Abstract
Mitophagy is the process of selective mitochondrial degradation via autophagy, which has an important role in mitochondrial quality control. Very little is known, however, about the molecular mechanism of mitophagy. A genome-wide yeast mutant screen for mitophagy-defective strains identified 32 mutants with a block in mitophagy, in addition to the known autophagy-related (ATG) gene mutants. We further characterized one of these mutants, ylr356wΔ that corresponds to a gene whose function has not been identified. YLR356W is a mitophagy-specific gene that was not required for other types of selective autophagy or macroautophagy. The deletion of YLR356W partially inhibited mitophagy during starvation, whereas there was an almost complete inhibition at post-log phase. Accordingly, we have named this gene ATG33. The new mutants identified in this analysis will provide a useful foundation for researchers interested in the study of mitochondrial homeostasis and quality control.
INTRODUCTION
The mitochondrion is an organelle that carries out a number of important metabolic processes such as fatty acid oxidation, the citric acid cycle, and oxidative phosphorylation. Mitochondrial oxidative phosphorylation supplies a large amount of energy that contributes to a range of cellular activities. However, this organelle is also the major source of cellular reactive oxygen species (ROS) that cause damage to mitochondrial lipid, DNA and proteins, and the accumulation of these types of damage are related to aging, cancer, and neurodegenerative diseases (Wallace, 2005). Thus, intensive analyses of mitochondrial DNA repair and damaged protein degradation mechanisms have been carried out (Larsson and Clayton, 1995; Rep and Grivell, 1996; Bogenhagen, 1999). In addition, it has long been assumed that autophagy is the pathway for mitochondrial recycling, and various theories suggest that a specific targeting of damaged mitochondria to vacuoles or lysosomes occurs by autophagy (Abeliovich and Klionsky, 2001). Very recently, several studies suggest that selective mitochondrial degradation via autophagy (mitophagy) might play an important role for mitochondrial quality control (Priault et al., 2005; Mijaljica et al., 2007; Nowikovsky et al., 2007; Zhang et al., 2007; Twig et al., 2008). However, the molecular mechanism of mitophagy is poorly understood.
Macroautophagy is the bulk (i.e., nonspecific) degradation of cytoplasmic components that allows cells to respond to various types of stress and to adapt to changing nutrient conditions (Klionsky, 2005; Yorimitsu and Klionsky, 2007). In contrast to macroautophagy, the cytoplasm-to-vacuole targeting (Cvt) pathway, pexophagy (specific autophagy of peroxisomes), and mitophagy are categorized as selective types of autophagy. These processes have specific cargos comprised of the Cvt complex (precursor aminopeptidase I (prApe1) and α-mannosidase (Ams1), along with receptor and adaptor proteins), peroxisomes and mitochondria, respectively (Shintani et al., 2002; Dunn et al., 2005; Farre et al., 2008; Kanki and Klionsky, 2008). Studies in the yeast Saccharomyces cerevisiae and other fungi have enabled the identification of several molecular factors essential for autophagy (Yorimitsu and Klionsky, 2005). At present, there are 32 genes that are primarily involved in autophagy-related (Atg) pathways. Most of the ATG genes are required for both macroautophagy and selective autophagy, but some are required only for specific types of autophagy (Kanki and Klionsky, 2008). For example, Atg19, a receptor protein for the Cvt pathway, binds the Cvt complex, and then interacts with Atg11, an adaptor protein for selective autophagy, and recruits them to the phagophore assembly site (PAS), where the sequestering cytosolic vesicles are generated (Shintani et al., 2002). Similarly, during pexophagy in Pichia pastoris, Atg30 localizes to peroxisomes, where it is bound by Atg11, allowing recruitment of the peroxisomes to the PAS (Farre et al., 2008). Atg11 is also required for mitochondrial degradation during starvation or in post-log phase, suggesting that mitochondria are selected by Atg11 for autophagic degradation (Kanki and Klionsky, 2008). Recently, we identified Atg32 as a mitochondrial protein that interacts with Atg11 and is required specifically for mitophagy (Kanki et al., 2009; Okamoto et al., 2009); however, the detailed mechanism of mitophagy has not been determined.
To figure out the molecular mechanism of selective mitochondria autophagy, we recently established a method to monitor this process (Kanki and Klionsky, 2008). Using this method, we screened a yeast knockout library for strains that are deficient in mitophagy. Among 4667 strains, we found 32 strains that showed a complete or partial block of mitophagy, in addition to the ATG gene knockout strains. We also screened these mutants to ascertain the functionality of macroautophagy and the Cvt pathway. Nine of the strains showed defects in all autophagic pathways, whereas the other 23 strains were normal for the Cvt pathway, but defective to varying extents for macroautophagy and mitophagy. We further characterized the product of one of the genes, YLR356W, whose function has not been previously identified. The Ylr356w protein localized to mitochondria, and the deletion of YLR356W resulted in an almost complete inhibition of mitophagy at post-log phase.
MATERIALS AND METHODS
Strains and Media
The yeast strains used in this study are listed in Supplemental Table S1. Yeast cells were grown in rich medium (YPD; 1% yeast extract, 2% peptone, 2% glucose), lactate medium (YPL; 1% yeast extract, 2% peptone, 2% lactate), synthetic minimal medium with glucose (SMD; 0.67% yeast nitrogen base, 2% glucose, amino acids, and vitamins), synthetic minimal medium with lactate (SML; 0.67% yeast nitrogen base, 2% lactate, amino acids, and vitamins), synthetic minimal medium with oleic acid (YTO; 0.67% yeast nitrogen base without amino acids, 0.1% Tween-40, and 0.1% oleic acid), or synthetic minimal medium with galactose (SMGal; 0.67% yeast nitrogen base, 2% galactose, amino acids, and vitamins). Starvation experiments were performed in synthetic minimal medium lacking nitrogen (SD-N: 0.17% yeast nitrogen base without amino acids, 2% glucose; SL-N: 0.17% yeast nitrogen base without amino acids, 2% lactate).
Mitophagy Screening
For the first round of screening, a yeast knockout strain library (BY4739 or BY4742 background) was analyzed. To express Om45-GFP, a DNA fragment encoding green fluorescent protein (GFP) was integrated at the 3′ end of OM45 by a PCR-based integration method (Longtine et al., 1998). Cells grown on SMD plates were shifted to YPL medium and cultured for 3 d (50 ± 5 h), and the vacuolar GFP fluorescence was observed by fluorescence microscopy. If there was no vacuolar GFP signal, or a weak signal, the mitochondrial GFP signal and the cell growth were also recorded.
For the secondary screening, the Om45-GFP-expressing strains that showed a weak or absent vacuolar GFP signal were cultured in YPL medium for 12 h and then shifted to SD-N medium. The cells were collected after 6 h, and the cell lysates equivalent to A600 = 0.2 U of cells were subjected to immunoblotting analysis.
Plasmids and Antibodies
The mitoPho8Δ60 expressing plasmid [ADH1-COXIV-PHO8Δ60(406)] was derived from ADH1-COXIV-PHO8Δ60(313) described previously (Campbell and Thorsness, 1998) by digesting with PvuI and inserting the ADH1-COXIV-PHO8Δ60 fragment into the pRS406 vector.
Monoclonal anti-YFP antibody clone JL-8 (Clontech, Mountain View, CA) and anti-Ape1 antiserum (Shintani et al., 2002) were used for immunoblotting. Monoclonal anti-myc and anti-porin antibodies were from Molecular Probes/Invitrogen (Eugene, OR), and anti-Pgk1 antiserum was a kind gift of Dr. Jeremy Thorner (University of California, Berkeley).
Assays for Autophagy and Pexophagy
For monitoring bulk autophagy, the alkaline phosphatase activity of Pho8Δ60 was carried out as described previously (Noda et al., 1995). The Pex14-GFP processing assay to monitor pexophagy has been described previously (Reggiori et al., 2005).
MitoPho8Δ60 Assay
For monitoring mitophagy, the mitochondrially-targeted Pho8Δ60-expressing strains were cultured in YPL medium for 12 h and then shifted to SD-N or SL-N medium. The cells were collected after 4 h, and the alkaline phosphatase activity of Pho8Δ60 was carried out as described previously (Noda et al., 1995).
Cell Fractionation and Submitochondrial Localization
Cells expressing Tim23-myc and Ylr356w tagged with protein A (PA) were converted to spheroplasts with Zymolyase (Zymo Research, Orange, CA), suspended in homogenization buffer (0.6 M mannitol, 20 mM HEPES, pH 7.4, and proteinase inhibitors) and homogenized in a Potter homogenizer on ice. The cell homogenate was centrifuged at 600 × g for 10 min at 4°C to remove the nucleus and unbroken cells. The supernatant fraction was then centrifuged at 6500 × g for 10 min at 4°C. The pellet was collected as the mitochondrial fraction. Isolated mitochondria was suspended in ice-cold suspension medium (0.6 M mannitol, 20 mM HEPES, pH 7.4) or hypotonic buffer (10 mM Tris-HCl, pH 7.4, and 1 mM EDTA) and treated with proteinase K (200 μg/ml) for 30 min on ice with or without 0.5% Triton X-100. The proteinase K reaction was stopped by adding 10% trichloroacetic acid (TCA). TCA precipitated proteins were washed with acetone and subjected to immunoblotting.
Fluorescence Microscopy
Yeast cells expressing fluorescent protein–fused chimeras were grown to midlog phase or starved in the indicated media. To label the vacuolar membrane or mitochondria, cells were incubated in medium containing 20 μg/ml N-(3-triethylammoniumpropyl)-4-(p-diethylaminophenylhexatrienyl) pyridinium dibromide (FM 4-64; Molecular Probes, Eugene, OR) or 1 μM MitoFluor Red 589 (Molecular Probes) at 30°C for 30 min, respectively. After being washed with medium, the cells were incubated in medium at 30°C for 30–60 min. Fluorescence microscopy observation was carried out as described previously (Monastyrska et al., 2008).
Electron Microscopy
The pep4Δ strain (TKY28) was cultured in YPL medium to midlog phase and then shifted to SD-N and cultured for 6 h or was cultured in YPL medium to stationary phase (for 50 h). Cells were frozen in a KF80-freezing device (Reichert-Jung, Vienna, Austria). Transmission electron microscopy was performed according to the procedures described previously (Baba, 2008).
RESULTS
A Genome-Wide Screen for Yeast Mitophagy Mutants
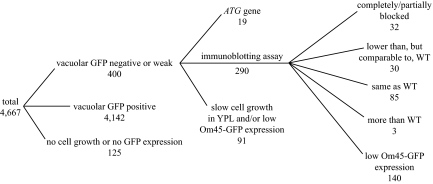
Mitophagy can be induced by culturing yeast strains in a medium with a nonfermentable carbon source such as lactate (i.e., YPL; Tal et al., 2007; Kanki and Klionsky, 2008) or ethanol and glycerol to post-log phase. The GFP-tagged on the C terminus of the mitochondrial outer membrane protein Om45 (Om45-GFP) accumulates in the vacuole, when mitophagy is induced (Kanki and Klionsky, 2008). To identify mitochondrial autophagy-related genes, we used a MATα yeast knockout library and chromosomally tagged the C terminus of Om45 with GFP in each strain. After the strains were cultured in YPL medium for 3 d to allow growth to the post-log phase, they were observed for vacuolar GFP fluorescence. Among 4667 strains examined (Figure 1), 4142 strains showed a clear level of vacuolar GFP, and 400 strains showed either no, or a very weak, vacuolar GFP signal (some examples are shown in Supplemental Figure S1A). We could not examine 125 strains because of their displaying a growth defect in YPD medium (some of the library strains grew poorly even in YPD) or because of a difficulty in the Om45-GFP tagging.
Figure 1.
Schematic diagram of the mitophagy screen and the resulting number of mutants.
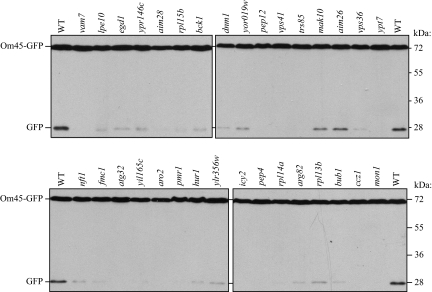
The vacuolar GFP-negative or weak strains included 19 autophagy-related (ATG) gene knockout strains. We screened all ATG gene knockout strains for mitophagy separately (see below); these strains were examined apart from the other mutants uncovered in the present screen. In addition to post-logarithmic-phase growth in lactate medium, mitophagy can be induced when cells are shifted from YPL to nitrogen starvation medium (SD-N), and the level of mitophagy can be semiquantitatively monitored by measuring the amount of GFP processed from Om45-GFP in the vacuole using immunoblotting (Kanki and Klionsky, 2008). For this GFP processing analysis, we required a certain volume of cells and an adequate level of Om45-GFP expression; we excluded 91 strains that showed very slow growth in YPL or very low Om45-GFP expression based on fluorescence microscopy. Among the remaining 290 strains that we screened by GFP processing, 32 strains showed a complete or partial block of mitophagy (Figure 2), 30 strains showed lower, but substantial, GFP processing compared with the wild-type strain, 85 strains showed the same level of GFP processing as the wild type, and 140 strains showed lower Om45-GFP expression; in these latter strains we could not determine from this analysis whether the lower amount of GFP processing was a result of a block in mitophagy or was due to a low level of Om45-GFP (an example is shown in Supplemental Figure S1B). Three strains, with deletions of VMA13, TFP1 (VMA1), or YOR331C (which overlaps with VMA4) showed more than a twofold increase in GFP processing compared with the wild type (our unpublished results). The results for all 4667 strains are listed in Supplemental Table S2 and summarized in Figure 1.
Figure 2.
Screen for defects in mitophagy. Wild-type (WT; BY4742) and the indicated mutant strains expressing Om45-GFP (top and bottom) were cultured in YPL medium for 12 h and then starved in SD-N for 6 h. The cell lysates equivalent to A600 = 0.2 U of cells were subjected to immunoblot analysis with anti-YFP antibody. The position of full-length Om45-GFP and free GFP are indicated.
Analysis for Defects in Macroautophagy and the Cvt Pathway
To further characterize the 32 newly identified mutant strains that showed a clear defect in mitochondrial degradation, we decided to monitor the nonspecific macroautophagy activity using the Pho8Δ60 alkaline phosphatase assay (Noda et al., 1995). Pho8Δ60 is a truncated form of the vacuolar alkaline phosphatase. Deletion of the native signal sequence causes the precursor protein to remain in the cytosol, and it is only delivered to the vacuole by an autophagic mechanism. On delivery, the C-terminal propeptide is removed, resulting in activation of the zymogen, which can be measured enzymatically. We introduced Pho8Δ60 into these 32 knockout strains and measured the Pho8Δ60-dependent alkaline phosphatase activity in both growing and 4-h starvation conditions (Supplemental Figure S2A). For the initial analysis, to simplify the strain construction, we did not delete the PHO13 gene, which encodes a cytosolic alkaline phosphatase. This resulted in a higher level of background activity during growing conditions; however, after 4-h starvation the alkaline phosphatase activity was significantly increased in wild-type cells relative to growing conditions, so there was an adequate signal-to-noise ratio. We found eight strains that showed a complete block of nonspecific autophagy and four strains that showed a partial block. We also screened these mutants for defects in the biosynthetic Cvt pathway, a selective type of autophagy used for delivery of the resident hydrolase Ape1 to the vacuole (Klionsky and Emr, 2000), by monitoring the processing of prApe1 (Supplemental Figure S2B). Eight strains showed a complete block of the Cvt pathway and one strain showed a partial block. We summarized the results for macroautophagy, the Cvt pathway and mitophagy in Supplemental Table S3.
Analysis of 23 Novel Mutants
Among these 32 mitophagy-related genes identified from the screen, nine of them are related with vacuolar protein sorting, membrane fusion machinery, or normal vacuolar function. As expected based on published data, deletion of these genes resulted in a partial or complete block in all autophagy-related pathways, and we did not pursue a further analysis of the associated gene products. The remaining 23 gene products are involved in diverse cellular processes. In particular, Atg32 is a mitochondrially-localized receptor required for starvation-dependent and post-log phase growth mitophagy (Kanki et al., 2009; Okamoto et al., 2009). In addition, eight of these 23 genes encode mitochondrially-related proteins. Accordingly, we decided to extend our analysis of these 23 mutants.
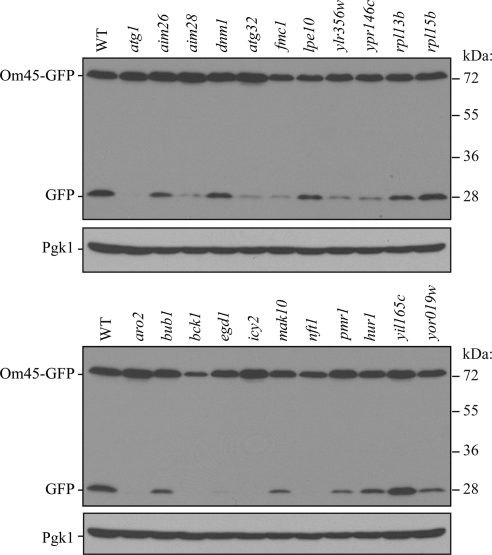
First, to verify the screen results, we decided to delete these 23 genes in another yeast strain background to eliminate potential strain-dependent phenotypes and to verify that the mitophagy defect was due to the correct gene deletion. We used the SEY6210 strain that had the integrated Om45-GFP tag, and conducted the same processing assay (Figure 3). Two mutants, rpl15bΔ and yil165cΔ, showed comparable GFP processing with the wild type, whereas other mutants showed partial or no GFP processing, consistent with the previous screen result. In two cases, for rpl14a and arg82, we were unable to generate the appropriate strains.
Figure 3.
Om45-GFP processing analysis of novel mutants. Wild-type (WT; TKYM22) and the indicated mutant strains expressing Om45-GFP (top and bottom) were cultured in YPL medium for 12 h and then starved in SD-N for 6 h. The cell lysates equivalent to A600 = 0.2 U of cells were subjected to immunoblot analysis with anti-YFP or anti-Pgk1 (loading control) antibodies or antiserum, respectively.
To provide a second method of mitophagy analysis, we also integrated GFP at the 3′ end of the IDH1 locus of the corresponding deletion strains and carried out an Idh1-GFP processing assay; Idh1 is a mitochondrial matrix protein, and its delivery to the vacuole should mirror that of Om45-GFP (Supplemental Figure S3). We found that the rpl15bΔ and aim28Δ mutant strains showed normal GFP-processing compared with the wild type, whereas the other mutant strains displayed reduced or no GFP-processing. In two cases, for ypr146cΔ and lpe10Δ, we were unable to generate the appropriate strains.
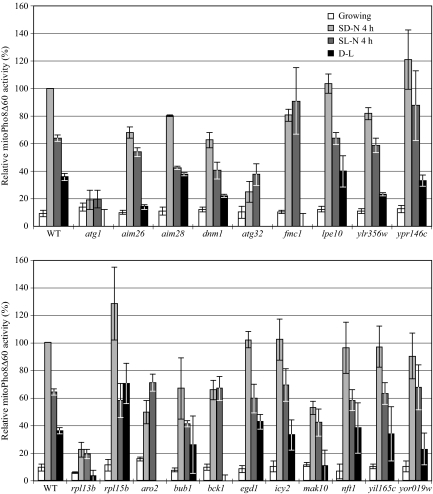
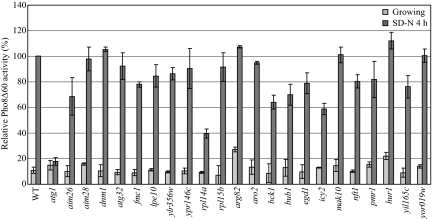
In some cases, we noted discrepancies between the results for the Om45-GFP and Idh1-GFP processing assays. In addition, neither of these assays is quantitative. Therefore, we modified a previously described alkaline phosphatase assay that uses a mitochondrially-targeted Pho8Δ60 (mitoPho8Δ60) construct (Campbell and Thorsness, 1998) and assayed the deletion mutants for mitophagy activity. The mitoPho8Δ60 can only be delivered into the vacuole after the autophagic degradation of mitochondria. Cells expressing mitoPho8Δ60 were cultured in YPL to midlog phase and shifted to SD-N or SL-N for 4 h, and mitoPho8Δ60-dependent alkaline phosphatase activity was measured. The YPL to SD-N shift will induce selective autophagic mitochondria degradation as well as nonselective autophagy, whereas the YPL to SL-N shift will only induce bulk autophagy (Kanki and Klionsky, 2008); thus, the SD-N minus SL-N value represents the activity of selective autophagic mitochondrial degradation. For the wild-type strain, we observed 36% higher alkaline phosphatase activity during the SD-N shift than was seen after the shift to SL-N (Figure 4). In the atg1Δ strain, the alkaline phosphatase activities during both starvation conditions represent the background level. In the atg32Δ strain, we observed a significant decrease (75%) of mitoPho8Δ60 activity during the SD-N shift compared with the wild type, which showed that Atg32 is required for efficient selective autophagic mitochondria degradation. We also observed a 63% decrease of alkaline phosphatase activity during the SL-N shift compared with the wild type, possibly because of the absence of Atg32-dependent mitochondrial degradation that occurs through nonspecific autophagy (which is likely to still require Atg32 to be efficient). We were unable to generate the mitoPho8Δ60 strains for deletions of ARG82, RPL14A, PMR1, and HUR1. For the remaining 19 mutants, eight strains (icy2Δ, rpl15bΔ, nft1Δ, yil165cΔ, lpe10Δ, egd1Δ, aim28Δ, and ypr146cΔ) showed mitophagy activity comparable to that of the wild type, whereas the 11 other strains displayed a significant to complete mitophagy defect (Figure 4). The potential reasons for the differences between the Om45-GFP processing assay results and those of the mitoPho8Δ60 assay are considered in the Discussion.
Figure 4.
MitoPho8Δ60 analysis of novel mutants. Wild-type (WT; KWY20) and the indicated mutant strains (top and bottom) expressing mitoPho8Δ60 were grown in YPL and shifted to SD-N and SL-N for 4 h. Samples were collected, and protein extracts were assayed for mitoPho8Δ60 activity. The value for the wild-type strain was set to 100%, and the other values were normalized. Values lower than zero are depicted as zero.
Next, to examine potential effects on nonspecific macroautophagy, we used the GFP-Atg8 processing assay. This assay relies on the same principle as the Om45-GFP processing assay, but the marker protein is a component of the autophagosome; substantial generation of free GFP from GFP-Atg8 is only seen during nonspecific autophagy. All 23 mutant strains essentially showed normal GFP-Atg8 processing (Supplemental Figure S4), demonstrating they do not have substantial defects in nonspecific macroautophagy. To extend the analysis, we generated pho8::pho8Δ60 pho13Δ strains in the SEY6210 background for each mutant and monitored them using the more quantitative Pho8Δ60 activity assay as a second method for analyzing potential macroautophagy defects (Figure 5); we were unable to generate the Pho8Δ60 strain for rpl13bΔ. Two strains (rpl14aΔ and icy2Δ) displayed a more than 40% decrease of Pho8Δ60 activity, seven strains (aim26Δ, fmc1Δ, bck1Δ, bub1Δ, egd1Δ, nft1Δ, and yil165cΔ) showed a slightly reduced Pho8Δ60 activity (<30% decrease), and the other strains were essentially normal.
Figure 5.
Wild-type (WT; WLY176) and the indicated mutant strains expressing Pho8Δ60 were grown in YPD and shifted to SD-N for 4 h. Samples were collected, and protein extracts were assayed for Pho8Δ60 activity. The value for the wild-type strain was set to 100%, and the other values were normalized.
Finally, we determined the subcellular localization of these proteins by chromosomally tagging them with GFP and observed them with fluorescence microscopy (Supplemental Figure S5). Atg32 has already been reported as being localized to the mitochondria. For the other 22 proteins, eight of them (Aim26, Aim28, Dnm1, Fmc1, Lpe10, Ypr146c, and Ylr356w) displayed a mitochondrial localization pattern. The localization of Ylr356w was further studied as described below. The subcellular localization of most proteins under nitrogen starvation condition was basically the same as during vegetative growth (data not show). The localization information is summarized in Supplemental Table S3.
Characterization of Ylr356w
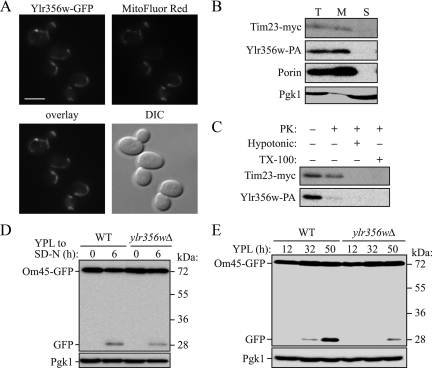
The mitophagy-related genes that we found from the screen include eight genes of unknown function. Among them, we initially focused on YLR356W. The Ylr356w protein is reported to localize to mitochondria (Huh et al., 2003), and we obtained consistent data between our different detection methods for both mitophagy and autophagy. Thus, we decided to characterize this gene as a candidate for a novel ATG gene. In agreement with the previous report, we found that Ylr356w tagged with GFP is mitochondrially-localized (Supplemental Figure S5, Figure 6A); however, the overexpressed chimera is not fully functional. We detected a similar mitochondrial localization using a chromosomally tagged GFP construct, but the fluorescence signal was extremely weak (our unpublished data). Accordingly, we further examined the mitochondrial localization of Ylr356w using a biochemical approach. A strain expressing protein A–tagged Ylr356w (Ylr356w-PA) and Myc-tagged Tim23 (inner membrane marker) was fractionated by differential centrifugation. Mitochondrial porin and Tim23-myc were enriched in the mitochondrial (6500 × g) fraction, along with Ylr356w-PA (Figure 6B), whereas the cytosolic marker Pgk1 was mostly in the supernatant fraction. Next, the isolated mitochondria were treated with proteinase K before or after hypotonic treatment or in the presence of Triton X-100. Although Tim23-myc was protected from proteinase K before hypotonic treatment or in the absence of detergent, Ylr356w-PA was degraded, suggesting that this protein localizes on the mitochondrial outer membrane (Figure 6C). As our data showed, macroautophagy and the Cvt pathway were essentially normal in ylr356wΔ strains in both the BY4742 and SEY6210 backgrounds (Figure 5, Supplemental Figures S2, S4, and S6A). There was a substantial decrease of Om45-GFP processing for starvation-induced mitophagy (Figure 3), and the mitoPho8Δ60 assay revealed a 36% decrease of mitophagy activity (Figure 4) in the ylr356wΔ strain. We also monitored pexophagy, another type of selective autophagy, using the Pex14-GFP processing assay. Pex14 is a peroxisomal membrane protein, and processing of Pex14-GFP to release free GFP can be used for monitoring peroxisome degradation in the vacuole (Reggiori et al., 2005). The ylr356wΔ strain displayed processing of Pex14-GFP at a level similar to that of the wild-type strain after shifting cells from oleic acid medium to starvation medium, whereas the atg1Δ mutant showed a complete block (Supplemental Figure S6B). From these findings, we conclude that YLR356W is a mitophagy-specific gene that is not required for macroautophagy or other types of selective autophagy.
Figure 6.
Characterization of Ylr356w. (A) A strain expressing Ylr356w-GFP under the control of the GAL1 promoter (TKYM201) was cultured in YPD medium to midlog phase and shifted to YPGal medium for 4 h. Cells were labeled with the mitochondrial marker MitoFluor Red 589. The localization of GFP and MitoFluor Red were visualized by fluorescence microscopy. DIC, differential interference contrast. Bar, 5 μm. (B) Mitochondria were purified from a strain expressing chromosomally tagged Tim23-myc and Ylr356w-PA as described in Materials and Methods. Equal amounts of the total cell homogenate (T), mitochondrial (M), and supernatant (S) fractions were loaded and detected with antibodies to myc and porin, a purified antibody that recognizes PA, or with antiserum to Pgk1. (C) Isolated mitochondria were treated with proteinase K (PK) with or without hypotonic or Triton X-100 treatment. Samples were TCA-precipitated and subjected to immunoblotting using the appropriate antibodies. (D) Wild-type (WT; TKYM22) and ylr356wΔ strains expressing Om45-GFP were cultured in YPL medium for 12 h and then starved in SD-N for 6 h. The cell lysates equivalent to A600 = 0.2 U of cells were subjected to immunoblot analysis with anti-YFP and anti-Pgk1 (loading control) antibody or antiserum, respectively. (E) Wild-type (WT; TKYM22) and ylr356wΔ strains expressing Om45-GFP were cultured in YPL medium for the indicated times. Cell lysates equivalent to A600 = 0.2 U of cells were subjected to immunoblot analysis with anti-YFP antibodies and anti-Pgk1 antiserum.
Considering that we observed relatively minor vacuolar GFP fluorescence from the ylr356wΔ (BY4742 background) strain expressing Om45-GFP at the post-log phase (day 3 in YPL medium) in our initial screen, although we obtained a relatively slight decrease (36% based on the mitoPho8Δ60 assay [Figure 4] and 33% based on the Om45-GFP processing assay [Figure 6D]) of mitophagy activity during starvation, we considered the possibility that Ylr356w might play different roles in these two mitophagy-inducing conditions. Thus, we repeated the analysis of Om45-GFP fluorescence of the ylr356wΔ strain in the SEY6210 background. After 45 h in YPL medium, the ylr356wΔ strain again showed only faint vacuolar GFP fluorescence (Supplemental Figure S6C), and Om45-GFP processing was mostly blocked (93 and 80% decrease compared with the wild type at 32 and 50 h in YPL, respectively; Figure 6E). One possibility to explain the mitophagy defect in the ylr356wΔ strain in the post-log phase was that the severe block in Om45-GFP processing was due to a growth defect in YPL medium that prevented the cells from reaching the mitophagy-inducing post-log phase. Accordingly, we checked the growth of the ylr356wΔ strain. This strain showed the same growth both on YPD and YPL plates and was similar to the wild-type strain (Supplemental Figure S6D). Thus, we concluded that Ylr356w was required primarily for mitophagy induced at the post-log phase and played a less significant role during starvation-induced mitophagy. On the basis of the cumulative analyses of the phenotypes of the ylr356wΔ strain, we named this gene ATG33.
Screen of ATG Knockout Strains for Mitophagy Defects
Several ATG genes are required for mitophagy (Kissova et al., 2004, 2007; Tal et al., 2007; Zhang et al., 2007; Kanki and Klionsky, 2008), and our first mitophagy screen revealed 18 ATG genes that may be required for this process (one of the additional strains with a mitophagy defect is deleted for YMR158W-A, a gene that overlaps with ATG16, thus implicating 19 ATG genes in total). Thus, we decided to screen all 28 ATG knockout strains that play a role in autophagy-related processes in S. cerevisiae using the Om45-GFP processing analysis. ATG genes that play a fundamental role in autophagy such as that encoding the protein kinase Atg1 and its binding partner Atg13, the genes for the ubiquitin-like protein modification systems (ATG3, 4, 5, 7, 8, 10, 12, and 16) and those that encode components that are involved in supplying membrane to the phagophore (ATG2 and ATG9) were essential for mitophagy (Supplemental Figure S7). Genes that are required for the Cvt pathway but not macroautophagy (ATG20, 21 and 24), or for macroautophagy but not the Cvt pathway (ATG17, 29, and 31) were also required for efficient mitophagy. Finally, the gene encoding ATG11, which is a common adaptor for selective types of autophagy was required for mitophagy as shown previously (Kanki and Klionsky, 2008), whereas the genes for the Cvt cargo-specific receptor ATG19 (Shintani et al., 2002), a vacuolar permease ATG22 (Yang et al., 2006), and an autophagy gene that is not required in S. cerevisiae ATG26 (Cao and Klionsky, 2007) were not required for mitophagy.
DISCUSSION
A Mitochondria Degradation Screen Identified 32 Mitophagy-Related Genes
The initial screen for mitophagy was carried out in 96-well plates. Thus, the screen was performed in a blind manner as the gene names of each strain were hidden during the screen. Among 25 ATG genes that are required for mitophagy (Supplemental Figure S7), our knockout library included 21 ATG knockout strains (atg2Δ, 4Δ, and 10Δ were not included in our BY4739 or BY4742 background library, and atg14Δ was incorrect). Based on our initial screen, 17 of them (18 if we include the strain where the deletion overlaps with ATG16) were identified as positive for a defect in mitophagy, and three of them were negative (atg17Δ, 24Δ, and 31Δ). If it is assumed that these ATG knockout strains serve as positive controls, our initial screen sensitivity was 81–86% (17 or 18/21). In other words, we may have missed 14–19% of the positive strains during our initial screen; however, considering that atg17Δ and atg31Δ have only partial defects, our detection rate may have been closer to 95%. We cannot calculate the sensitivity of the secondary screen, because the ATG genes were screened by the Om45-GFP–processing assay (the secondary screen for the other mutants). The fact that at least one novel ATG gene, YLR356W/ATG33, was identified from our screen further supports its utility.
Our screen identified eight membrane trafficking-related genes (CCZ1, MON1, PEP12, TRS85, VAM7, VPS36, VPS41, and YPT7) that are required for mitophagy. All of them are also required for both macroautophagy and the Cvt pathway. The requirement of some of these genes for autophagy has been reported previously (Wichmann et al., 1992; Sato et al., 1998; Wurmser et al., 2000; Meiling-Wesse et al., 2002, 2005; Wang et al., 2003; Nazarko et al., 2005), and the requirement of membrane-trafficking pathways for autophagy has also been reported (Ishihara et al., 2001; Reggiori et al., 2004). It is widely believed that defects in membrane-trafficking pathways affect the lipid supply that is needed for extension of the phagophore, the initial sequestering compartment that generates the autophagosome. The two genes identified here that are involved in membrane trafficking that have not been reported previously as affecting autophagy, PEP12 and VPS36, presumably do so for the same reason.
We identified nine mitophagy-related genes whose functions were not previously known (AIM26, AIM28, ATG32, HUR1, ICY2, YIL165C, YLR356W, YOR019W, and YPR146C). HUR1 overlaps with PMR1, which encodes a cation P-type ATPase in the Golgi complex. Thus, the phenotype of the hur1Δ strain may result from a knockout of the PMR1 gene. The further characterization of the other eight gene products may provide substantial insight into the mechanism of mitophagy. In particular, Ylr356w localized to mitochondria and may play an important role in determining whether a particular mitochondrial segment is destined for degradation by autophagy. Thus, we initially focused on the YLR356W gene and the corresponding protein.
DNM1 encodes mitochondrial dynamin-related GTPase that is required for mitochondrial fission. The fragmentation of mitochondria is a prerequisite for mitophagy in mammalian cells (Twig et al., 2008) and the dnm1Δ strain inhibits the mitophagy induced by mdm38 conditional knockout in yeast (Nowikovsky et al., 2007). The identification of the dnm1Δ strain from our screen further confirmed the importance of mitochondrial fission for mitophagy.
Different Methods to Monitor Mitophagy
In this article, we used the Om45-GFP processing and mitoPho8Δ60 assays to measure mitophagy activity. Importantly, with the mitoPho8Δ60 assay, we can measure mitophagy in a quantitative manner. Both assays showed the expected results for the wild-type, atg1Δ, and atg32Δ strains, demonstrating they are adequate for measuring mitophagy. In some cases, however, we obtained different results between these two methods. One potential problem with the mitoPho8Δ60 assay is that the marker protein may not be properly targeted to the mitochondria in each mutant strain, especially in mutants that may have a defect in the mitochondrial protein import system. This would cause an apparent mitophagy defect, and correct localization should be confirmed in each case.
YLR356W/ATG33 Is Required Primarily for Mitophagy Induced at the Post-Log Phase
Although a genome-wide screen for protein localization revealed that Ylr356w localizes to mitochondria (Huh et al., 2003), there have not been any other reports about this protein. Ylr356w is composed of 197 amino acids and is predicted to have four transmembrane domains. This protein is conserved within fungi, but not in higher eukaryotes. YLR356W is a mitophagy-specific gene that is not required for macroautophagy or other types of selective autophagy (Figures 5 and 6, Supplemental Figures S2B, S4, and S6). Although ylr356wΔ blocked mitophagy to half the level of the wild type during starvation, it blocked mitophagy almost completely at the post-log phase (Figure 6, D and E). This finding suggests that Ylr356w may be required to detect or present aged mitochondria for mitophagy when cells have reached the post-log phase.
The Induction of Mitophagy during Starvation and at the Post-Log Phase in Yeast
Although mitochondria depolarized by an uncoupler such as CCCP (carbonyl cyanide m-chlorophenylhydrazone) are degraded by autophagy in mammalian cells (Narendra et al., 2008; Sandoval et al., 2008), we did not observe mitochondrial degradation in wild-type yeast under similar conditions (our unpublished results). Thus, in our experience, nitrogen starvation or culturing cells to the post-log phase in a nonfermentable carbon source medium are the only reliable methods that can induce mitophagy efficiently in a wild-type yeast strain. A previous study suggests that mitophagy in S. cerevisiae occurs by a microautophagic process when cells are grown under nonfermentable conditions (Kissova et al., 2007). Thus, we considered the possibility that mitophagy might happen by different mechanisms depending on the inducing conditions. We attempted to examine the mode of autophagic sequestration occurring in SD-N versus post-log phase growth through electron microscopy (Supplemental Figure S8). In both conditions we could detect mitochondria within double-membrane vesicles, suggesting a macroautophagic mechanism; however, we cannot rule out the possibility of a microautophagic process.
It is thought that mitophagy is induced to adapt the cell to conditions where the cell energy requirement is decreased, and accordingly the cell needs to reduce the amount of mitochondria when reaching the post-log phase in nonfermentable medium (Kanki and Klionsky, 2008), although there is little direct evidence for this hypothesis. On the other hand, macroautophagy is induced at the post-log phase (Wang et al., 2001), presumably because cells are starved at this growing phase. Thus, it has been unclear whether mitophagy is induced at the post-log phase through some specific mechanism or simply as a result of cellular starvation. The specific requirement of Ylr356w for mitophagy primarily at the post-log phase suggests that there are some differences between the pathways of mitophagy induced during starvation versus post-log phase growth. Because only a fraction of the total mitochondrial pool is degraded by mitophagy at the post-log phase (compare full-length Om45-GFP and the processed GFP band in Figure 6E, WT), it is reasonable to propose that aged or damaged mitochondria are selected for degradation. We propose that Ylr356w may contribute to this selection process, although future experiments will be needed to confirm this hypothesis.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health Grant GM53396 (D.J.K.) and Japan Society for the Promotion of Science Postdoctoral Fellowships for Research Abroad (T.K.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-03-0225) on September 30, 2009.
REFERENCES
- Abeliovich H., Klionsky D. J. Autophagy in yeast: mechanistic insights and physiological function. Microbiol. Mol. Biol. Rev. 2001;65:463–479. doi: 10.1128/MMBR.65.3.463-479.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M. Electron microscopy in yeast. Methods Enzymol. 2008;451:133–149. doi: 10.1016/S0076-6879(08)03210-2. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F. Repair of mtDNA in vertebrates. Am. J. Hum. Genet. 1999;64:1276–1281. doi: 10.1086/302392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell C. L., Thorsness P. E. Escape of mitochondrial DNA to the nucleus in yme1 yeast is mediated by vacuolar-dependent turnover of abnormal mitochondrial compartments. J. Cell Sci. 1998;111:2455–2464. doi: 10.1242/jcs.111.16.2455. [DOI] [PubMed] [Google Scholar]
- Cao Y., Klionsky D. J. Atg26 is not involved in autophagy-related pathways in Saccharomyces cerevisiae. Autophagy. 2007;3:17–20. doi: 10.4161/auto.3371. [DOI] [PubMed] [Google Scholar]
- Dunn W. A., Jr, Cregg J. M., Kiel J.A.K.W., van der Klei I. J., Oku M., Sakai Y., Sibirny A. A., Stasyk O. V., Veenhuis M. Pexophagy: the selective autophagy of peroxisomes. Autophagy. 2005;1:75–83. doi: 10.4161/auto.1.2.1737. [DOI] [PubMed] [Google Scholar]
- Farre J. C., Manjithaya R., Mathewson R. D., Subramani S. PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev. Cell. 2008;14:365–376. doi: 10.1016/j.devcel.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Ishihara N., Hamasaki M., Yokota S., Suzuki K., Kamada Y., Kihara A., Yoshimori T., Noda T., Ohsumi Y. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol. Biol. Cell. 2001;12:3690–3702. doi: 10.1091/mbc.12.11.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T., Klionsky D. J. Mitophagy in yeast occurs through a selective mechanism. J. Biol. Chem. 2008;283:32386–32393. doi: 10.1074/jbc.M802403200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T., Wang K., Cao Y., Baba M., Klionsky D. J. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev. Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissova I., Deffieu M., Manon S., Camougrand N. Uth1p is involved in the autophagic degradation of mitochondria. J. Biol. Chem. 2004;279:39068–39074. doi: 10.1074/jbc.M406960200. [DOI] [PubMed] [Google Scholar]
- Kissova I., Salin B., Schaeffer J., Bhatia S., Manon S., Camougrand N. Selective and non-selective autophagic degradation of mitochondria in yeast. Autophagy. 2007;3:329–336. doi: 10.4161/auto.4034. [DOI] [PubMed] [Google Scholar]
- Klionsky D. J. The molecular machinery of autophagy: unanswered questions. J. Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D. J., Emr S. D. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson N. G., Clayton D. A. Molecular genetic aspects of human mitochondrial disorders. Annu. Rev. Genet. 1995;29:151–178. doi: 10.1146/annurev.ge.29.120195.001055. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., III, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Meiling-Wesse K., Barth H., Thumm M. Ccz1p/Aut11p/Cvt16p is essential for autophagy and the cvt pathway. FEBS Lett. 2002;526:71–76. doi: 10.1016/s0014-5793(02)03119-8. [DOI] [PubMed] [Google Scholar]
- Meiling-Wesse K., Epple U. D., Krick R., Barth H., Appelles A., Voss C., Eskelinen E.-L., Thumm M. Trs85 (Gsg1), a component of the TRAPP complexes, is required for the organization of the preautophagosomal structure during selective autophagy via the Cvt pathway. J. Biol. Chem. 2005;280:33669–33678. doi: 10.1074/jbc.M501701200. [DOI] [PubMed] [Google Scholar]
- Mijaljica D., Prescott M., Devenish R. J. Different fates of mitochondria: alternative ways for degradation? Autophagy. 2007;3:4–9. doi: 10.4161/auto.3011. [DOI] [PubMed] [Google Scholar]
- Monastyrska I., He C., Geng J., Hoppe A. D., Li Z., Klionsky D. J. Arp2 links autophagic machinery with the actin cytoskeleton. Mol. Biol. Cell. 2008;19:1962–1975. doi: 10.1091/mbc.E07-09-0892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D., Tanaka A., Suen D. F., Youle R. J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarko T. Y., Huang J., Nicaud J. M., Klionsky D. J., Sibirny A. A. Trs85 is required for macroautophagy, pexophagy and cytoplasm to vacuole targeting in Yarrowia lipolytica and Saccharomyces cerevisiae. Autophagy. 2005;1:37–45. doi: 10.4161/auto.1.1.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T., Matsuura A., Wada Y., Ohsumi Y. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 1995;210:126–132. doi: 10.1006/bbrc.1995.1636. [DOI] [PubMed] [Google Scholar]
- Nowikovsky K., Reipert S., Devenish R. J., Schweyen R. J. Mdm38 protein depletion causes loss of mitochondrial K+/H+ exchange activity, osmotic swelling and mitophagy. Cell Death Differ. 2007;14:1647–1656. doi: 10.1038/sj.cdd.4402167. [DOI] [PubMed] [Google Scholar]
- Okamoto K., Kondo-Okamoto N., Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev. Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- Priault M., Salin B., Schaeffer J., Vallette F. M., di Rago J. P., Martinou J. C. Impairing the bioenergetic status and the biogenesis of mitochondria triggers mitophagy in yeast. Cell Death Differ. 2005;12:1613–1621. doi: 10.1038/sj.cdd.4401697. [DOI] [PubMed] [Google Scholar]
- Reggiori F., Monastyrska I., Shintani T., Klionsky D. J. The actin cytoskeleton is required for selective types of autophagy, but not nonspecific autophagy, in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 2005;16:5843–5856. doi: 10.1091/mbc.E05-07-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Wang C.-W., Nair U., Shintani T., Abeliovich H., Klionsky D. J. Early stages of the secretory pathway, but not endosomes, are required for Cvt vesicle and autophagosome assembly in Saccharomyces cerevisiae. Mol. Biol. Cell. 2004;15:2189–2204. doi: 10.1091/mbc.E03-07-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep M., Grivell L. A. The role of protein degradation in mitochondrial function and biogenesis. Curr. Genet. 1996;30:367–380. doi: 10.1007/s002940050145. [DOI] [PubMed] [Google Scholar]
- Sandoval H., Thiagarajan P., Dasgupta S. K., Schumacher A., Prchal J. T., Chen M., Wang J. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T. K., Darsow T., Emr S. D. Vam7p, a SNAP-25-like molecule, and Vam3p, a syntaxin homolog, function together in yeast vacuolar protein trafficking. Mol. Cell. Biol. 1998;18:5308–5319. doi: 10.1128/mcb.18.9.5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T., Huang W.-P., Stromhaug P. E., Klionsky D. J. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev. Cell. 2002;3:825–837. doi: 10.1016/s1534-5807(02)00373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal R., Winter G., Ecker N., Klionsky D. J., Abeliovich H. Aup1p, a yeast mitochondrial protein phosphatase homolog, is required for efficient stationary phase mitophagy and cell survival. J. Biol. Chem. 2007;282:5617–5624. doi: 10.1074/jbc.M605940200. [DOI] [PubMed] [Google Scholar]
- Twig G., et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace D. C. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.-W., Stromhaug P. E., Kauffman E. J., Weisman L. S., Klionsky D. J. Yeast homotypic vacuole fusion requires the Ccz1-Mon1 complex during the tethering/docking stage. J. Cell Biol. 2003;163:973–985. doi: 10.1083/jcb.200308071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Wilson W. A., Fujino M. A., Roach P. J. Antagonistic controls of autophagy and glycogen accumulation by Snf1p, the yeast homolog of AMP-activated protein kinase, and the cyclin-dependent kinase Pho85p. Mol. Cell. Biol. 2001;21:5742–5752. doi: 10.1128/MCB.21.17.5742-5752.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichmann H., Hengst L., Gallwitz D. Endocytosis in yeast: evidence for the involvement of a small GTP-binding protein (Ypt7p) Cell. 1992;71:1131–1142. doi: 10.1016/s0092-8674(05)80062-5. [DOI] [PubMed] [Google Scholar]
- Wurmser A. E., Sato T. K., Emr S. D. New component of the vacuolar class C-Vps complex couples nucleotide exchange on the Ypt7 GTPase to SNARE-dependent docking and fusion. J. Cell Biol. 2000;151:551–562. doi: 10.1083/jcb.151.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Huang J., Geng J., Nair U., Klionsky D. J. Atg22 recycles amino acids to link the degradative and recycling functions of autophagy. Mol. Biol. Cell. 2006;17:5094–5104. doi: 10.1091/mbc.E06-06-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T., Klionsky D. J. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12:1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T., Klionsky D. J. Eating the endoplasmic reticulum: quality control by autophagy. Trends Cell Biol. 2007;17:279–285. doi: 10.1016/j.tcb.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Qi H., Taylor R., Xu W., Liu L. F., Jin S. The role of autophagy in mitochondria maintenance: characterization of mitochondrial functions in autophagy-deficient S. cerevisiae strains. Autophagy. 2007;3:337–346. doi: 10.4161/auto.4127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.