Abstract
The variant surface glycoprotein (VSG) of bloodstream form Trypanosoma brucei (Tb) is a critical virulence factor. The VSG glycosylphosphatidylinositol (GPI)-anchor strongly influences passage through the early secretory pathway. Using a dominant-negative mutation of TbSar1, we show that endoplasmic reticulum (ER) exit of secretory cargo in trypanosomes is dependent on the coat protein complex II (COPII) machinery. Trypanosomes have two orthologues each of the Sec23 and Sec24 COPII subunits, which form specific heterodimeric pairs: TbSec23.1/TbSec24.2 and TbSec23.2/TbSec24.1. RNA interference silencing of each subunit is lethal but has minimal effects on trafficking of soluble and transmembrane proteins. However, silencing of the TbSec23.2/TbSec24.1 pair selectively impairs ER exit of GPI-anchored cargo. All four subunits colocalize to one or two ER exit sites (ERES), in close alignment with the postnuclear flagellar adherence zone (FAZ), and closely juxtaposed to corresponding Golgi clusters. These ERES are nucleated on the FAZ-associated ER. The Golgi matrix protein Tb Golgi reassembly stacking protein defines a region between the ERES and Golgi, suggesting a possible structural role in the ERES:Golgi junction. Our results confirm a selective mechanism for GPI-anchored cargo loading into COPII vesicles and a remarkable degree of streamlining in the early secretory pathway. This unusual architecture probably maximizes efficiency of VSG transport and fidelity in organellar segregation during cytokinesis.
INTRODUCTION
Trypanosoma brucei spp. are phylogenically ancient parasitic protozoa, responsible for African trypanosomiasis (sleeping sickness) in humans and the veterinary disease Nagana in cattle. Transmitted by the tse-tse fly (Glossina ssp.) vector, T. brucei have a digenetic life cycle alternating between the bloodstream form (BSF) in vertebrate hosts and the procyclic insect form (PCF) and other forms in the fly. As an adaptation to their respective environments, each stage elaborates a unique, densely packed glycosylphosphatidylinositol (GPI)-anchored protein surface coat. In BSF trypanosomes, this is composed of the homodimeric variant surface glycoprotein (VSG), whereas PCF cells express monomeric procyclin (Cross, 1975; Roditi and Clayton, 1999). Approximately 10% of total protein synthesis in BSF cells is devoted to the expression of a single VSG variant (∼107 copies/cell), and switching expression to antigenically distinct VSGs enables the parasite to avoid the host immune response. This process, called antigenic variation, is critical to the survival of the parasite; thus, VSG is the lynchpin to pathogenesis in the immunocompetent mammalian host (Horn and Barry, 2005). Also, VSG was the first protein shown to be GPI anchored, and GPI structure and biosynthesis were first determined in trypanosomes (Ferguson, 1999). Consequently, trypanosomes and VSG have provided a longstanding model system for investigation of GPI function in eukaryotic cells.
VSG is synthesized in the endoplasmic reticulum (ER), where N-linked glycans and the GPI anchor are attached and dimerization occurs (Bangs et al., 1986; Ferguson et al., 1986). It is subsequently transported rapidly through the Golgi, where N-glycans and the GPI anchor are processed, to the flagellar pocket where exocytosis allows incorporation into the surface coat (Bangs et al., 1986, 1988; Duszenko et al., 1988). The GPI anchor strongly influences VSG trafficking in the secretory and endocytic pathways. Soluble GPI-minus VSG is delayed in ER exit relative to native controls and is missorted to the lysosome during post-Golgi transport (Triggs and Bangs, 2003). Conversely, addition of a GPI anchor can rescue biosynthetic cargo destined for lysosomal delivery (Schwartz et al., 2005). Finally, the GPI anchor is critical for sorting and recycling of VSG, which is constantly taken up with cargo endocytosed for nutritional purposes (Grünfelder et al., 2003).
In eukaryotic cells, ER exit is generally mediated by coat protein complex II (COPII)-coated vesicles, which form at discrete ER exit sites (ERES) (Hughes and Stephens, 2008). COPII coat formation is initiated by Sec12, an ER-localized guanine nucleotide exchange factor, which induces a guanosine diphosphate (GDP)-to-guanosine triphosphate (GTP) switch on Sar1, a cytosolic GTPase (Nakano and Muramatsu, 1989; Barlowe and Schekman, 1993; Futai et al., 2004). Activated Sar1:GTP undergoes a conformational change, embedding into the ER membrane and recruiting the Sec23/Sec24 heterodimer via specific interactions with Sec23 (Bi et al., 2002; Fath et al., 2007). This “prebudding complex” is responsible for capturing and incorporating membrane secretory cargo into the forming vesicle (Barlowe et al., 1994; Kuehn et al., 1998). Cargo capture is thought to involve interaction between specific motifs in the cytoplasmic domains of cargo molecules and the Sec24 subunit and to a lesser extent with Sec23 (Miller et al., 2003, 2005; Mossessova et al., 2003; Cai et al., 2007; Farhan et al., 2007; Mancias and Goldberg, 2007). Subsequently, the Sec13/Sec31 heterotetramer is recruited, concentrating the prebudding complexes, and stimulating membrane deformation and subsequent vesicle scission from the ER (Matsuoka et al., 1998).
In Saccharomyces cerevisiae, GPI anchors facilitate loading of cargo into COPII vesicles. This process is influenced by de novo synthesis of ceramide (Horvath et al., 1994; Sutterlin et al., 1997) and is mediated by sphingolipid- and ergosterol-rich lipid rafts (Bagnat et al., 2000). This phenomenon is not found in either trypanosomes or mammalian cells (Sutterwala et al., 2007). ER exit of GPI-anchored cargo in yeast is also dependent on the p24 protein family. These small transmembrane proteins have cytoplasmic domains for interaction with COPII coats, and they associate in combinatorial oligomers, presumably to form lumenal cargo recognition domains (Jenne et al., 2002). Disruption of p24 expression impacts loading of GPI-anchored cargo into COPII vesicles in both yeast and mammals (Jenne et al., 1995; Schimmoler et al., 1995; Belden and Barlowe, 1996; Marzioch et al., 1999; Takida et al., 2008; Castillion et al., 2009). Trypanosomes have at least six orthologues of p24 (Supplemental Table S2), but the role of these proteins, and more broadly, the role of COPII vesicles in ER exit has yet to be studied systematically.
In this work, we investigate the role of the COPII machinery in ER exit in BSF T. brucei with special regard to GPI-anchored cargo. We use conditional expression of a TbSar1 dominant-negative mutant and RNA interference (RNAi) silencing of TbSec23 and TbSec24. Trypanosomes have two distinct orthologues each of TbSec23 and TbSec24, and we biochemically characterize their associations into functional heterodimers. In addition, using TbSec23.2 as an ERES marker we characterize the architecture of the early secretory pathway in relationship to the Golgi and to unique cytoskeletal elements in close association with the flagellum. Our results suggest a selective model for ER exit of GPI-anchored cargo and highlight a unique architecture of the early secretory pathway in these unusual eukaryotes.
MATERIALS AND METHODS
Maintenance of Trypanosomes
The Lister 427 strain of bloodstream form T. brucei brucei (expressing VSG221, herein referred to as BS221) were grown in HMI-9 medium supplemented with 10% fetal bovine serum (FBS) and 10% Serum Plus (SAFC Biosciences, Lenexa, KS) at 37°C in humidified 5% CO2 (Hirumi and Hirumi, 1994). The Lister 427 Strain 13-90 double marker bloodstream cell line (BS-DM) was grown in HMI-9 medium supplemented with 20% Tet system-approved FBS (Clontech, Mountain View, CA; Atlanta Biologicals, Lawrenceville, GA). BS-DM cells constitutively express T7 RNA polymerase and tetracycline repressor under neomycin and hygromycin selection, respectively (Wirtz et al., 1999). BS-DM cells were used as host cell lines for expression of the individual inducible constructs described below.
Construction of Inducible and Epitope-tagged Cell Lines
Genomic DNA from BS221 cells was used as template for all polymerase chain reactions (PCRs). All constructs, fusions, and mutations were confirmed by sequencing before transfection. Transfections of BSF trypanosomes were done by Amaxa nucleofection as described previously (Burkard et al., 2007), and stable clonal transformants were selected with phleomycin (1.6 μg/ml), neomycin (5 μg/ml), hygromycin (10 μg/ml), or puromycin (2 μg/ml) as appropriate.
The TbSar1 open reading frame (Tb05.5K5.150, nt 1-586) was amplified from genomic (g)DNA with an in frame fusion of the T7 epitope tag (MASMTGGQQMG) at the C terminus, immediately before the stop codon. This PCR product was cloned into the tetracycline-inducible pLew100 vector (Wirtz et al., 1999) by using EcoRI/BamHI to form pLew100:Sar1WT. The dominant-negative mutant of TbSar1 (TbSar1DN) was made by converting threonine to asparagine at residue 34 (T34N), using the QuikChange multisite-directed mutagenesis kit (Stratagene, La Jolla, CA) and pLew100:Sar1WT as template. Both the pLew100:Sar1WT and pLew100:Sar1DN constructs were linearized with NotI, electroporated into DM-BS cells, and clonal cell lines selected for with phleomycin. Expression of the exogenous epitope-tagged TbSar1 proteins was induced with 1 μg/ml tetracycline.
For construction of all RNAi plasmids, an ∼1- to 2-kbp section from the start of each open reading frame (ORF) was cloned into the inducible double-stranded RNA (dsRNA) vector p2T7Ti (LaCount et al., 2000) by using appropriate restriction sites. Inserts were TbSec23.1 (Tb927.8.3660, nt 18-2101), TbSec23.2 (Tb10.6k152840, nt 9-1724), TbSec24.1 (Tb927.3.121, nt 17-1093), and TbSec24.2 (Tb927.3.5420, nt 171-2101) (Supplemental Table S1). Constructs were linearized with NotI and TbSec RNAi vectors were introduced separately into DM-BS cells and clonal cell lines selected for with phleomycin. Cloning of the BiPN:GPI reporter into pXS5neo has been described previously (Schwartz et al., 2005). After replacement of neomycin resistance cassette with the puromycin cassette, the construct was linearized with XhoI, stably transfected into the TbSec23.1 and TbSec23.2 RNAi cell lines, and clonal transformants were selected with puromycin.
For localization and pull-down studies, the TbSec23 paralogues were each in situ epitope tagged with a hemagglutinin (HA) tag (YPYDVPDYA), and likewise the TbSec24 paralogues were each fused to a Ty tag (EVHTNQDPLD), by homologous recombination at 3′ end of each ORF. This allows for the analysis of the tagged gene products at expression levels approximating that of the endogenous proteins (Shen et al., 2001). For each construct, ∼500 base pairs upstream of the 3′ end of the ORF were PCR amplified with the tag sequence in frame immediately before the stop codon. These were then cloned upstream of the appropriate antibiotic resistance cassette (TbSec23s, neo; TbSec24s, and hyg) in pXS5 (Triggs and Bangs, 2003) by using KpnI/XmaI or KpnI/EcoRI. Likewise, an ∼500-base pair PCR product from the immediate 3′-untranslated region (UTR) sequence of each gene was placed in the matching vector downstream of the resistance cassette by using PacI/SacI. Each completed construct, containing a 3′ segment of the ORF with an in frame epitope tag at the C terminus, followed by the aldolase intragenic region, the antibiotic resistance cassette and the 3′-UTR, was digested with KpnI/SacI and stably transfected into BS221 cells. In total, four clonal cell lines were developed, each with a different combination of tagged TbSec23 and TbSec24 components: TbSec23.1:HA/TbSec24.1:Ty, TbSec23.2:HA/TbSec24.1:Ty, TbSec23.1:HA/TbSec24.2:Ty TbSec23.2:HA/TbSec24.2:Ty. In frame chromosomal fusions were confirmed by PCR from gDNA and sequencing. In situ epitope tagging of the putative glycosyltransferase TbGT15:Ty has been described previously (Sutterwala et al., 2008).
Northern Analysis
Total RNA was extracted from mid-log phase parasites by using TRIzol reagent (Promega) according to the manufacturer's instructions. PCR-generated probes (TbSec23.1, nt 939-1743; TbSec23.2, 1025-1857; TbSec24.1, nt 1028-1900; and TbSec24.2, nt 1025-1900) were labeled with [32P]dCTP with a random priming kit (Stratagene) and desalted on ProbeQuant 96 G-50 columns (GE Healthcare, Piscataway, NJ). RNA (5 μg/lane) was fractionated on 1% agarose/17.5% formaldehyde gels and transferred to Zeta Probe nylon membranes (Bio-Rad Laboratories, Hercules, CA) by capillary action. Membrane-bound RNA was photo cross-linked at 1200 μJ (1 min). Membranes were prehybridized for 4 h at 42°C (5× SSPE, 5× Denhardt's solution, 50% formamide, 0.5% SDS, and 100 μg/ml denatured salmon testes DNA), and denatured probes were added in fresh, prewarmed hybridization buffer. After hybridization (42°C overnight), membranes were washed once in 2× SSC, 0.1% SDS (20 min at room temperature [RT]), and twice in 0.2× SSC, 0.1% SDS (15 min for 42°C). To normalize, blots were stripped (3 times at 80°C for 30 min; 2× SSC and 0.5% SDS) and reprobed with a tubulin probe. In all cases, hybridization was analyzed by phosphorimaging using a Typhoon Storm 860 system (GE Healthcare) with native ImageQuant software.
Metabolic Radiolabeling and Immunoprecipitation
Pulse-chase metabolic radiolabeling with [35S]methionine/cysteine of trypanosomes and immunoprecipitation of labeled reporters have been detailed previously (Triggs and Bangs, 2003; Peck et al., 2008). Unless stated otherwise, lysates for immunoprecipitation were prepared in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1.0% NP-40, 0.5% deoxycholate, and 0.1% SDS). Pulse times were 2 min (VSG), 5 min (BiPN:GPI), 10 min (TbbCATL), or 15 min (p67). VSG transit to the surface was analyzed as described previously (Bangs et al., 1986; Sutterwala et al., 2008). In brief, radiolabeled cells were hypotonically lysed, allowing for conversion of surface, membrane-bound VSG to soluble VSG by endogenous GPI-phospholipase C (PLC). Cell-associated and soluble VSG were then separated by centrifugation and immunoprecipitated under standard conditions. For TbSar1WT and TbSar1DN expression, control and tetracycline-induced cells (0 or 2 h) were radiolabeled for 1 h, with continued tetracycline stimulation as appropriate. TbSar1 polypeptides and VSG were immunoprecipitated from cell lysates with anti-T7 or anti-VSG. All immunoprecipitates were separated on 12% SDS-polyacrylamide gel electrophoresis (PAGE) gels and visualized via the Typhoon system (GE Healthcare). Quantification of phosphorimages was done by ImageQuant software as described previously (Tazeh and Bangs, 2007).
Sequential immunoprecipitation experiments have been detailed previously (Bangs et al., 1996). In brief, parasites were radiolabeled with [35S]methionine/cysteine for 2.5 h. Cells were lysed in TEN buffer (50 mM Tris-HCl, 150 mM NaCl, and 5 mM EDTA, pH 7.5) with 0.5% NP-40, protease inhibitor cocktail (2 μg/ml each of leupeptin, antipain, pepstatin, and chymostatin), 0.1 mM tosyllysine chloromethyl ketone, and 0.5 mM phenylmethylsulfonyl fluoride. Lysates were cleared of debris by microcentrifugation (5 min at 13,000 rpm) and immunoprecipitated overnight with either anti-HA or anti-Ty antibody bound to protein A-Sepharose beads. Precipitates were washed twice in TEN buffer. Bound material was solubilized in 1% SDS (200 μl) and reconstituted to standard RIPA conditions (1.0 ml final). Secondary immunoprecipitates were done with the alternate antibody (anti-HA or anti-Ty as appropriate) bound to protein A-Sepharose beads (6 h at 4°C). Beads were washed twice in TEN buffer, and bound material was solubilized in sample buffer, fractionated on a 12% SDS-PAGE gel, and imaged as described above.
Antibodies
Rabbit anti-BiP, rabbit anti-VSG221, monoclonal mouse anti-p67 (mAb218), and rabbit anti-TbbCATL have all been described previously (Bangs et al., 1993; Alexander et al., 2002; Peck et al., 2008). Mouse anti-HA (Invitrogen, Carlsbad CA), rabbit anti-HA (Covance Research Products, Berkeley, CA), and mouse anti-Ty (UAB Hybridoma Facility, Birmingham, AL) were used as in previous publications (Sutterwala et al., 2008). Rabbit anti-T7 (Bethyl Laboratories, Montgomery, TX) was used at the recommended dilutions. Rabbit anti-Golgi reassembly stacking protein (GRASP) (He et al., 2004), and mouse monoclonal antibody (mAb) L3B2 anti-FAZ (Kohl et al., 1999) were generously provided by Graham Warren (Max F. Perutz Laboratories, Vienna, Austria) and Keith Gull (University of Oxford, Oxford, United Kingdom), respectively.
Immunofluorescence Microscopy
Immunostaining of formaldehyde-fixed parasites was done as described previously, with minor alterations (Sutterwala et al., 2008). After settling onto poly-l-lysine slides, cells were incubated (1 h at RT) with blocking buffer: phosphate-buffered saline (PBS), 10% normal goat serum (NGS), 1% Triton X-100, and 1% bovine serum albumin. Cells were stained with primary antibodies diluted in blocking buffer, washed, and stained with the appropriate secondary antibodies and 4,6-diamidino-2-phenylindole (DAPI) (500 ng/ml). After washing, slides were mounted with PBS:glycerol (1:1). Serial image stacks (0.2-μm Z-increment) were collected with capture times from 100 to 500 ms (100× PlanApo oil immersion 1.4 numerical aperture) on a motorized Axioplan IIi (Carl Zeiss, Jena, Germany) equipped with a rear-mounted excitation filter wheel, a triple pass (DAPI/fluorescein isothiocyanate/Texas Red) emission cube, differential interference contrast (DIC) optics, and a Orca AG charge-coupled device camera (Hamamatsu, Bridgewater, NJ). All images were collected with OpenLab 4.0 software (Improvision, Lexington, MA), and individual channel stacks were deconvolved by a constrained iterative algorithm, pseudocolored, and merged using Velocity 5.0 software (Improvision). This arrangement allows pixel precise localization in x, y and z dimensions (Supplemental Figure S1).
Transmission Electron Microscopy
For immunolocalization at the ultrastructural level, parasites were fixed in 4% paraformaldehyde/0.05% glutaraldehyde in phosphate-buffered saline for 1 h at 4°C. Samples were then embedded in 10% gelatin and infiltrated overnight with 2.3 M sucrose and 20% polyvinyl pyrrolidone in piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) and MgCl2 at 4°C. Samples were trimmed, frozen in liquid nitrogen, and sectioned with an Ultracut UCT cryo-ultramicrotome (Leica Microsystems, Bannockburn, IL). Fifty-nanometer sections were blocked with 5% FBS and 5% NGS for 30 min and subsequently incubated with mouse anti-BiP (1/50) and rabbit anti-HA (1/200; Sigma-Aldrich, St. Louis, MO) antibodies for 1 h at room temperature. Sections were then washed in block buffer and probed with goat anti-mouse (12-nm colloidal gold-conjugated) and goat anti-rabbit (18-nm colloidal gold-conjugated) antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA) for 1 h at room temperature. Sections were washed in PIPES buffer followed by a water rinse and stained with 0.3% uranyl acetate/2% methylcellulose. Samples were viewed with a 1200EX transmission electron microscope (JEOL USA, Peabody, MA). Parallel controls omitting primary antibodies were consistently negative at the concentration of colloidal gold-conjugated secondary antibodies used in these studies.
RESULTS
Identification of Trypanosomal COPII Machinery
Trypanosome COPII coat orthologues were identified by querying the T. brucei genome database (www.sanger.ac.uk/Projects/T_brucei) with the respective S. cerevisiae open reading frames (Supplemental Table S1). Single orthologues were found for Sar1 and Sec31, whereas two paralogous genes exist for Sec23 (TbSec23.1 and TbSec23.2), Sec24 (TbSec24.1 and TbSec24.2), and Sec13 (TbSec13.1 and TbSec13.2) (Supplemental Table S1). We have chosen an unbiased, numbered nomenclature for the T. brucei orthologues (i.e., TbSec23.1, etc.) because the functional relationship to other orthologues over such large phylogenetic distances is dubious. To begin our studies on the early secretory pathway in African trypanosomes, we chose to focus on TbSar1, TbSec23, and TbSec24, because Sar1 initiates the process of COPII vesicle formation and because the Sec23/Sec24 heterodimer is critical to cargo recruitment into the budding vesicle. Alignment of the paralogous TbSec23 and TbSec24 open reading frames reveals that these sequences are modestly similar to each other (59 and 44%, respectively).
Secretory Transport Is COPII Dependent in Trypanosomes
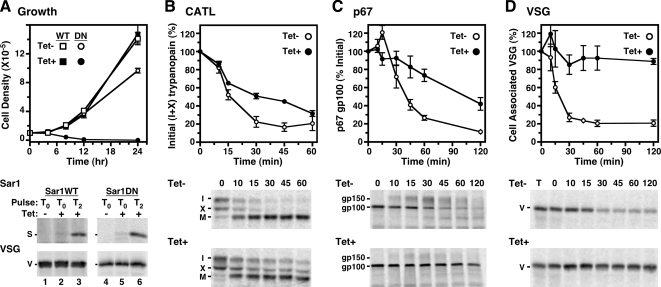
In yeast and mammals, trafficking of all classes of secretory cargo is COPII-vesicle dependent. To confirm this in trypanosomes, we generated a dominant-negative mutant of TbSar1. A single amino acid change in Sar1 (T34N in Pichia pastoris; T39N in mammals) creates a GDP-locked dominant-negative mutant, with reduced affinity for GTP. Expression of this mutant inhibits further rounds of COPII vesicle formation (Kuge et al., 1994; Connerly et al., 2005). Using this approach, we made the equivalent TbSar1 mutation (T34N), creating the dominant-negative, GDP-locked form of TbSar1 (TbSar1DN). This construct and a matched wild-type control, both of which contain a C-terminal T7-epitope tag, were introduced into BSF trypanosomes under a tetracycline-inducible promoter, and conditional synthesis was monitored by metabolic radiolabeling and specific immunoprecipitation with anti-T7 antibody (Figure 1A, bottom). Expression of both TbSar1WT and TbSar1DN was detected as early as 1 h of induction (lanes 2 and 5) and increased at 3 h (lanes 3 and 6). Expression of TbSar1WT had no effect on cell growth, but strikingly TbSar1DN expression resulted in arrest of cell growth after only 4 h of induction, followed rapidly by cell death (Figure 1A, top). This catastrophic effect indicates that TbSar1 activity is critical for cell viability.
Figure 1.
TbSar1DN expression is lethal and delays forward protein trafficking in T. brucei. (A–D) Transgenic cell lines containing the inducible TbSar1WT (squares, A only) and TbSar1DN (circles, all panels) constructs were cultured without (Tet−, open symbols) or with tetracycline (Tet+, closed symbols) to induce expression of the wild-type and dominant-negative TbSar1 proteins. (A) Growth of the TbSar1WT and TbSar1DN cell lines were monitored (top, mean ± SD for triplicate cultures), and cells were metabolically radiolabeled (bottom) for 1 h in the absence (lanes 1 and 4) or presence of tetracycline after 0 h (lanes 2 and 5) or 2 h (lanes 3 and 6) of prior tetracycline induction. As indicated, lysates were immunoprecipitated with anti-T7 for TbSar1 proteins (S, top images) or anti-VSG (V, bottom images) as a labeling/loading control. Precipitates were analyzed by SDS-PAGE (107 cell equivalents/lane) and phosphorimaging. (B–D) Control (Tet−) and induced (Tet+, 2.5 h) TbSar1DN cells were pulse-chase radiolabeled as described in Materials and Methods, and trafficking of endogenous TbbCATL (B), p67 (C), and VSG (D) was followed by immunoprecipitation and SDS-PAGE (5 × 106 cell equivalents/lane, TbbCATL and p67; 3 × 105 cell equivalents/lane, VSG). Phosphorimages of representative gels are presented for each reporter (bottom). The rates of turnover (top) were quantified (mean ± SEM for triplicate independent experiments) as disappearance of the initial forms for TbbCATL (I + X) and for p67 (gp100) and as loss of cell-associated VSG during hypotonic lysis as a percentage of total VSG in a time zero whole cell lysate (T).
To confirm that cargo egress from the ER is COPII dependent in trypanosomes, we determined the effect of TbSar1DN expression on the intracellular transport of three well-characterized, endogenous secretory reporters: TbbCATL (soluble), p67 (type I transmembrane), and VSG (GPI anchored). After TbSar1DN induction, ER exit was monitored by pulse-chase analysis using distinct transport-dependent processing events for each reporter. TbbCATL,1 a lysosomal cathepsin-L like protease (Caffrey and Steverding, 2009), is synthesized in the ER as an immature proenzyme (∼53 kDa, I) and as an additional smaller “precursor” form (∼50 kDa, X) (Figure 1B, Tet−). The exact nature of precursor X is not clear, but our own observations (Sutterwala and Bangs, unpublished) indicate that it is derived from the TbbCATL open reading frame. On arrival in the lysosome, both forms are converted by proteolytic cleavage to mature active TbbCATL (43 kDa, M). After TbSar1DN induction, there was a 1.9-fold delay in the processing of the immature forms of TbbCATL (I+X) (Table 1). p67, a lysosomal associated membrane protein-like type I transmembrane protein, is synthesized as a highly N-glycosylated 100-kDa ER glycoform (gp100). Poly-N-acetyllactosamine modification of the N-glycans in the Golgi results in formation of the 150-kDa glycoform (gp150) (Figure 1C, Tet−), which is subsequently degraded upon arrival in the lysosome (Alexander et al., 2002; Peck et al., 2008). TbSar1DN expression reduced the processing of gp100 to gp150, indicating a direct inhibition of ER exit (Figure 1C, Tet+). The overall rate of p67 transport as measured by loss of gp100 decreased 2.6-fold after TbSar1DN induction (Table 1). Finally, we monitored transport of VSG, a GPI-anchored homodimer. GPI anchor attachment occurs immediately after translocation into the ER, and VSG is then rapidly transported to the cell surface via the Golgi and flagellar pocket (Bangs et al., 1985, 1986; Ferguson et al., 1986; Duszenko et al., 1988). The assay for VSG transport relies on the onset of susceptibility to hydrolysis by endogenous GPI-PLC during hypotonic lysis, concurrent with arrival at the cell surface. Newly synthesized VSG en route to the flagellar pocket is resistant to the action of GPI-PLC under these conditions. After TbSar1DN expression, VSG trafficking was drastically reduced (Figure 1D, bottom, Tet− vs. Tet+), with the rate of transport decreasing ∼12-fold (Figure 1D and Table 1). Collectively, the severe effect of TbSar1DN expression on VSG transport, in addition to the significant delays seen for both TbbCATL and p67 trafficking, indicates that ER exit of newly synthesized trypanosomal secretory cargo is critically dependent on TbSar1 and the COPII machinery. We attempted to visualize these delayed reporters within the ER but were unsuccessful, probably because the window of time between TbSar1DN induction and cell death is too short to allow sufficient accumulation of newly synthesized proteins for immunodetection.
Table 1.
Kinetics (t1/2) of reporter transporta
| Reporter | Tet | TbSar1DN | TbSec23.1 | TbSec23.2 | TbSec24.1 | TbSec24.2 |
|---|---|---|---|---|---|---|
| TbbCATLb | − | 16.5 ± 2.3 | 10.5 ± 0.5 | 14.5 ± 0.3 | 11.5 ± 0.5 | 11.0 ± 0.8 |
| + | 31.2 ± 3.6 (1.9) | 10.2 ± 0.9 (1.0) | 19.5 ± 0.8 (1.3) | 16.0 ± 2.5 (1.4) | 13.0 ± 0.8 (1.2) | |
| p67c | − | 40.7 ± 3.9 | 43.8 ± 3.3 | 39.7 ± 4.2 | 36.5 ± 2.3 | 37.0 ± 3.2 |
| + | 107.7 ± 9.3 (2.6) | 44.2 ± 7.1 (1.0) | 58.3 ± 3.4 (1.5) | 51.5 ± 3.6 (1.4) | 41.0 ± 4.5 (1.1) | |
| VSGd,e | − | 18.9 ± 2.6 | 27.5 ± 2.2 | 23.5 ± 4.4 | 20.7 ± 4.2 | 23.2 ± 5.3 |
| + | 219.7 ± 8.5 (11.6) | 52.5 ± 2.8 (1.9) | 128.7 ± 12.7 (5.5) | 81.8 ± 1.3 (4.0) | 30.0 ± 5.3 (1.3) | |
| BiPN:GPIf | − | N.D. | 19.1 ± 0.4 | 20.7 ± 0.8 | N.D. | N.D. |
| + | 28.8 ± 3.9 (1.5) | 55.1 ± 9.4 (2.7) | ||||
N.D., not determined.
aKinetics of transport were quantified, as defined below, in control (Tet−) cells and after induction (Tet+) of dominant-negative TbSar1 or specific COPII subunit RNAi, as indicated. Data were analyzed in Prism 4 (GraphPad Software, San Diego, CA). Half-times (t1/2) in minutes were determined graphically and are presented as means ± SEM for triplicate determinations. -Fold differences are indicated in parentheses.
bTbbCATL was measured as loss of initial species (I + X).
cp67 was measured as loss of initial gp100 ER glycoform.
dVSG was measured as loss of cell-associated VSG after hypotonic lysis.
eHalf-times for VSG transport in induced TbSar1DN and TbSec23.2 RNAi cells were extrapolated and should be considered approximate.
fBiPN:GPI was measured as loss of initial immature form (I).
Silencing of TbSec23 and TbSec24 Paralogues Is Lethal in BSF Trypanosomes
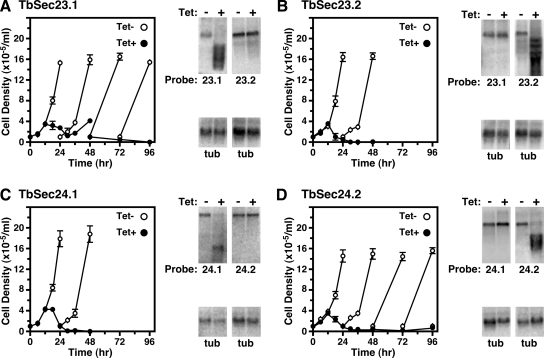
The requirement of TbSar1 for viability and efficient cargo transport led us to investigate the role of the TbSec23 and TbSec24 isoforms. Inducible dsRNA constructs targeting each subunit were introduced individually into BSF trypanosomes and in each case cellular viability was severely affected by RNAi silencing (Figure 2, A–D). Sustained growth arrested as early as 12 h postinduction, and cell death followed beginning at 18–24 h. Consequently, all subsequent RNAi phenotypic analyses were performed at 12 h of dsRNA induction. Northern blot analyses of total RNA isolated from control and silenced cells indicated that TbSec23.1 and TbSec23.2 transcript levels were specifically reduced to 33.2 ± 5.8 and 35.3 ± 6.3%, respectively (mean ± SEM, n = 3), without significantly affecting transcript levels from the paralogous gene (Figure 2, A and B). Similarly, induction of dsRNA targeting TbSec24.1 or TbSec24.2 specifically depleted transcript levels to 30.9 ± 3 0.2 and 23.5 ± 11.4%, respectively (mean ± SEM, n = 3) (Figure 2, C and D). Interestingly, however, knockdown of each TbSec24 transcript led to a modest increase of the paralogous transcript (TbSec24.1 up ∼70%; TbSec24.2 up ∼40%), suggesting that cells attempt to compensate for the depletion of one TbSec24 subunit message with the upregulation of the other. Collectively, these data indicate that BSF cell viability is dependent on the expression of all TbSec23 and TbSec24 subunits. Consistent with this conclusion is the repeated failure of all attempts to achieve double-targeted gene disruptions for each of the four COPII subunits (data not shown).
Figure 2.
TbSec23 and TbSec24 Subunits are essential for cell growth. Transgenic cell lines containing TbSec23.1 (A), TbSec23.2 (B), TbSec24.1 (C), and TbSec24.2 (D) RNAi constructs were cultured without (Tet−, open circles) or with tetracycline (Tet+, closed circles) to initiate specific dsRNA synthesis. In each case, cell growth was monitored for triplicate cultures (left), and cultures were diluted to the starting cell density every 24 h if density exceeded 5 × 105 cells/ml. RNA was extracted at 12 h of induction for Northern analyses (right). RNA (5 μg/lane) from control and RNAi-silenced cell lines were run in parallel and probed for RNAi target TbSec23 or TbSec24 transcript levels. As a control for specificity matched blots were probed for the alternate TbSec transcript, e.g., TbSec23.2 control for TbSec23.1 knockdown. All blots were stripped and reprobed for tubulin (tub). Blots were visualized by phosphorimaging, and relative levels of knockdown for triplicate independent experiments were quantified and normalized to tubulin.
VSG Transport Is Preferentially Dependent on TbSec23.2 and TbSec24.1
The presence of duplicate isoforms of TbSec23 and TbSec24 raises the question of whether they are functionally redundant or whether they play distinct cargo-specific roles in ER exit. To investigate this issue, we analyzed the trafficking of our suite of endogenous secretory reporters after specific RNAi silencing for each COPII subunit. At best minor defects in trafficking of TbbCATL or p67 were observed when any of the four COPII subunits were ablated; -fold decreases in transport rates ranged from 1.0- to 1.5-fold for all combinations of subunits and reporters (Supplemental Figure S2 and Table 1). Likewise, minor defects were seen in VSG trafficking for TbSec23.1 and TbSec24.2 ablation, 1.9- and 1.3-fold, respectively (Figure 3, A and D, and Table 1). In contrast, dramatic and roughly equivalent reductions in VSG trafficking were seen after silencing of TbSec23.2 (5.5-fold) or TbSec24.1 (4.0-fold) (Figure 3, B and C, and Table 1). Together, these data suggest that the paralogous TbSec23 and TbSec24 subunits are redundant for TbbCATL and p67 trafficking. In contrast, VSG is preferentially dependent on TbSec23.2 and TbSec24.1 for efficient egress from the ER.
Figure 3.
RNAi of TbSec23.2 or TbSec24.1 delays VSG transport. Specific dsRNA synthesis was induced for 12 h in the TbSec RNAi cell lines as indicated, and the transport of newly synthesized VSG was assayed as in Figure 1D (3 × 105 cell equivalents/lane). Phosphorimages of representative matched gels from control (Tet−) and silenced (Tet+) cells are presented (A–D, bottom). Loss of cell-associated VSG during hypotonic lysis was quantified and is presented as mean ± SEM for five independent experiments (A–D, top).
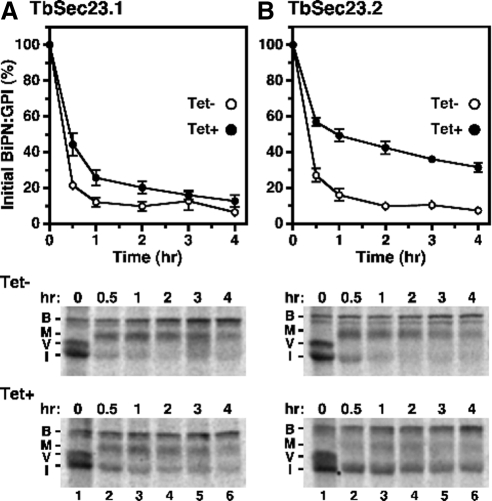
BiPN:GPI Trafficking Is Preferentially Dependent on TbSec23.2
To determine whether the TbSec23.2/24.1 dependence of VSG trafficking is VSG-specific, or rather applies to all GPI-anchored cargo, we monitored the progression of BiPN:GPI, a single GPI-anchored reporter that was constitutively expressed in the TbSec23.1 and TbSec23.2 RNAi cell lines. We have shown previously that attachment of a GPI anchor to the N-terminal ATPase domain of BiP, an ER-localized molecular chaperone, led to a mixed fate of shedding from the cell surface and degradation in the lysosome (Schwartz et al., 2005). BiPN:GPI is initially synthesized as an ∼55-kDa immature form (I) in the ER, which is subject to GPI carbohydrate modification during transit of the Golgi, forming a higher molecular weight mature glycoprotein (M). The fate of BiPN:GPI in the TbSec23.1 and TbSec23.2 cell lines after RNAi silencing was determined by pulse-chase analysis and immunoprecipitation with anti-BiP antibody (Figure 4, A and B). Endogenous BiP (B) was detected in all samples, indicating equivalent loading (lanes 1–6). In addition, as described previously (Bangs et al., 1996), endogenous newly synthesized VSG (V), which transiently associates with BiP during folding, was detected in equivalent amounts in all time zero samples (lanes 1). Depletion of TbSec23.1 message had a minor effect on the transport kinetics of BiPN:GPI as judged by the relative rates of disappearance of the immature ER glycoform (Figure 4A, lanes 2–6, and Table 1). In contrast, knockdown of TbSec23.2 dramatically impaired ER export of the BiPN:GPI precursor and maturation in the Golgi, with the rate of transport delayed by ∼2.7-fold (Figure 4B, lanes 2–6, and Table 1). These data indicate that similar to VSG, ER egress of BiPN:GPI is preferentially dependent on TbSec23.2, suggesting that all GPI-anchored cargo are dependent on this COPII subunit.
Figure 4.
RNAi of TbSec23.2 delays ER export of BiPN:GPI. BiPN:GPI was constitutively expressed in the TbSec23.1 and TbSec23.2 RNAi cell lines. After induction of specific dsRNA synthesis for 12 h, BiPN:GPI progression was monitored by pulse-chase radiolabeling, immunoprecipitation with anti-BiP antibody, and SDS-PAGE. The rate of disappearance of the BiPN:GPI immature (I) form was quantified (mean ± SEM for four independent experiments) for the TbSec23.1 (A) and TbSec23.2 (B) transgenic RNAi cell lines (top). Phosphorimages of representative gels (5 × 106 cell equivalents/lane) from Tet− and Tet+ cultures are shown in the bottom panels. BiP, B; mature BiPN:GPI, M; VSG, V; and immature BiPN:GPI, I.
TbSec23.1/TbSec24.2 and TbSec23.2/TbSec24.1 Form Obligate Physical Pairs
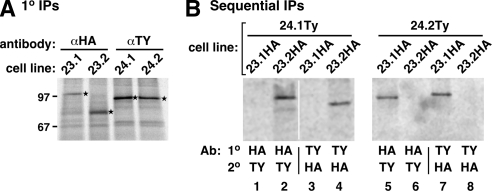
The matched impact of TbSec23.2 and TbSec24.1 silencing on VSG transport (Figure 3 and Table 1) could be explained if these two subunits form exclusive and obligate heterodimers within the COPII coat. In this scenario, knockdown of either subunit would have the same overall effect. Such associations have been seen in other systems and can have functional consequences for selective trafficking of secretory cargo (Shimoni et al., 2000). To investigate this possibility in trypanosomes, we analyzed the physical associations between the TbSec23 and TbSec24 subunits by epitope tagging and pull-down experiments. Four BSF cell lines were developed with single HA-tagged TbSec23 subunits and single Ty-tagged TbSec24 subunits in all possible combinations: TbSec23.1:HA/TbSec24.1:Ty, TbSec23.1:HA/TbSec24.2:Ty, TbSec23.2:HA/TbSec24.1:Ty, and TbSec23.2:HA/TbSec24.2:Ty. All four doubly HA/Ty-tagged cell lines were metabolically radiolabeled, and standard lysates (containing NP-40, deoxycholate, and SDS) were subjected to immunoprecipitation with either anti-HA or anti-Ty antibodies (Figure 5A). In each case, epitope-tagged polypeptides of the predicted size and immunoreactivity were detected, confirming that the tagged proteins are expressed appropriately.
Figure 5.
TbSec23.1/TbSec24.2 and TbSec23.2/TbSec24.1 form obligate physical pairs. TbSec subunits were in situ epitope tagged (TbSec23.1 and TbSec23.2, HA tag; TbSec24.1 and TbSec24.2, Ty tag), in all possible HA/Ty double combinations, by homologous recombination as described in Materials and Methods. (A) Tagged cell lines were pulse radiolabeled (2 h) and lysed in standard RIPA buffer. Labeled polypeptides were specifically immunoprecipitated from cell extracts with anti-epitope antibodies as indicated and analyzed by SDS-PAGE (107 cell equivalents/lane) and phosphorimaging. Bands corresponding to the expected size of each TbSec component (TbSec23.1:HA, 106.6 kDa; TbSec23.2:HA, 83.3 kDa; TbSec24.1:Ty, 108.1 kDa; and TbSec24.2:Ty, 100.8 kDa) are indicated (stars). These bands were not immunoprecipitated from untagged control cell lines (data not shown). (B) Physical interactions were determined by sequential immunoprecipitations using the doubly HA/Ty-tagged cell lines as described in Materials and Methods. Each cell line was radiolabeled (2 h), lysed under nondenaturing conditions, and immunoprecipitated first with either anti-HA or anti-Ty antibodies as indicated (1°). Specific immunoprecipitates were solubilized under denaturing conditions and reprecipitated with the alternate antibody (2°, anti-Ty or anti-HA). Final precipitates were analyzed by SDS-PAGE (107 cell equivalents/lane) and phosphorimaging. The vertical white line between lanes 2 and 3 indicates intervening lanes that were digitally excised for the sake of presentation (all remaining lanes from same exposure).
To investigate the physical pairing of individual TbSec23 and TbSec24 subunits into heterodimers, radiolabeled lysates were prepared under gentler conditions (NP-40 only) to preserve molecular interactions and were then subjected to immunoprecipitation with anti-HA or anti-Ty. These primary pull-downs (Figure 5B, 1°) were solubilized in SDS, readjusted to standard lysate conditions, and then immunoprecipitated again with the other antibody, either anti-HA or anti-TY as appropriate (Figure 5B, 2°). The results were unequivocal. TbSec23.1:HA was detected in secondary pull-downs only when coexpressed with TbSec24.2:Ty (lane 7 vs. lane 3) and vice versa (lane 5 vs. lane 6). Likewise, TbSec23.2:HA was detected in secondary pull-downs only when coexpressed with TbSec24.1:Ty (lane 4 vs. lane 8) and vice versa (lane 2 vs. lane 1). No evidence was detected for coprecipitation of Sec23.1:HA and Sec24.1:Ty (lanes 1 and 3) or for TbSec23.2:HA and TbSec24.2Ty (lanes 6 and 8). Collectively, these data indicate that the TbSec23 and TbSec24 subunits have discrete interactions, yielding two exclusive and obligate heterodimers: TbSec23.1/TbSec24.2 and TbSec23.2/TbSec24.1.
This obligate pairing of subunits can be explained by primary sequence differences between paralogues. Crystallographic studies in S. cerevisiae defined the interface surfaces involved in Sec23/Sec24 dimerization (Bi et al., 2002), and these regions in the trypanosome subunits display considerable variation (Supplemental Figure S3B). When the T. brucei sequences are modeled onto the S. cerevisiae crystal structure, subtle and compensatory differences in three critical contact surfaces are revealed (Supplemental Figure S4). These changes suggest a coevolution of matched T. brucei subunits such that dimerization is physically restricted, consistent with the pattern revealed by our biochemical analyses.
COPII-defined ERES Are in Proximity to the Golgi and the FAZ
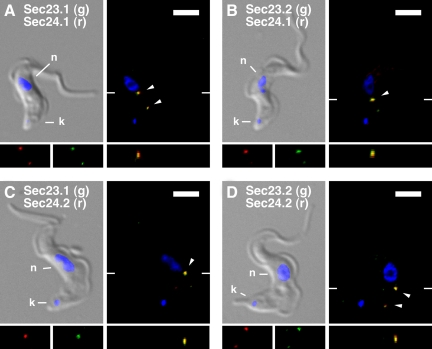
The existence of discrete TbSec23/TbSec24 heterodimers raises the issue of subcellular localization. Are there distinct ERES defined by each pair, or do they all colocalize in the same sites for ER exit? To address these questions, we performed immunofluorescence imaging with the four double-tagged TbSec23:HA/TbSec24:Ty cell lines. Each cell line was stained with an anti-HA and anti-Ty antibody to label the indicated TbSec23 and TbSec24 subunits (Figure 6, A–D). Despite their exclusive physical interactions, each TbSec23:HA/TbSec24:Ty combination, and by extension all four TbSec subunits, colocalized precisely in three dimensions to one or two discrete sites in the postnuclear region of postcytokinesis BSF trypanosomes (G1 phase cells with a single kinetoplast and nucleus). These findings indicate that recently divided BSF trypanosomes have one or two ERES per cell, with uniform composition in regard to COPII components. Interestingly, these sites are also consistently proximal to the flagellum in a region called the flagellar attachment zone (FAZ) (Figure 7). This apparent juxtaposition with the FAZ suggests that the ERES may be organized on the FAZ-associated ER (FAZ:ER), an extension of smooth ER that interdigitates longitudinally with a quartet of subpellicular microtubules that subtend the flagellum along the entire length of the cell body (Figure 9) (Bastin et al., 2000; Vaughan et al., 2008).
Figure 6.
All TbSec23 and TbSec24 subunits colocalize in T. brucei. As indicated (A–D) the doubly HA/Ty epitope-tagged cell lines were fixed, permeabilized, and immunostained with anti-HA (staining TbSec23.1:HA or TbSec23.2:HA) and anti-Ty (staining TbSec24.1:Ty or TbSec24.2:Ty). After staining with appropriate secondary antibodies (anti-HA, green and anti-Ty, red), cells were imaged by epifluorescence microscopy. For each pair, merged DAPI/DIC images with stained nucleus (n) and kinetoplast (k) indicated are presented (top left). Corresponding three-channel summed stack projections are presented (top right) with discrete sites (1 or 2 per cell) at which each pair of TbSec subunits colocalize indicated (arrowheads). Single channel red and green images in the region of colocalization are presented (bottom left), as are images of the x-z transect through the major region of colocalization (bottom right). The planes of the x-z transects are indicated by marginal hatch marks in the three channel x-y images. Bars, 5 μm.
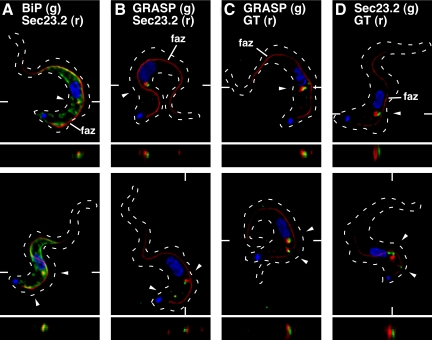
Figure 7.
Localization of ERES components relative to the flagellar attachment zone, ER, and Golgi. Epitope-tagged cell lines were fixed/permeabilized and stained with combinations of anti-tag and anti-native marker protein antibodies as appropriate. For each set of staining conditions, representative three-channel summed stack projections of cells containing one (top row) or two (bottom row) ERES:Golgi junctions are presented (arrowheads). Cells were also stained with mAb L3B2 (anti-FAZ filament, red) to reveal orientation relative to the longitudinal FAZ (indicated in top row only) and with DAPI to mark the central nucleus and posterior kinetoplast. TbSec23.2HA-expressing cells were stained with anti-HA (red) to detect the tagged ERES marker and with either anti-BiP (A) or anti-GRASP (B) to localize these endogenous ER and Golgi matrix markers, respectively (green). Cells expressing TbGT15:Ty alone (C) or simultaneously with TbSec23.2:HA (D) were stained with anti-Ty (red) to detect the tagged glycosyltransferase and with either anti-GRASP (C) or anti-HA (D) to detect the Golgi matrix and ERES, respectively (green). Dashed outlines mark the cellular boundary as obtained from matched DIC images (data not shown). The planes of z transects through selected ERES:Golgi junctions (shown below each image) are indicated by marginal hatch marks in the three channel x-y images.
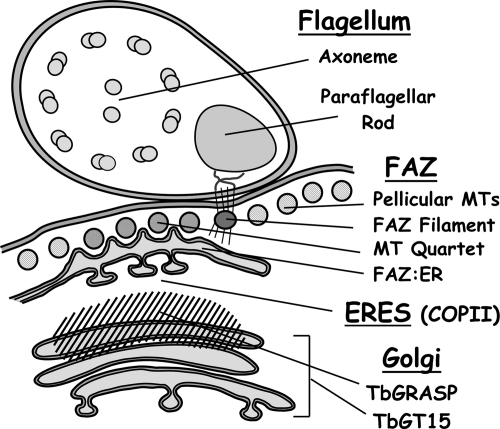
Figure 9.
Diagram of ERES:Golgi junctions in BSF trypanosomes. A partial cross section of a cell in the postnuclear region transecting the junction of an ERES and Golgi complex is represented. Major structures include the flagellum containing the 9 + 2 axoneme and paraflagellar rod; the FAZ, including pellicular microtubules, FAZ filament, antiparallel microtubule quartet, and the FAZ:ER; the ERES defined by COPII subunits; and the Golgi defined by TbGT15 and TbGRASP. Diagram inspired by Bastin et al., 2000 and Hill, 2003).
The ERES in procyclic trypanosomes have been shown to be in proximity to the Golgi, as defined by TbGRASP (He et al., 2004), and we wished to visualize this architecture in BSF trypanosomes. We began by localizing the ERES in TbSec23.2:HA cells in relationship to the ER with anti-BiP antibody, and in relationship to the Golgi with either an anti-TbGRASP antibody (He et al., 2004) or an anti-Ty antibody to detect a Ty-tagged glycosyltransferase (TbGT15:Ty). We have previously validated TbGT15:Ty as a Golgi marker (Sutterwala et al., 2008). In addition, the FAZ was simultaneously localized as a subcellular reference point in all images with a mAb (L3B2) to the FAZ filament (Kohl et al., 1999). Typical images for all of these antibody combinations in BSF cells containing one (top) or two (bottom) ERES are presented in Figure 7. TbSec23.2:HA colocalized with one or two distinct lobes of the ER, which were in turn closely aligned with the FAZ filament along the entire cell body (Figure 7A), confirming these sites as true ERES, and the FAZ:ER as the site of ERES assembly. Closely juxtaposed with the ERES, but distal to the FAZ, was a zone defined by TbGRASP (Figure 7B), which closely abutted the ERES, but when viewed in the z-dimension was laterally distinct. When imaged relative to TbGT15:Ty, the TbGRASP-defined zone was typically juxtaposed and overlapping (Figure 7C). Notably, of 150 Golgi scored for TbGT15:Ty and TbGRASP staining, only ∼18% showed precise colocalization; the remaining ∼82% showed only partial overlap between the two markers; and in the later case, the TbGRASP signal was always proximal to the ERES and FAZ. Finally, when TbGT15:Ty was imaged relative to the ERES, it defined a distinct structure that was distal to the FAZ and showed no lateral overlap with the ERES (Figure 7D). Collectively, these data indicate that the ERES and Golgi are closely associated in BSF trypanosomes and suggest that the entire ERES:Golgi junction, whether there is one or two per cell, is closely associated with the FAZ:ER in the region between the centrally located nucleus and the posterior flagellar pocket.
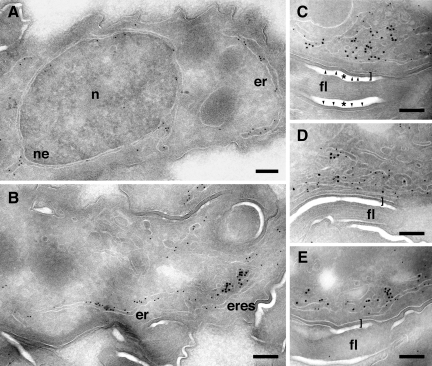
To confirm that the ERES is assembled on the FAZ:ER, we performed immunoelectron microscopy on the TbSec23.2:HA cell line with anti-BiP to detect ER elements and with anti-HA to detect COPII-containing structures (Figure 8). BiP staining was associated with both the nuclear envelope (Figure 8A) and peripheral ER elements (Figure 8, A and B), whereas HA staining was clustered in association with peripheral ER elements (Figure 8B). In each case, little staining was seen elsewhere over the cell body confirming the specificity of the staining patterns. TbSec23.2:HA staining was typically observed as clusters consistently associated with underlying peripheral ER elements (Figure 8, B–E) and usually in close association with the flagellum (Figure 8, C–E). These results are fully consistent with images acquired by fluorescence microscopy and confirm the FAZ:ER as the site of ERES formation.
Figure 8.
Ultrastructural localization of ERES. The TbSec23.2:HA cell line was fixed and prepared for immunoelectron microscopy as described in Materials and Methods. Sections were stained with anti-BiP (12-nm gold) and anti-HA (18-nm gold) to detect ER and ERES respectively. (A and B) Images showing specificity of staining with both reagents. Anti-BiP staining is predominantly associated with peripheral reticular membranes and nuclear envelope (A and B), and anti-HA staining is clustered in discrete sites associated with peripheral ER membranes (B). (C and D) Images of individual ERES demonstrating proximity to the flagellum and confirming the association of ERES with the FAZ:ER. Bars (A–E), 0.25 μm. Nucleus, n; nuclear envelope, ne; peripheral ER, er; ER exit sites, eres; and flagellum, fl. Double-thick surface coats at the interface of cell body and flagellar membranes are indicated by brackets. Asterisks indicate fixation-induced lacunae flanking the flagellum and bounded by the flagellar membrane (arrowheads in C only).
DISCUSSION
In this work, we address the mechanistic basis of GPI-dependent trafficking in the early secretory pathway of African trypanosomes. In yeast, the COPII machinery mediates ER exit of all classes of secretory cargo, including soluble, polytopic membrane, and GPI-anchored proteins (Schimmoler et al., 1995; Kuehn et al., 1996). Conditional expression of a dominant-negative form of TbSar1 confirmed that the COPII machinery affects similar functions in trypanosomes. The failure of newly synthesized p67 to be processed to mature glycoforms, indicates that the TbSar1DN block is in ER-to-Golgi transport. The extremely rapid onset of growth arrest and subsequent cell death upon induction of TbSar1DN synthesis was striking. These results confirm that TbSar1, and by inference the entire COPII machinery, is functional and directly involved with the forward transport of secretory cargo from the ER in trypanosomes, a conclusion supported by the RNAi phenotypes seen with the four TbSec23/24 subunits.
Multiple and redundant COPII subunits within a species are common. The human genome contains two Sar1, Sec23, and Sec31 paralogues each and four Sec24 genes (Shen et al., 1993; Paccaud et al., 1996; Tang et al., 1999; Stankewich et al., 2006). The Sec24 subunits are redundant and nonessential, with overlapping abilities to recognize transport signals (Lang et al., 2006; Wendeler et al., 2006). S. cerevisiae has three Sec24 paralogues (Sec24, Lst1, and Iss1). Only Sec24 is essential, but loss of Lst1 significantly impacts forward trafficking of the plasma membrane ATPase Pma1 (Kurihara et al., 2000; Shimoni et al., 2000). P. pastoris has two paralogues of Sec23 (PpSec23 and PpShl23) and Sec24 (PpSec24 and PpLst1); yet, only PpSec23 and PpSec24 are required for viability. Interestingly, PpSec23 forms heterodimers with both Sec24 subunits, but PpShl23 only interacts with PpLst1 forming a redundant and nonessential heterodimer (Esaki et al., 2006).
Trypanosomes stand in contrast to each of these eukaryotic systems in that both TbSec23 and TbSec24 genes are essential. Furthermore, the TbSec23.1/TbSec24.2 and TbSec23.2/TbSec24.1 subunits form obligate and exclusive heterodimers. There is considerable sequence variation in the trypanosomal subunits (Supplemental Figure S3B) in the regions known to form the dimer interface (Bi et al., 2002) (Supplemental Figure S4), which likely form the basis for dimer exclusivity. These discrete dimers are redundant for the forward trafficking of soluble (TbbCATL) and transmembrane (p67) cargo. However, trafficking of GPI-anchored cargo is selectively dependent on the TbSec23.2/TbSec24.1 pair. The need for bloodstream form trypanosomes to synthesize and transport a full compliment of VSG (107 molecules per cell per 6 h cell cycle) may explain the requirement for this heterodimer, but why is the TbSec23.1/TbSec24.2 heterodimer essential? Perhaps it is a simple matter of capacity to handle the total flux of all newly synthesized secretory cargo from the ER. Alternatively, this heterodimer may be preferentially required for transport of some as of yet unidentified cargo. These possibilities are not mutually exclusive.
There is precedent in S. cerevisiae for GPI-selective cargo loading at discrete ERES and into distinct COPII vesicle populations (Muniz et al., 2001; Castillion et al., 2009). In trypanosomes, defined Sec23/Sec24 heterodimers present two alternatives for the organization of ER exit. There may be a single class of COPII vesicles, containing both heterodimers and responsible for transport of all cargo. In this case, recruitment of GPI-anchored cargo into nascent vesicles would be mediated by the TbSec23.2/TbSec24.1 dimer. The alternative is two distinct vesicles populations, each containing one Sec23/Sec24 pair or the other. GPI-anchored cargo would be dependent on the TbSec23.2/TbSec24.1 vesicles, and non-GPI cargo would be accommodated by either class.
Whatever the model, a mechanism must exist for coupling GPI-anchored cargo to the cytoplasmic TbSec23.2/TbSec24.1 dimer. Recruitment of secretory cargo, including GPI-anchored proteins, to nascent COPII vesicles can be mediated by p24s, a family of small type I transmembrane adaptor proteins abundant in COPII vesicles (Jenne et al., 1995; Dominquez et al., 1998). p24s have cytoplasmic motifs for interaction with COPII components and associate in oligomers with combinatorial potential for recognition of secretory cargo (Jenne et al., 2002). Yeast has eight p24 orthologues; vertebrates have as many as 10 (Schimmoler et al., 1995; Belden and Barlowe, 1996; Marzioch et al., 1999; Strating et al., 2009). Individual disruption of p24s in yeast, and of p23 in mammalian cells, selectively impacts ER exit of GPI-anchored cargo (Jenne et al., 1995; Schimmoler et al., 1995; Belden and Barlowe, 1996; Marzioch et al., 1999; Takida et al., 2008; Castillion et al., 2009). The T. brucei genome contains six putative p24 orthologues based on homology searches with Emp24 (Supplemental Table S2), and it may be that these proteins mediate GPI recruitment into TbSec23.2/TbSec24.1-containing vesicles. There is also sequence variation between the TbSec24 isoforms in the B-site region (Supplemental Figure S3C), which in ScSec24 is a binding site for exported membrane proteins, including p24s (Miller et al., 2003; Mossessova et al., 2003). B-site variation, in conjunction with cargo-specific combinations of trypanosomal p24 proteins, could account for the selective dependence of GPI-anchored cargo on the TbSec23.2/TbSec24.1 heterodimer.
The physical organization of the early secretory pathway in BSF trypanosomes differs markedly from other model organisms. In postcytokinesis cells, there are one or two ERES closely associated with the Golgi, consistent with the localization of TbSec13 in PCF trypanosomes (He et al., 2004). Nothing is known about ERES:Golgi organization in other kinetoplastids. However, S. cerevisiae typically has 20 or more ERES throughout the ER, with ill-defined Golgi associations, whereas Pichia has fewer ERES that are clearly associated with defined Golgi complexes (Rossanese et al., 1999; Stephens, 2003). In contrast, animal cells have hundreds of ERES, and competing models of Golgi organization (Lippencott-Schwartz et al., 2000; Shorter and Warren, 2002).
Other features of ERES:Golgi organization in BSF trypanosomes merit comment (Figure 9). First and foremost is the exclusive formation of ERES on the FAZ:ER and thus its association with unique elements of the flagellar cytoskeleton. A bilobe structure thought to be involved in nucleating Golgi formation is also closely juxtaposed to the FAZ in PCF cell (Morriswood et al., 2009), but our results indicate for the first time that it is the entire ERES:Golgi complex that is associated with the trypanosome cytoskeleton, probably via the FAZ:ER. This may reflect the need to ensure fidelity in organellar segregation during cytokinesis. The flagellum and the FAZ are intimately involved in this process and probably provide the scaffold for partitioning the ERES:Golgi into each daughter cell. The role of the bilobe structure in this process in BSF trypanosomes remains to be defined. Second is the overlapping localization of TbGRASP with the Golgi, in contrast with PCF trypanosomes in which TbGRASP colocalizes precisely with other Golgi markers (He et al., 2004; Ho et al., 2006). GRASP orthologues act as cytosolic matrix components to maintain Golgi structure (Barr et al., 1997; Feinstein and Linstedt, 2008), and a simple explanation for the observed distribution is that TbGRASP defines an earlier compartment(s) of the BSF Golgi devoid of the TbGT15 glycosyltransferase. However, GRASP may have other roles in secretory trafficking (Kinseth et al., 2007), including physical interactions with the Sec23/Sec24 dimer (Behnia et al., 2007). It may be that TbGRASP has additional functions in trypanosomes such as maintaining the ERES:Golgi junction and/or in facilitating efficient ER-to-Golgi transport. Finally, our results provide preliminary insights into the replication of the entire ERES:Golgi junction in BSF trypanosomes. Approximately 80% of postcytokinesis cells (1 kinetoplast/1 nucleus) have two Golgi (n = 150; Schwartz and Bangs, unpublished observations), indicating that Golgi duplication is an early event in the cell cycle, as was observed for PCF trypanosomes (He et al., 2004). When a single ER:Golgi junction is present it is located forward near the nucleus, whereas in cells with two junctions, the second is proximal to the flagellar pocket and is frequently less prominent, suggesting that it is the new Golgi (Figures 6 and 7). Similar organization is found in PCF cells, but the old and new ERES:Golgi junctions are spatially much closer along the longitudinal axis of the cell. This may be a simple consequence of repositioning of the flagellar pocket closer to the nucleus in PCF (Matthews, 1999), thereby compressing the organization of the early secretory pathway. Alternatively, these morphological differences may influence replication of these secretory organelles in a stage-specific manner.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to laboratory members past and present, to Dr. Ron Peck and Kevin Schwartz for contribution to the early phase of this work, and to Dr. Sebastian Bednarek for thoughtful discussion and comments. We are particularly indebted to Dr. Wandy Beatty (Washington University-St. Louis) for assistance with electron microscopy and to Dr. James Keck (University of Wisconsin-Madison) for assistance with molecular modeling. We also thank Drs. Graham Warren and Keith Gull for generous gifts of antibodies. This work was supported by U.S. Public Health Service grant R01 AI-35739 (to J.D.B.). E.S.S. was supported by National Institutes of Health Microbial Pathogenesis and Host Responses training grant (T32 AI-055397) to University of Wisconsin-Madison.
Glossary
Abbreviations used:
- COPII
coat protein complex II
- ERES
endoplasmic reticulum exit site(s)
- FAZ
flagellar attachment zone
- GPI
glycosylphosphatidylinositol
- VSG
variant surface glycoprotein.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-07-0542) on September 16, 2009.
1In our prior publications, we have used the term “trypanopain” to refer to TbbCATL as a generic name for this cathepsin L orthologue in all T. brucei subspecies. In accord with the recent proposal of Caffrey and Steverding (2009), we now adopt this universal nomenclature.
REFERENCES
- Alexander D. L., Schwartz K. J., Balber A. E., Bangs J. D. Developmentally regulated trafficking of the lysosomal membrane protein p67 in Trypanosoma brucei. J. Cell Sci. 2002;115:3252–3262. doi: 10.1242/jcs.115.16.3253. [DOI] [PubMed] [Google Scholar]
- Bagnat M., Keranen S., Shevchenko A., Shevchenko A., Simons K. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc. Natl. Acad. Sci. USA. 2000;97:3254–3259. doi: 10.1073/pnas.060034697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangs J. D., Andrews N. W., Hart G. W., Englund P. T. Post-translational modification and intracellular transport of a trypanosome variant surface glycoprotein. J. Cell Biol. 1986;103:255–263. doi: 10.1083/jcb.103.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangs J. D., Brouch E. M., Ransom D., Roggy J. L. A soluble secretory reporter system in Trypanosoma brucei. J. Biol. Chem. 1996;271:18387–18393. doi: 10.1074/jbc.271.31.18387. [DOI] [PubMed] [Google Scholar]
- Bangs J. D., Doering T., Englund P., Hart G. Biosynthesis of a variant surface glycoprotein of Trypanosoma brucei. J. Biol. Chem. 1988;263:17697–17705. [PubMed] [Google Scholar]
- Bangs J. D., Herald D., Krakow J. L., Hart G. W., Englund P. T. Rapid processing of the carboxyl terminus of a trypanosome variant surface glycoprotein. Proc. Natl. Acad. Sci. USA. 1985;82:3207–3211. doi: 10.1073/pnas.82.10.3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangs J. D., Uyetake L., Brickman M. J., Balber A. E., Boothroyd J. C. Molecular cloning and cellular localization of a BiP homologue in Trypanosoma brucei. Divergent ER retention signals in a lower eukaryote. J. Cell Sci. 1993;105:1101–1113. doi: 10.1242/jcs.105.4.1101. [DOI] [PubMed] [Google Scholar]
- Barlowe C., Orci L., Yeung T., Hosobuchi M., Hamamoto S., Salama N., Rexach M., Ravazzola M., Amherdt M., Schekman R. COPII: a membrane coat formed by SEC proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 1994;77:895–907. doi: 10.1016/0092-8674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- Barlowe C., Schekman R. Sec12 encodes a guanine-nucleotide exchange factor essential for transport vesicle budding from the ER. Nature. 1993;365:347–350. doi: 10.1038/365347a0. [DOI] [PubMed] [Google Scholar]
- Barr F. A., Puype M., Vanderkerckhove J., Warren G. GRASP65, a protein involved in the stacking of Golgi cisternae. Cell. 1997;91:253–262. doi: 10.1016/s0092-8674(00)80407-9. [DOI] [PubMed] [Google Scholar]
- Bastin P., Pullen T. J., Moreira-Leite F. F., Gull K. Inside and outside of the trypanosome flagellum: a multifuntional organelle. Microbes Infect. 2000;2:1865–1874. doi: 10.1016/s1286-4579(00)01344-7. [DOI] [PubMed] [Google Scholar]
- Behnia R., Barr F. A., Flanagan J. J., Barlowe C., Munro S. The yeast orthologue of Grasp65 forms a complex with a coiled-coil protein that contributes to ER to Golgi traffic. J. Cell Biol. 2007;176:255–261. doi: 10.1083/jcb.200607151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belden W. J., Barlowe C. Erv25p, a component of COPII-coated vesicles, forms a complex with Emp24p that is required for efficient endoplasmic reticulum to Golgi transport. J. Biol. Chem. 1996;271:26939–26946. doi: 10.1074/jbc.271.43.26939. [DOI] [PubMed] [Google Scholar]
- Bi X., Corpina R., Goldberg J. Structure of the Sec23/24-Sar1 pre-budding complex of the COPII vesicle coat. Nature. 2002;419:271–277. doi: 10.1038/nature01040. [DOI] [PubMed] [Google Scholar]
- Burkard G., Fragoso C. M., Roditi I. Highly efficient stable transformation of bloodstream forms of Trypanosoma brucei. Mol. Biochem. Parasitol. 2007;153:220–223. doi: 10.1016/j.molbiopara.2007.02.008. [DOI] [PubMed] [Google Scholar]
- Caffrey C. R., Steverding D. Kinetoplastid papain-like cysteine peptidases. Mol. Biochem. Parasitol. 2009;167:12–19. doi: 10.1016/j.molbiopara.2009.04.009. [DOI] [PubMed] [Google Scholar]
- Cai H., Yu S., Menon S., Cai Y., Lazarova D., Fu C., Reinisch K., Hay J. C., Ferro-Novick S. TRAPPI tethers COPII vesicles by binding the coat subunit Sec23. Nature. 2007;445:941–944. doi: 10.1038/nature05527. [DOI] [PubMed] [Google Scholar]
- Castillion G. A., Watanabe R., Taylor M., Schwabe T.M.E., Riezman H. Concentration of GPI-anchored proteins upon ER exit in yeast. Traffic. 2009;10:186–200. doi: 10.1111/j.1600-0854.2008.00857.x. [DOI] [PubMed] [Google Scholar]
- Connerly P. L., Esaki M., Montegna E. A., Strongin D. E., Levi S., Soderholm J., Glick B. S. Sec16 is a determinant of transitional ER organization. Curr. Biol. 2005;15:1439–1447. doi: 10.1016/j.cub.2005.06.065. [DOI] [PubMed] [Google Scholar]
- Cross G. A. Identification, purification and properties of clone-specific glycoprotein antigens constituting the surface coat of Trypanosoma brucei. Parasitology. 1975;71:393–417. doi: 10.1017/s003118200004717x. [DOI] [PubMed] [Google Scholar]
- Dominquez M., Dejgaard K., Füllekerug J., Dahan S., Fazel A., Paccaud J.-P., Thomas D. Y., Bergeron J.J.M., Nilsson T. gp25L/emp24/p24 protein family members of the cis Golgi network bind both COP I and II coatomer. J. Cell Biol. 1998;140:751–765. doi: 10.1083/jcb.140.4.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duszenko M., Ivanov I. E., Ferguson M. A., Plesken H., Cross G. A. Intracellular transport of a variant surface glycoprotein in Trypanosoma brucei. J. Cell Biol. 1988;106:77–86. doi: 10.1083/jcb.106.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esaki M., Liu Y., Glick B. The budding yeast Pichia pastoris has a novel Sec23p homolog. FEBS Lett. 2006;580:5215–5221. doi: 10.1016/j.febslet.2006.08.058. [DOI] [PubMed] [Google Scholar]
- Farhan H., Reiterer V., Korkhov V., Schmid J., Freissmuth M., Sitte H. Concentrative export from the endoplasmic reticulum of the γ-aminobutyric acid transporter 1 requires binding to Sec24D. J. Biol. Chem. 2007;282:7679–7689. doi: 10.1074/jbc.M609720200. [DOI] [PubMed] [Google Scholar]
- Fath S., Mancias J. D., Bi X., Goldberg J. Structure and organization of coat proteins in the COPII cage. Cell. 2007;129:1325–1336. doi: 10.1016/j.cell.2007.05.036. [DOI] [PubMed] [Google Scholar]
- Feinstein T. N., Linstedt A. D. GRASP55 regulates Golgi ribbon formation. Mol. Biol. Cell. 2008;19:2696–2707. doi: 10.1091/mbc.E07-11-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson M. A. The structure, biosynthesis and functions of glycosylphosphatidylinositol anchors, and the contributions of trypanosome research. J. Cell Sci. 1999;112:2799–2809. doi: 10.1242/jcs.112.17.2799. [DOI] [PubMed] [Google Scholar]
- Ferguson M. A., Duszenko M., Lamont G. S., Overath P., Cross G. A. Biosynthesis of Trypanosoma brucei variant surface glycoproteins, N-glycosylation and addition of a phosphatidylinositol membrane anchor. J. Biol. Chem. 1986;26:356–362. [PubMed] [Google Scholar]
- Futai R., Hamamoto S., Orci L., Schekman R. GTP/GDP exchange by Sec12p enables COPII vesicle bud formation on synthetic liposomes. EMBO J. 2004;23:4146–4155. doi: 10.1038/sj.emboj.7600428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünfelder C. G., Engstler M., Wieise F., Schwarz H., Stierhof Y. D., Morgan G. W., Field M. C., Overath P. Endocytosis of a glycosylphosphatidylinositol-anchored protein via clathrin-coated vesicles, sorting by default in endosomes and exocytosis via RAB11-positive carriers. Mol. Biol. Cell. 2003;14:2029–2040. doi: 10.1091/mbc.E02-10-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C. Y., Ho H. H., Malsam J., Chalouni C., West C. M., Ullu E., Toomre D., Warren G. Golgi duplication in Trypanosoma brucei. J. Cell Biol. 2004;165:313–321. doi: 10.1083/jcb.200311076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K. L. Biology and mechanism of trypanosome cell motility. Eukaryot. Cell. 2003;2:200–208. doi: 10.1128/EC.2.2.200-208.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirumi H., Hirumi K. Axenic culture of African trypanosome bloodstream forms. Parasitol. Today. 1994;10:81–84. doi: 10.1016/0169-4758(94)90402-2. [DOI] [PubMed] [Google Scholar]
- Ho H. H., He C. Y., de Graffenried C. L., Murrells L. J., Warren G. Ordered Assembly of the Duplicating Golgi in Trypanosoma brucei. Proc. Nat. Acad. Sci. USA. 2006;103:7676–7681. doi: 10.1073/pnas.0602595103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn D., Barry J. D. The central roles of telomeres and subtelomeres in antigenic variation in African trypanosomes. Chromosome Res. 2005;13:525–533. doi: 10.1007/s10577-005-0991-8. [DOI] [PubMed] [Google Scholar]
- Horvath A., Sutterlin C., Manning-Krieg U., Movva N. R., Riezman H. Ceremide synthesis enhances transport of GPI-anchored proteins to the Golgi apparatus in yeast. EMBO J. 1994;13:3687–3695. doi: 10.1002/j.1460-2075.1994.tb06678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes H., Stephens D. J. Assembly, organization, and function of the COPII coat. Histochem. Cell Biol. 2008;129:129–151. doi: 10.1007/s00418-007-0363-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenne N., Frey K., Brugger B., Wieland F. T. The absence of Emp24p, a component of ER-derived COPII-coated vesicles, causes a defect in transport of selected proteins to the Golgi. EMBO J. 1995;14:1329–1339. doi: 10.1002/j.1460-2075.1995.tb07119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenne N., Frey K., Brugger B., Wieland F. T. Oligomeric state and stoichiometry of p24 proteins in the early secretory pathway. J. Biol. Chem. 2002;277:46504–46511. doi: 10.1074/jbc.M206989200. [DOI] [PubMed] [Google Scholar]
- Kinseth M. A., Anjard C., Fuller D., Guizzunti G., Loomis W. F., Malhotra V. The Golgi-associated protein GRASP is required for unconventional protein secretion during development. Cell. 2007;130:524–534. doi: 10.1016/j.cell.2007.06.029. [DOI] [PubMed] [Google Scholar]
- Kohl L., Sherwin T., Gull K. Assembly of the paraflagellar rod and the flagellum attachment zone complex during the Trypanosoma brucei cell cycle. J. Eukaryot. Microbiol. 1999;46:105–109. doi: 10.1111/j.1550-7408.1999.tb04592.x. [DOI] [PubMed] [Google Scholar]
- Kuehn M. J., Herrmann J. M., Schekman R. COPII-cargo interactions direct protein sorting into ER-derived transport vesicles. Nature. 1998;391:187–190. doi: 10.1038/34438. [DOI] [PubMed] [Google Scholar]
- Kuehn M. J., Schekman R., Ljungdahl P. Amino acid permeases require COPII components and the ER resident membrane protein Shr3 for packaging into transport vesicles in vitro. J. Cell Biol. 1996;135:585–595. doi: 10.1083/jcb.135.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuge O., Dascher C., Orci L., Rowe T., Amherdt M., Plutner H., Ravazzola M., Tanigawa G., Rothman J. E., Balch W. E. Sar1 promotes vesicle budding from the endoplasmic reticulum but not Golgi compartments. J. Cell Biol. 1994;125:51–65. doi: 10.1083/jcb.125.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara T., Hamamoto S., Gimeno R., Kaiser C., Schekman R., Yoshihisa T. Sec24p and Iss1p function interchangeably in transport vesicle formation from the endoplasmic reticulum in Saccharomyces cerevisiae. Mol. Biol. Cell. 2000;11:983–998. doi: 10.1091/mbc.11.3.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCount D. J., Bruse S., Hill K. L., Donelson J. E. Double-stranded RNA interference in Trypanosoma brucei using head-to-head promoters. Mol. Biochem. Parasitol. 2000;111:67–76. doi: 10.1016/s0166-6851(00)00300-5. [DOI] [PubMed] [Google Scholar]
- Lang M. R., Lapierre L. A., Frotscher M., Goldenring J. R., Knapik E. W. Secretory COPII coat component Sec23a is essential for craniofacial chondrocyte maturation. Nat. Gene. 2006;38:1198–1203. doi: 10.1038/ng1880. [DOI] [PubMed] [Google Scholar]
- Lippencott-Schwartz J., Roberts T. H., Hirschberg K. Secretory protein trafficking and organelle dynamics in living cells. Annu. Rev. Cell Dev. Biol. 2000;16:557–589. doi: 10.1146/annurev.cellbio.16.1.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancias J. D., Goldberg J. The transport signal on Sec22 for packaging into COPII coated vesicles is a conformational epitope. Traffic. 2007;6:403–414. doi: 10.1016/j.molcel.2007.03.017. [DOI] [PubMed] [Google Scholar]
- Marzioch M., Henthorn D. C., Herrmann J. M., Wilson R., Thomas D. Y., Bergeron J.J.M., Solari R.C.E., Rowley A. Erp1p and Erp2p, partners for Emp24p and Erv25p in a yeast p24 complex. Mol. Biol. Cell. 1999;10:1923–1938. doi: 10.1091/mbc.10.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka K., Orci L., Amherdt M., Bednarek S. Y., Hamamoto S., Schekman R., Yeung T. COPII-coated vesicle formation reconstituted with purified coat proteins and chemically defined liposomes. Cell. 1998;93:263–275. doi: 10.1016/s0092-8674(00)81577-9. [DOI] [PubMed] [Google Scholar]
- Matthews K. R. Developments in the differentiation of Trypanosoma brucei. Parasitol. Today. 1999;15:76–80. doi: 10.1016/s0169-4758(98)01381-7. [DOI] [PubMed] [Google Scholar]
- Miller E. A., Beilharz T. H., Malkus P. N., Lee M., Hamamoto S., Orci L., Schekman R. Multiple cargo binding sites on the COPII subunit Sec24p ensure capture of diverse membrane proteins into transport vesicles. Cell. 2003;114:497–509. doi: 10.1016/s0092-8674(03)00609-3. [DOI] [PubMed] [Google Scholar]
- Miller E. A., Liu Y., Barlowe C., Schekman R. ER-Golgi transport defects are associated with mutations in the Sed5p-binding domain of the COPII coat subunit, Sec24p. Mol. Biol. Cell. 2005;16:3719–3726. doi: 10.1091/mbc.E05-03-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morriswood B., He C. Y., Sealey-Cardona M., Yelinek J., Pypaert M., Warren G. The bilobe structure of Trypanosoma brucei contains a MORN-repeat protein. Mol. Biochem. Parasitol. 2009;167:95–103. doi: 10.1016/j.molbiopara.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Mossessova E., Bickford L., Goldberg J. SNARE selectivity of the COPII coat. Cell. 2003;114:483–495. doi: 10.1016/s0092-8674(03)00608-1. [DOI] [PubMed] [Google Scholar]
- Muniz M., Morsomme P., Riezman H. Protein sorting upon exit from the endoplasmic reticulum. Cell. 2001;104:313–320. doi: 10.1016/s0092-8674(01)00215-x. [DOI] [PubMed] [Google Scholar]
- Nakano A., Muramatsu M. A novel GTP-binding protein, Sar1p, is involved in transport from the endoplasmic reticulum to the Golgi apparatus. J. Cell Biol. 1989;109:2677–2691. doi: 10.1083/jcb.109.6.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paccaud J. P., Reith W., Carpentier J. L., Ravazzolla M., Amherdt M., Schekman R. W., Orci L. Cloning and functional characterization of mammalian homologues of the COPII component Sec23. Mol. Biol. Cell. 1996;7:1535–1546. doi: 10.1091/mbc.7.10.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck R. F., Shiflett A. M., Schwartz K. J., McCann A., Hajduk S. L., Bangs J. D. The LAMP-like protein p67 plays an essential role in the lysosome of African trypanosomes. Mol. Microbiol. 2008;68:933–946. doi: 10.1111/j.1365-2958.2008.06195.x. [DOI] [PubMed] [Google Scholar]
- Roditi I., Clayton C. An unambiguous nomenclature for the major surface glycoproteins of the procyclic form of Trypanosoma brucei. Mol. Biochem. Parasitol. 1999;103:99–100. doi: 10.1016/s0166-6851(99)00124-3. [DOI] [PubMed] [Google Scholar]
- Rossanese O. W., Soderholm J., Bevis B. J., Sears I. B., O'Connor J., Williamson E. K., Glick B. S. Golgi structure correlates with transitional endoplasmic reticulum organization in Pichia pastoris and Saccharomyces cerevisiae. J. Cell Biol. 1999;145:69–81. doi: 10.1083/jcb.145.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmoler F., Singer-Kruger B., Schroder S., Kruger U., Barlowe C., Riezman H. The absence of Emp24p, a component of ER-derived COPII-coated vesicles, causes a defect in transport of selected proteins to the Golgi. EMBO J. 1995;14:1329–1339. doi: 10.1002/j.1460-2075.1995.tb07119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz K. J., Peck R. F., Tazeh N. N., Bangs J. D. GPI valence and the fate of secretory membrane proteins in African trypanosomes. J. Cell Sci. 2005;118:5499–5511. doi: 10.1242/jcs.02667. [DOI] [PubMed] [Google Scholar]
- Shen K. A., Hammond C. M., Moore H. H. Molecular analysis of SAR1-related cDNAs from a mouse pituitary cell line. FEBS Lett. 1993;335:380–385. doi: 10.1016/0014-5793(93)80423-r. [DOI] [PubMed] [Google Scholar]
- Shen S., Arhin G. K., Ullu E., Tschudi C. In vivo epitope tagging of Trypanosoma brucei genes using a one step PCR strategy. Mol. Biochem. Parasitol. 2001;113:171–173. doi: 10.1016/s0166-6851(00)00383-2. [DOI] [PubMed] [Google Scholar]
- Shimoni Y., Kurihara T., Ravazzola M., Amherdt M., Orci L., Schekman R. Lst1 and Sec24p cooperate in sorting of the plasma membrane ATPase into COPII vesicles in Saccharomyces cerevisiae. J. Cell Biol. 2000;151:973–984. doi: 10.1083/jcb.151.5.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J., Warren G. Golgi architecture and inheritance. Annu. Rev. Cell Dev. Biol. 2002;18:379–420. doi: 10.1146/annurev.cellbio.18.030602.133733. [DOI] [PubMed] [Google Scholar]
- Stankewich M. C., Stabach P. R., Morrow J. S. Human Sec31B: a family of new mammalian orthologues of yeast Sec31p that associate with the COPII coat. J. Cell Sci. 2006;119:958–969. doi: 10.1242/jcs.02751. [DOI] [PubMed] [Google Scholar]
- Stephens D. J. De novo formation, fusion and fission of mammalian COPII-coated endoplasmic reticulum exit sites. EMBO J. 2003;4:210–217. doi: 10.1038/sj.embor.embor736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strating J. R., van Bakel N. N., Leunissen J. A., Martens G. J. A comprehensive overview of the vertebrate p24 family: identification of a novel tissue-specifically expressed member. Mol. Biol. Evol. 2009;26:1707–1714. doi: 10.1093/molbev/msp099. [DOI] [PubMed] [Google Scholar]
- Sutterlin C., Doering T. L., Schimmoler F., Schroder S., Riezman H. Specific requirements for the ER to Golgi transport of GPI-achored proteins in yeast. J. Cell Sci. 1997;110:2703–2714. doi: 10.1242/jcs.110.21.2703. [DOI] [PubMed] [Google Scholar]
- Sutterwala S. S., Creswell C. H., Sanyal S., Menon A. K., Bangs J. D. De novo sphingolipid synthesis is essential for viability, but not for transport of glycosylphosphatidylinositol anchored proteins, in African trypanosomes. Eukaryot. Cell. 2007;6:454–464. doi: 10.1128/EC.00283-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutterwala S. S., Hsu F. F., Sevova E. S., Schwartz K. J., Zhang K., Key P., Turk J., Beverley S. M., Bangs J. D. Developmentally regulated sphingolipid synthesis in African trypanosomes. Mol. Microbiol. 2008;70:281–296. doi: 10.1111/j.1365-2958.2008.06393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takida S., Maeda Y., Kinoshita T. Mammalian GPI-anchored proteins require p24 proteins for their efficient transport from the ER to the plasma membrane. Biochem. J. 2008;409:555–562. doi: 10.1042/BJ20070234. [DOI] [PubMed] [Google Scholar]
- Tang B., Kausalya J., Low D.Y.H., Lock M. L., Hong W. A family of mammalian proteins homologous to yeast Sec24p. Biochem. Biophys. Res. Commun. 1999;258:679–684. doi: 10.1006/bbrc.1999.0574. [DOI] [PubMed] [Google Scholar]
- Tazeh N. N., Bangs J. D. Multiple motifs regulate trafficking of the LAMP-like protein p67 in the ancient eukaryote Trypanosoma brucei. Traffic. 2007;8:1007–1017. doi: 10.1111/j.1600-0854.2007.00588.x. [DOI] [PubMed] [Google Scholar]
- Triggs V. P., Bangs J. D. Glycosylphosphatidylinositol-dependent protein trafficking in bloodstream stage Trypanosoma brucei. Eukaryot. Cell. 2003;2:76–83. doi: 10.1128/EC.2.1.76-83.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan S., Kohl L., Ngai I., Wheeler R. J., Gull K. A repetitive protein essential for the flagellum attachment zone filament structure and function in Trypanosoma brucei. Protist. 2008;159:127–136. doi: 10.1016/j.protis.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Wendeler M. W., Paccaud J. P., Hauri H. P. Role of Sec24 isoforms in selective export of membrane proteins from the endoplasmic reticulum. EMBO Rep. 2006;8:258–264. doi: 10.1038/sj.embor.7400893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz E., Leal S., Ochatt C., Cross G. A tightly regulated inducible expression system for conditional gene knockouts and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 1999;99:89–1001. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.