, KLP59D RNAi had a small, statistically insignificant impact on kMT plus ends during metaphase (Figure 4, B and D), suggesting that plus-end polymerization is almost normal. Why this imbalance does not induce aberrant spindle elongation or obvious MT buckling and instead reduces spindle length in treated cells is unclear. However, at least in some systems, spindle MT bundles including kMTs are composed of a tiled array of shorter, continually moving MTs (Mastronarde et al., 1993; Burbank et al., 2006; Yang et al., 2007). Such an organization could conceivably buffer an overall imbalance of kMT polymerization and depolymerization at spindle equators and poles.
KLP59D Stimulates Pacman- and Flux-driven Anaphase A
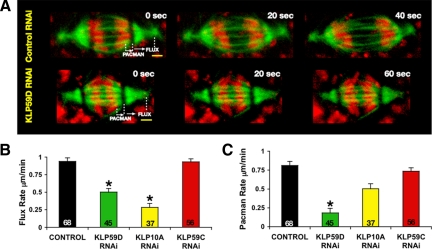
KLP59D's role in promoting Flux provides a potential mechanism for its contribution to anaphase A. To test whether KLP59D 1) suppresses Flux in anaphase spindles and/or 2) also contributes to Pacman-based anaphase A, we measured both Pacman and Flux by photobleaching lines across anaphase spindles containing eGFP-α-tubulin– and Hoechst-labeled chromosomes (Figure 5A). Pacman motility was measured directly as the rate at which disjoined chromatids moved relative to the photobleached lines; anaphase Flux velocity was measured directly as the rate at which bleached lines approached the poles. In controls, both processes occurred simultaneously and at almost equal rates: anaphase Flux and Pacman rates averaged 0.9 ± 0.05 and 0.8 ± 0.05 μm/min, respectively (Figure 5, B and C; Supplemental Movie S3). As expected, KLP59D RNAi significantly slowed anaphase Flux (to 0.5 ± 0. 05 μm/min; p < 0.001; Figure 5, A and B; Supplemental Movie S4).
Figure 5.
KLP59D stimulates Pacman-Flux–based anaphase A. (A) Flux and Pacman rates were measured from anaphase spindles of live RNAi-treated cells. Selected frames from time-lapse recordings are shown. Scale bars, 2 μm. (B and C) Rates of Flux and Pacman measured in control versus kinesin-13 (KLP59D, KLP10A, and KLP59C) RNAi spindles. Treatments significantly different from the control; *p < 0.001. N is indicated in the bars.
KLP59D RNAi also strongly inhibited Pacman-based anaphase motion. Relative to controls, KLP59D RNAi decreased Pacman motility nearly fourfold (to 0.18 ± 0.04 μm/min; p < 0.001; Figure 5, A and C; Supplemental Movie S4). Therefore, KLP59D participates in both mechanisms underlying anaphase A. The combined losses of Flux and Pacman motility after KLP59D RNAi can account entirely for the decreased rate of chromatid-to-pole movement (Supplemental Table S1). In KLP59D-depleted cells, the sum of the residual Flux (0.5 μm/min) and Pacman (0.2 μm/min) motilities matches the observed anaphase A rate (0.75 μm/min). This effect is markedly different from that obtained by depleting either KLP10A, which affects only Flux, or KLP59C, which impacts neither Pacman nor Flux (Figure 5, B and C; Supplemental Table S1).
Centrosomal KLP59D Localizes KLP10A to Poles
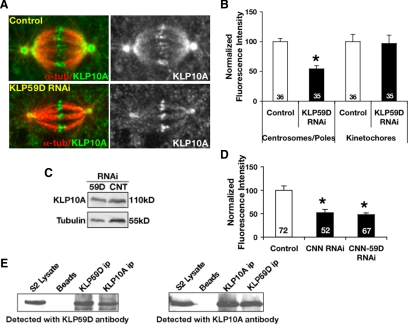
KLP59D's association with centrosomes but not spindle poles is, on its surface, difficult to reconcile with its role in depolymerizing pole focused MT minus ends and stimulation of Flux. As mentioned above, a second Drosophila kinesin-13, KLP10A, also stimulates Flux but strongly accumulates on poles. Further investigation suggested that KLP59D's role in Flux is tightly coupled to KLP10A. In particular, we found that KLP59D RNAi reduced the quantity of KLP10A on centrosomes and spindle poles by roughly 50% (Figure 6, A and B). This occurred without a coincident alteration of KLP10A's localization to other spindle structures (e.g., centromeres/kinetochores) or the total KLP10A titer in cells (Figure 6, B and C). Although such an effect could result from decreased MT density at poles, which in turn could diminish KLP10A localization, we noted 1) no difference in average tubulin immunofluorescence at the poles of KLP59D RNAi-treated cells, 2) that the ratio of tubulin:KLP10A immunofluorescence intensities at poles is nearly doubled by KLP59D RNAi (control RNAi, 1.6 ± 0.1; KLP59D RNAi, 3.8 ± 0.4), and 3) that KLP59D RNAi similarly reduced the levels of centrosome-associated KLP10A when MTs were depolymerized by colchicine (Supplemental Figure S3, A and B). KLP59D's influence on pole-associated KLP10A appears to be mediated by the centrosome because eliminating centrosomes by centrosomin RNAi (Megraw et al., 1999; Mahoney et al., 2006) also reduced KLP10A levels at spindle poles by ∼50% (Figure 6D). Codepletion of centrosomin and KLP59D induced no further reduction in pole associated KLP10A (Figure 6D). KLP10A RNAi did not substantially alter the level of centrosome or kinetochore-associated KLP59D (data not shown).
Figure 6.
KLP59D targets KLP10A to centrosomes/spindle poles. (A) Representative examples of RNAi-treated cells fixed, immunostained with anti-KLP10A (green) and anti-α-tubulin (red), and imaged using identical methods (left). Right panel, only KLP10A labeling. (B) Average KLP10A immunofluorescence intensities at spindle poles/centrosomes and kinetochores. Values are normalized so that control RNAi intensities are 100%. KLP10A levels decrease ∼50% upon KLP59D RNAi; p < 0.001. (C) Western blots of KLP59D RNAi cell lysate demonstrate a slight increase (1.3-fold) in total KLP10A compared with control (CNT). (D) Levels of pole-associated KLP10A in cells treated with centrosomin (CNN) RNAi or co-RNAi with KLP59D. Values are normalized so that control intensities are 100%. (E) Western blots of cross-immunoprecipitations of KLP59D and KLP10A from S2 cell lysates. Beads, protein G beads only; ip, immunoprecipitate.
KLP59D RNAi also reduced the levels at spindle poles of the NuMA-like protein abnormal spindles (Asp; Saunders et al., 1997; Supplemental Figure S4, A and B). A targeting relationship between Asp and KLP10A had been suggested previously (Morales-Mulia and Scholey, 2005). Asp is thought to form an insoluble pole matrix that zips together MT minus ends and maintain the attachment between centrosomes and spindle poles and has been proposed as a mechanism to immobilize pole associated KLP10A (Rogers et al., 2005). Collectively, these data suggest that KLP59D's impact on Flux is largely indirect and mediated through its targeting of pole-associated KLP10A and, perhaps Asp.
Finally, we found that KLP59D and KLP10A coimmunoprecipitate from S2 cell lysates suggesting that they associate in vivo (Figure 6E). An association of this kind could provide a physical basis for the results described above. We note that two MT-destabilizing kinesin-8s from Schizosaccharomyces pombe, Klp5 and Klp6, have been reported to physically associate and function as heterodimers (Garcia et al., 2002; West et al., 2002). The transport motor kinesin II also functions as a heteromeric complex consisting of two different motor polypeptides (Cole et al., 1993). However, we do not believe this to be the case with KLP59D and KLP10A, because they do not colocalize at poles or kinetochores and show only low-affinity direct binding in vitro (data not shown) Moreover, unlike Klp5 and Klp6, KLP10A and KLP59D do not bind solely to one another because immunoprecipitation of transgenic GFP-KLP59D (or GFP-KLP10A) from cell lysates recovers both endogenous KLP59D and KLP10A (Supplemental Figure S4, C and D). As mentioned above, KLP10A and KLP59D have distinct impacts on Pacman, further supporting the contention that they do not function solely as part of the same complex. We found no evidence that mixtures of these proteins displayed any altered MT-destabilizing activity (data not shown).
KLP59D Interacts with the Microtubule Lattice Distinctly from other Drosophila Kinesin-13 Family Members
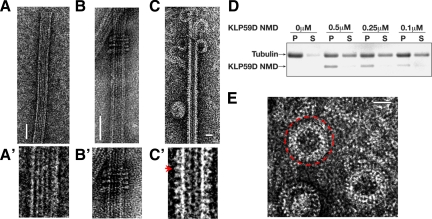
To conclude our studies, we used EM to examine features of KLP59D's structural interaction with MTs. Some kinesin-13 family members, including KLP10A, KLP59C, and the vertebrate MCAK have been shown to oligomerize into rings or spirals around MTs in the presence of the nonhydrolyzable ATP analogue AMP-PNP (Moores et al., 2006; Tan et al., 2006). Whereas the functional relevance, if any, of these structures remains unclear, they may represent a class-specific intermediate of kinesin-13s enzymatic activity. They have also been proposed as a means to couple kinetochore motility to MT depolymerization. However, we found that KLP59D uniformly decorated the MT lattice and formed no or very few rings around MTs (Figure 7, C and C′), even though KLP59D is a bona fide depolymerizer of MTs in vitro (Figure 7D, Supplemental Figure S5). Truncated neck+motor KLP59D constructs formed rings on only 3% of the MTs examined (N = 178), whereas similar KLP10A constructs formed rings in 20% of the MTs (Tan et al., 2006). We also observed many isolated rings (i.e., not associated with MTs; Figure 7E), which are likely depolymerization products consisting of curved protofilaments with bound kinesin-13s (Moores et al., 2002). The increased lattice decoration and decreased formation of MT associated rings makes KLP59D different from the other Drosophila kinesin-13s but similar to the behavior reported for Plasmodium kinesin-13 (Moores et al., 2002, 2006). In context with the data reported above, these findings argue against a requirement for kinesin-13 rings in Pacman-based chromosome motion.
Figure 7.
Electron microscopy analysis of KLP59D. (A) Electron micrographs of negatively stained MTs. (A′) Higher magnification of A. (B) KLP10A NMD forms MT associated rings. (B′) Higher magnification of B. (C) KLP59D neck/motor domain (NMD) decorates the MT lattice and generates protofilament “peels” at MT ends in the presence of AMPPNP. (C′) Higher magnification of B. KLP59D motors visible as the small protrusions from the MT lattice (marked with red arrow). (D) KLP59D disassembles MTs in vitro. Taxol-stabilized MTs were incubated with the indicated concentrations of purified KLP59D NMD in the presence of ATP. After centrifugation, pellet (P) and supernatant (S) samples were analyzed by SDS-PAGE and Coomassie-stained. (E) Enlargement of isolated rings; one ring is marked with a red circle. Scale bars, 50 nm.
DISCUSSION
The results of this study reveal KLP59D to be unique among Drosophila kinesin-13 family members in several regards. It is the only known kinesin-13, which targets to centrosomes but not spindle poles and the only Drosophila kinesin-13 positioned on outer kinetochores. KLP59D is also the first regulator of MT dynamics with a demonstrated role in both Pacman- and Flux-based anaphase A. Finally our data suggest that KLP59D's interaction with MTs is distinct from most other kinesin-13s in that it is incapable of oligomerizing into MT-associated rings.
Collectively, our findings support a functional model wherein KLP59D contributes to Pacman and Flux in S2 cells via very different mechanisms (Figure 8). KLP59D's role in Pacman likely involves its direct depolymerization of kMT plus ends because it 1) is a potent MT depolymerizing protein in vitro, 2) is the only Drosophila family kinesin-13 required for Pacman in this cell type, and 3) targets to outer kinetochores positioning it appropriately to directly interact with kMT plus ends (Maiato et al., 2006). We note that although KLP59D remains associated with kinetochores throughout mitosis, it appears to have little influence on kMT plus-end behaviors outside of anaphase and thus is probably subject to some form of cell cycle regulation. KLP59D's primary sequence contains several predicted cdk1 phosphorylation motifs (unpublished results).
Figure 8.
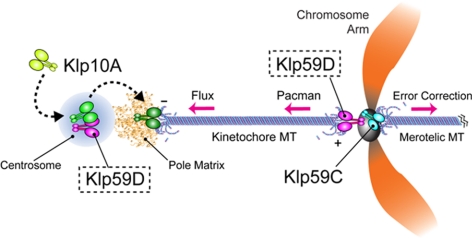
Model: KLP59D stimulates both anaphase Pacman- and Flux-based motility. KLP59D contributes indirectly to poleward Flux by binding and recruiting KLP10A to the spindle poles via the centrosomes. We speculate that KLP10A is modified during its transit through the centrosome, enabling it to function optimally as a MT depolymerizer at the spindle pole. KLP59D directly depolymerizes kinetochore MT plus ends during anaphase, thereby driving Pacman motility. In S2 cells centromeric KLP59C corrects aberrant chromosome attachments.
This raises some confusion because a different kinesin-13, KLP59C, stimulates Pacman in Drosophila early embryos (Rogers et al., 2004). Our favored solution is that KLP59D replaces KLP59C as the Pacman MT depolymerase in S2 cells. It has been proposed that the different requirements of the particularly rapid early embryonic mitoses, in which anaphase A movements are roughly an order of magnitude faster than in S2 cells, present challenges that favor distinct molecular components (Civelekoglu-Scholey et al., 2006). A pertinent example of this is cytoplasmic dynein, which is utilized differently in different cell types and has its most pronounced impact on anaphase A in Drosophila early embryos (Sharp et al., 2000; Howell et al., 2001; Civelekoglu-Scholey et al., 2006). Along these lines, KLP59C's ability to suppress MT rescues (Mennella et al., 2005) may afford it a necessary role in the stimulation of Pacman in early embryos but not S2 cells.
We note that depletion of KLP59C from S2 cells results in a significant increase in the frequency of lagging chromosomes (chromosomes which remain at the spindle equator during anaphase), which is typical of chromosome malorientation and suggests that KLP59C corrects chromosome-MT attachment errors in this cell type (depletion of KLP59D or KLP10A does not impact lagging chromosome frequencies; Supplemental Figure S6). A similar function has been demonstrated for the vertebrate kinesin-13s MCAK and Kif2B (Kline-Smith et al., 2004; Bakhoum et al., 2008).
Intriguingly, unlike several other kinesin-13s examined to date including the two other Drosophila family members, KLP59D does not readily form MT-associated rings in vitro, likely because it lacks several charged amino acids recently demonstrated as essential for the formation of such structures (Tan et al., 2008). Phosphorylation within the motor domain of KLP10A was recently found to inhibit ring assembly, perhaps by altering the position of the domain containing these amino acids relative to the MT (Mennella et al., 2009). Considering this in context with our functional analyses of KLP59D, this seems to indicate that kinesin-13 rings are not required for Pacman-based chromosome movements. Kinetochores contain numerous additional MT-binding proteins that could maintain their association with depolymerizing kMT ends in the absence of any ring-like structures (Maiato et al., 2004; Cheeseman and Desai, 2008). This conclusion is also consistent with the results of recent EM analyses of the kinetochore–MT interface (Dong et al., 2007; McIntosh et al., 2008). Whether kinesin-13 rings are required for Pacman-based motion during the rapid embryonic mitoses (driven by KLP59C) and/or other activities such as the stimulation of Flux (driven by the ring-forming KLP10A) needs to be determined.
In contrast to KLP59D's apparently direct role in Pacman, its contribution to Flux is more complicated. KLP59D stimulates Flux by promoting the depolymerization of MT minus ends at poles. Yet, although KLP59D concentrates on centrosomes, which are generally positioned proximally to poles, it is absent from the pole itself, suggesting that it cannot directly depolymerize pole focused MT ends. Instead, our data suggest that KLP59D stimulates Flux by delivering a second kinesin-13, KLP10A, to poles via centrosomes. We propose that KLP59D recruits cytoplasmic KLP10A (and/or the pole-organizing protein, Asp) to centrosomes, which is subsequently modified, released, and supplied to the pole in an optimized state. Given that centrosomes contain numerous regulatory proteins including kinases and phosphatases (Andersen et al., 2003), phospho-modification of centrosome-associated KLP10A is a reasonable possibility. We note that centrosomes have been proposed to serve as nodes or waystations in protein processing and regulation (Rieder et al., 2001). A physical association KLP59D and KLP10A, suggested by their coimmunoprecipitation, could underlie their targeting relationship. However, any association (direct or indirect) would have to be transient because our preliminary analyses indicate that KLP10A normally turns over very rapidly from centrosomes. Additional biochemical analyses need to be performed to determine whether coimmunoprecipitation of KLP10A and KLP59D is representative of a functionally relevant interaction between these proteins.
We note that there is evidence to suggest that the majority of cytoplasmic kinesin-13s are maintained in an inactive state to prevent the nonspecific, global disassembly of cellular MTs (Mennella et al., 2005, 2009). Activation of KLP10A at centrosomes and subsequent release to the local environment may allow the protein to function specifically in Flux without perturbing other aspects of spindle MT dynamics (note that we are not suggesting that KLP10A is only activated at centrosomes).
Of course, we cannot rule out the possibility that KLP59D RNAi subtly alters the nucleation or dynamics of spindle MTs independently of KLP10A. Indeed, such an effect could explain why depletion of KLP59D causes spindle to shorten, whereas depletion of KLP10A increases spindle length (Buster et al., 2007). Any resulting disruption of spindle architecture could in turn influence the ability of KLP10A and Asp to target appropriately. We could identify no obvious disruption in spindle MT density or organization after KLP59D RNAi. However, such effects could occur below the detection limits of the techniques used.
KLP59D is not alone in impacting on the pole localization of KLP10A and Asp as depletion of the minus-end directed motor, cytoplasmic dynein, also mislocalizes these proteins within S2 cell spindles (Morales-Mulia and Scholey, 2005). Studies in vertebrate cells have also suggested that a complex of dynein and Asp-like protein, NuMA, are required for delivery of kinesin-13s to the poles (Gaetz and Kapoor, 2004). Is KLP59D also involved in this pathway? Close comparison of the impacts of KLP59D versus dynein RNAi reveals differences that strongly argue against such a scenario. In particular dynein depletion causes a redistribution of KLP10A from the poles to the half-spindle as well as the loss of Asp from the transition region between centrosomes and poles. Such effects were never observed in cells depleted of KLP59D. Our belief is that KLP59D's influence on the localization of KLP10A and Asp occurs by an independent and novel mechanism.
In conclusion, our results highlight the complex and cooperative relationships can exist between multiple members of the same kinesin subfamily in a single system. This echoes the general conclusions generated by earlier studies of the three vertebrate kinesin-13 family members, Kif2A, B, and C, which all contribute to mitosis but in somewhat different ways. Of these, Kif2B appears to be most similar to KLP59D because its inhibition induces the formation of monoastral spindles and strongly suppresses chromosome oscillations (Manning et al., 2007). Kif2B also lacks several charged amino acids involved in MT-associated ring formation, similarly to KLP59D (Tan et al., 2008). However, there are also clear differences in the activities of these proteins. In particular a recent study demonstrated that functional perturbation of Kif2B results in chromosome malorientation and lagging chromosomes (Bakhoum et al., 2009), which was not observed in our analyses of KLP59D. The degree to which the specific functional and physical relationships exhibited by Drosophila kinesin-13s (e.g., physical binding and targeting) are conserved remains unknown. Future studies in a variety of systems to compare and contrast the overall spectrum of activities performed by kinesin-13s should be illuminating.
Supplementary Material
ACKNOWLEDGMENTS
We thank A. Asenjo and V. Mennella for help in the MT-depolymerization assays. We also thank R. Vale (UCSF) for eGFP-α-tubulin expressing S2 cells, G. Karpen (Lawrence Berkeley National Laboratory) for Cid antibody, and D. Glover (University of Cambridge, United Kingdom) for Asp antibody. This work is supported by grants from the National Institutes of Health to D.J.S. and in part by Phillip Morris USA. D.J.S. is a scholar of the Leukemia and Lymphoma Society.
Abbreviations used:
- kMT
kinetochore-associated microtubule
- MT
microtubule
- NMD
neck-linker/motor domain.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-07-0557) on September 30, 2009.
REFERENCES
- Andersen J. S., Wilkinson C. J., Mayor T., Mortensen P., Nigg E. A., Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- Bakhoum S. F., Thompson S. L., Manning A. L., Compton D. A. Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat. Cell Biol. 2009;11:27–35. doi: 10.1038/ncb1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbank K. S., Groen A. C., Perlman Z. E., Fisher D. S., Mitchison T. J. A new method reveals microtubule minus ends throughout the meiotic spindle. J. Cell Biol. 2006;175:369–375. doi: 10.1083/jcb.200511112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buster D. W., Zhang D., Sharp D. J. Poleward tubulin flux in spindles: regulation and function in mitotic cells. Mol. Biol. Cell. 2007a;18:3094–3104. doi: 10.1091/mbc.E06-11-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I. M., Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- Civelekoglu-Scholey G., Sharp D. J., Mogilner A., Scholey J. M. Model of chromosome motility in Drosophila embryos: adaptation of a general mechanism for rapid mitosis. Biophys. J. 2006;90:3966–3982. doi: 10.1529/biophysj.105.078691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole D. G., Chinn S. W., Wedaman K. P., Hall K., Vuong T., Scholey J. M. Novel heterotrimeric kinesin-related protein purified from sea urchin eggs. Nature. 1993;366:268–270. doi: 10.1038/366268a0. [DOI] [PubMed] [Google Scholar]
- Dong Y., Vanden Beldt K. J., Meng X., Khodjakov A., McEwen B. F. The outer plate in vertebrate kinetochores is a flexible network with multiple microtubule interactions. Nat. Cell Biol. 2007;9:516–522. doi: 10.1038/ncb1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaetz J., Kapoor T. M. Dynein/dynactin regulate metaphase spindle length by targeting depolymerizing activities to spindle poles. J. Cell Biol. 2004;166:465–471. doi: 10.1083/jcb.200404015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M. A., Koonrugsa N., Toda T. Two kinesin-like Kin I family proteins in fission yeast regulate the establishment of metaphase and the onset of anaphase A. Curr. Biol. 2002;12:610–621. doi: 10.1016/s0960-9822(02)00761-3. [DOI] [PubMed] [Google Scholar]
- Goshima G., Vale R. D. Cell cycle-dependent dynamics and regulation of mitotic kinesins in Drosophila S2 cells. Mol. Biol. Cell. 2005;16:3896–3907. doi: 10.1091/mbc.E05-02-0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell B. J., McEwen B. F., Canman J. C., Hoffman D. B., Farrar E. M., Rieder C. L., Salmon E. D. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J. Cell Biol. 2001;155:1159–1172. doi: 10.1083/jcb.200105093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline-Smith S. L., Khodjakov A., Hergert P., Walczak C. E. Depletion of centromeric MCAK leads to chromosome congression and segregation defects due to improper kinetochore attachments. Mol. Biol. Cell. 2004;15:1146–1159. doi: 10.1091/mbc.E03-08-0581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney N. M., Goshima G., Douglass A. D., Vale R. D. Making microtubules and mitotic spindles in cells without functional centrosomes. Curr. Biol. 2006;16:564–569. doi: 10.1016/j.cub.2006.01.053. [DOI] [PubMed] [Google Scholar]
- Maiato H., DeLuca J., Salmon E. D., Earnshaw W. C. The dynamic kinetochore-microtubule interface. J. Cell Sci. 2004;117:5461–5477. doi: 10.1242/jcs.01536. [DOI] [PubMed] [Google Scholar]
- Maiato H., Hergert P. J., Moutinho-Pereira S., Dong Y., Vandenbeldt K. J., Rieder C. L., McEwen B. F. The ultrastructure of the kinetochore and kinetochore fiber in Drosophila somatic cells. Chromosoma. 2006;115:469–480. doi: 10.1007/s00412-006-0076-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maney T., Ginkel L. M., Hunter A. W., Wordeman L. The kinetochore of higher eucaryotes: a molecular view. Int. Rev. Cytol. 2000;194:67–131. doi: 10.1016/s0074-7696(08)62395-5. [DOI] [PubMed] [Google Scholar]
- Manning A. L., Ganem N. J., Bakhoum S. F., Wagenbach M., Wordeman L., Compton D. A. The kinesin-13 proteins Kif2a, Kif2b, and Kif2c/MCAK have distinct roles during mitosis in human cells. Mol. Biol. Cell. 2007;18:2970–2979. doi: 10.1091/mbc.E07-02-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde D. N., McDonald K. L., Ding R., McIntosh J. R. Interpolar spindle microtubules in PTK cells. J. Cell Biol. 1993;123:1475–1489. doi: 10.1083/jcb.123.6.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh J. R., Grishchuk E. L., Morphew M. K., Efremov A. K., Zhudenkov K., Volkov V. A., Cheeseman I. M., Desai A., Mastronarde D. N., Ataullakhanov F. I. Fibrils connect microtubule tips with kinetochores: a mechanism to couple tubulin dynamics to chromosome motion. Cell. 2008;135:322–333. doi: 10.1016/j.cell.2008.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw T. L., Li K., Kao L. R., Kaufman T. C. The centrosomin protein is required for centrosome assembly and function during cleavage in Drosophila. Development. 1999;126:2829–2839. doi: 10.1242/dev.126.13.2829. [DOI] [PubMed] [Google Scholar]
- Mennella V., Rogers G. C., Rogers S. L., Buster D. W., Vale R. D., Sharp D. J. Functionally distinct kinesin-13 family members cooperate to regulate microtubule dynamics during interphase. Nat. Cell Biol. 2005;7:235–245. doi: 10.1038/ncb1222. [DOI] [PubMed] [Google Scholar]
- Mennella V., Tan D. Y., Buster D. W., Asenjo A. B., Rath U., Ma A., Sosa H. J., Sharp D. J. Motor domain phosphorylation and regulation of the Drosophila kinesin 13, KLP10A. J. Cell Biol. 2009;186:481–490. doi: 10.1083/jcb.200902113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T. J., Salmon E. D. Mitosis: a history of division. Nat. Cell Biol. 2001;3:E17–E21. doi: 10.1038/35050656. [DOI] [PubMed] [Google Scholar]
- Moores C. A., Cooper J., Wagenbach M., Ovechkina Y., Wordeman L., Milligan R. A. The role of the kinesin-13 neck in microtubule depolymerization. Cell Cycle. 2006;5:1812–1815. doi: 10.4161/cc.5.16.3134. [DOI] [PubMed] [Google Scholar]
- Moores C. A., Yu M., Guo J., Beraud C., Sakowicz R., Milligan R. A. A mechanism for microtubule depolymerization by KinI kinesins. Mol. Cell. 2002;9:903–909. doi: 10.1016/s1097-2765(02)00503-8. [DOI] [PubMed] [Google Scholar]
- Morales-Mulia S., Scholey J. M. Spindle pole organization in Drosophila S2 cells by dynein, abnormal spindle protein (Asp), and KLP10A. Mol. Biol. Cell. 2005;16:3176–3186. doi: 10.1091/mbc.E04-12-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath U., Wang D., Ding Y., Xu Y. Z., Qi H., Blacketer M. J., Girton J., Johansen J., Johansen K. M. Chromator, a novel and essential chromodomain protein, interacts indirectly with the putative spindle matrix protein, Skeletor. J. Cell. Biochem. 2004;93:1033–1047. doi: 10.1002/jcb.20243. [DOI] [PubMed] [Google Scholar]
- Rieder C. L., Faruki S., Khodjakov A. The centrosome in vertebrates: more than a microtubule-organizing center. Trends Cell Biol. 2001;11:413–419. doi: 10.1016/s0962-8924(01)02085-2. [DOI] [PubMed] [Google Scholar]
- Rogers G. C., Rogers S. L., Schwimmer T. A., Ems-McClung S. C., Walczak C. E., Vale R. D., Scholey J. M., Sharp D. J. Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature. 2004;427:364–370. doi: 10.1038/nature02256. [DOI] [PubMed] [Google Scholar]
- Rogers G. C., Rogers S. L., Sharp D. J. Spindle microtubules in flux. J. Cell Sci. 2005;118:1105–1116. doi: 10.1242/jcs.02284. [DOI] [PubMed] [Google Scholar]
- Rogers G. C., Rusan N. M., Peifer M., Rogers S. L. A multicomponent assembly pathway contributes to the formation of acentrosomal microtubule arrays in interphase Drosophila cells. Mol. Biol. Cell. 2008;19:3163–3178. doi: 10.1091/mbc.E07-10-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders R. D., Avides M. C., Howard T., Gonzalez C., Glover D. M. The Drosophila gene abnormal spindle encodes a novel microtubule-associated protein that associates with the polar regions of the mitotic spindle. J. Cell Biol. 1997;137:881–890. doi: 10.1083/jcb.137.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp D. J., Rogers G. C., Scholey J. M. Cytoplasmic dynein is required for poleward chromosome movement during mitosis in Drosophila embryos. Nat. Cell Biol. 2000;2:922–930. doi: 10.1038/35046574. [DOI] [PubMed] [Google Scholar]
- Tan D., Asenjo A. B., Mennella V., Sharp D. J., Sosa H. Kinesin-13s form rings around microtubules. J. Cell Biol. 2006;175:25–31. doi: 10.1083/jcb.200605194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan D., Rice W. J., Sosa H. Structure of the kinesin13-microtubule ring complex. Structure. 2008;16:1732–1739. doi: 10.1016/j.str.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West R. R., Malmstrom T., McIntosh J. R. Kinesins klp5(+) and klp6(+) are required for normal chromosome movement in mitosis. J. Cell Sci. 2002;115:931–940. doi: 10.1242/jcs.115.5.931. [DOI] [PubMed] [Google Scholar]
- Wordeman L. Microtubule-depolymerizing kinesins. Curr. Opin. Cell Biol. 2005;17:82–88. doi: 10.1016/j.ceb.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Yang G., Houghtaling B. R., Gaetz J., Liu J. Z., Danuser G., Kapoor T. M. Architectural dynamics of the meiotic spindle revealed by single-fluorophore imaging. Nat. Cell Biol. 2007;9:1233–1242. doi: 10.1038/ncb1643. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.