Abstract
Yin-Yang 1 (YY1) is a ubiquitously expressed zinc finger transcription factor. It regulates a vast array of genes playing critical roles in development, differentiation, and cell cycle. Very little is known about the mechanisms that regulate the functions of YY1. It has long been proposed that YY1 is a phosphoprotein; however, a direct link between phosphorylation and the function of YY1 has never been proven. Investigation of the localization of YY1 during mitosis shows that it is distributed to the cytoplasm during prophase and remains excluded from DNA until early telophase. Immunostaining studies show that YY1 is distributed equally between daughter cells and rapidly associates with decondensing chromosomes in telophase, suggesting a role for YY1 in early marking of active and repressed genes. The exclusion of YY1 from DNA in prometaphase HeLa cells correlated with an increase in the phosphorylation of YY1 and loss of DNA-binding activity that can be reversed by dephosphorylation. We have mapped three phosphorylation sites on YY1 during mitosis and show that phosphorylation of two of these sites can abolish the DNA-binding activity of YY1. These results demonstrate a novel mechanism for the inactivation of YY1 through phosphorylation of its DNA-binding domain.
INTRODUCTION
Yin-Yang 1 (YY1) is a ubiquitously expressed transcription factor that has been implicated in the regulation of a large number of genes critical for basic processes of development, cell growth, differentiation, cell cycle, and even apoptosis. The essential role of YY1 is underscored by the fact that its deletion resulted in peri-implantation lethality in mice, and disruption of only one allele caused severe developmental abnormalities (Donohoe et al., 1999). A substantial amount of information has been compiled over the past decade about the wide variety of target genes regulated by YY1; however, the regulation of the various functions of YY1 remains enigmatic.
The expression of the human yy1 gene is under the control of a promoter that contains three Sp1-binding sites but neither a TATA box nor a CCAAT box, thus classifying YY1 among the constitutively expressed house keeping genes (Yao et al., 1998). Although higher YY1 expression levels have been detected in some types of cancers (Erkeland et al., 2003; Seligson et al., 2005; de Nigris et al., 2006), this type of regulation is not commonly observed for YY1. Other modes of regulation have been proposed including localization, cleavage, interaction with other proteins, and posttranslational modification. Regulation of YY1 through changes in subcellular localization of YY1 into the cytoplasm has been reported during early stages of development in Xenopus (Ficzycz et al., 2001), muscle cell differentiation (Delehouzee et al., 2005), G1/S transition of the mammalian cell cycle (Palko et al., 2004), and apoptosis (Krippner-Heidenreich et al., 2005). YY1 has also been shown to be cleaved under certain conditions of muscle cell differentiation (Walowitz et al., 1998) and apoptosis (Krippner-Heidenreich et al., 2005). Several proteins have been shown to physically interact with YY1 and regulate its DNA binding or transcriptional activity. For example, YY1 was shown to reside in a complex with the retinoblastoma protein (Rb) in the early stages of smooth muscle cell differentiation, preventing YY1 from binding to DNA (Petkova et al., 2001; Hiromura et al., 2003). Overexpression of the viral oncoprotein E1A results in relief of YY1 repression on the adeno-associated virus (AAV) P5 promoter. The interaction between E1A and YY1 switches YY1 from a repressor into an activator of gene expression on this promoter (Shi et al., 1991).
Several types of posttranslational modifications have been reported to occur on YY1, including acetylation (Yao et al., 2001; Takasaki et al., 2007), O-linked glycosylation (Hiromura et al., 2003), sumoylation (Deng et al., 2007), poly(ADP-ribosyl)ation (Oei et al., 1997; Oei et al., 1998), and S-nitrosation (Hongo et al., 2005).
The involvement of YY1 in proliferation has been proposed since its identification (Shi et al., 1991). Proof for this has been provided by accumulated data showing physical and functional interactions between YY1 and key regulators of the cell cycle including Rb (Petkova et al., 2001; Delehouzee et al., 2005), c-Myc (Riggs et al., 1993; Shrivastava et al., 1993, 1996), and p53 (Gronroos et al., 2004; Sui et al., 2004; Yakovleva et al., 2004). Moreover, YY1 has been shown to regulate genes in a cell cycle–dependent manner. At the G1/S transition of the cell cycle, YY1 plays a critical role in the up-regulation of the replication-dependent histone gene H3.2 (Eliassen et al., 1998) and the Cdc6 gene (cell division cycle 6 homolog; Schlisio et al., 2002). YY1 was also shown to up-regulate a set of genes needed upon exit from mitosis and entry into G1 (Elkon et al., 2003). Furthermore, RNAi-mediated inhibition of YY1 in HeLa cells resulted in proliferation arrest and severe cytokinesis defects (Affar el et al., 2006).
The regulation of YY1 at different stages of the cell cycle is poorly understood and has not been studied in mitosis. In this study, we investigate the distribution of YY1 during cell division and reveal a novel mechanism for the regulation of YY1 through phosphorylation.
MATERIALS AND METHODS
Cell Culture
HeLa S3 cells were grown at 37°C in 5% CO2 in DMEM (Cellgro, Herndon, VA) supplemented with 10% fetal bovine serum (FBS; Mediatech, Herndon, VA), 1% nonessential amino acids (Sigma, St. Louis, MO), and 1% penicillin-streptomycin (Mediatech). Cells were trypsinized and split into new plates at subconfluency every other day.
Plasmid Construction
pET-20b(+)-YY1.
The NcoI/EcoRI fragment encompassing the open reading frame of human YY1 was subcloned from pCMV-HA-YY1 (a gift from Dr. Bernhard Lüscher, Aachen University, Germany; Austen et al., 1997) into the NcoI/EcoRI site in the multiple cloning region of pET-20b(+) vector (Invitrogen, Carlsbad, CA).
pCS2(+)-Flag-YY1.
pCS2(+)-Flag was a generous gift from Dr. Yoichi Kato (Biomedical Sciences Department, Florida State University). pCS2(+)-Flag was constructed by inserting the Flag sequence in the BamHI site of pCS2(+) vector (Turner and Weintraub, 1994). For the construction of pCS2(+)-Flag-YY1, the NcoI/EcoRI fragment was excised from pET-20b(+)-YY1 after blunting the NcoI site and inserted into pCS2(+)-Flag digested with BamHI/EcoRI after blunting the BamHI site.
Mutagenesis
Point mutants of YY1 at serine 247, threonine 348, and threonine 378 residues to aspartic acid and alanine were generated using QuikChange II Site-Directed Mutagenesis Kit from Stratagene (La Jolla, CA). Mutagenesis was performed according to manufacturer's instructions, using the human YY1 open reading frame in pET-20b(+)-YY1 plasmid as a template. Primers were designed using the QuikChange Primer Design Program on the Stratagene Web site. All mutations were confirmed by sequencing. The mutated YY1 sequences were then subcloned from pET-20b(+) into pCS2(+)-Flag vector as described above.
Transfections and Stable Cell Line Generation
The plasmid of interest was transfected into HeLa S3 cells (grown as described earlier) using Lipofectamine transfection reagent (Invitrogen), according to manufacturer's instructions. For the generation of a HeLa cell line stably overexpressing Flag-YY1, HeLa S3 cells were cotransfected with pCS2(+)-Flag-YY1 and the selection marker pSV-Neo plasmid at a ratio of 15:1. Individual colonies of stable transfectant HeLa cells were selected by G418 addition as previously described (Hurt et al., 1991) and tested for Flag-YY1 expression. The stable cell line selected for this study is referred to here as HeLa-Flag-YY1.
Cell Synchronization
To synchronize HeLa cells at G1/S, a double-thymidine arrest was performed as previously described (Whitfield et al., 2000). For endogenous YY1 immunostaining in the different phases of mitosis, HeLa cells were grown on coverslips, blocked with double-thymidine arrest, and then released into fresh medium. Samples were collected at 7–9 h after release. To arrest cells in S-phase, single-thymidine arrest was performed by adding thymidine to a final concentration of 2 mM for 18 h. To synchronize cells at prometaphase, nocodazole (Sigma) was added to the medium to a final concentration of 50 ng/ml for 18 h. For the nocodazole arrest/release experiment, cells were synchronized with nocodazole, as described above. Mitotic cells were detached from the plate surface by tapping the plate and collected by aspiration. Cells were washed two times with PBS and then one time with medium and replated in fresh growth medium. Samples were collected at indicated times for preparation of whole-cell extracts (WCEs).
WCE Preparation
HeLa cells were washed three times with ice-cold PBS. Cells were then scraped in freshly prepared ice-cold lysis buffer (20 mM HEPES, pH 7.5, 150 mM NaCl, 0.5% Triton-X 100, 1 mM EDTA, 2.5 mM EGTA, 2 mM DTT, 1 mM sodium orthovanadate, 10 mM NaF, and 25 mM β-glycerophosphate), supplemented with a cocktail of protease inhibitors (Sigma). Cells were lysed on ice for 15 min. Lysates were then pipetted up and down several times to shear DNA and centrifuged at 18,000 × g for 15 min at 4°C.
Immunoprecipitation
Immunoprecipitation (IP) of YY1 was performed using anti-YY1 (C-20) rabbit polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA). WCEs were incubated with antibody for 3 h, rotating at 4°C. Protein A-agarose beads (Santa Cruz Biotechnology) were then added to the mixture and incubated for an additional hour. Immune complexes bound to the beads were collected by centrifugation at 500 × g at 4°C for 2 min and then washed three times with lysis buffer. SDS-PAGE buffer was added to the beads and boiled for 5 min. For IP of Flag-YY1 with anti-Flag antibody, the same procedure was followed, except that anti-Flag mouse mAb cross-linked to resin beads (Resin M2, Sigma) was added to the lysates and the mixture was incubated for 4 h at 4°C with rotation.
Purification of Flag-YY1 and Phosphatase Assay
HeLa-Flag-YY1 cells were blocked at prometaphase with nocodazole as described above. WCEs were prepared, followed by Flag-YY1 IP as described above. Resin M2-Flag-YY1 complex was washed three times with lysis buffer and one additional time with lysis buffer without phosphatase inhibitors. Flag-YY1 was eluted from the beads by addition of elution buffer (50 mM Tris, pH 7.5, 150 mM NaCl, and 200 μg/ml Flag-peptide) for 5–10 min at 4°C, with shaking. Equal aliquots of the purified Flag-YY1 were then incubated at 30°C with or without λ-phosphatase (New England BioLabs, Beverly, MA) for 30 min, in the presence or absence of phosphatase inhibitors (10 mM NaF and 25 mM β-glycerophosphate), before the electrophoretic mobility shift assay (EMSA).
Western Blotting
Protein samples were separated on SDS-PAGE gels and then transferred by electroblotting onto a nitrocellulose membrane (Hybond-C extra, GE Healthcare, Piscataway, NJ). The membrane was blocked for 30 min at RT in blocking solution (PBS, 0.5% Tween-20, and 5% nonfat dry milk). Mouse monoclonal (H-10), rabbit polyclonal (H-414) anti-YY1 antibodies, mouse monoclonal anti-cyclin B antibody, or mouse monoclonal anti-GAPDH (Santa Cruz Biotechnology) were added to the membrane, in blocking solution, for 2 h at room temperature (RT) or overnight at 4°C. The membrane was washed three times for 10 min with PBST. Horseradish peroxidase (HRP)-conjugated anti-mouse and anti-rabbit secondary antibodies (GE Healthcare, Waukesha, WI) were then added to the membrane in blocking solution and incubated for an hour at RT, after which it was washed three times as above. Specific protein bands were detected by the addition of SuperSignal West Pico Chemiluminescent Substrate (Pierce, Rockford, IL) for 5 min and exposure to X-ray film (Fuji Medical Systems, Stamford, CT).
EMSA
Double-stranded DNA oligonucleotides were end-labeled using T4-polynucleotide kinase (New England Biolabs) and [γ-32P]ATP (Perkin Elmer-Cetus, Boston, MA). EMSA conditions were as previously described (Eliassen et al., 1998), except that the native gels were 6% polyacrylamide and the dry gels were exposed to a phosphorimager screen and scanned using a Typhoon 9410 Imager (GE Healthcare). The DNA probes used were as follows: H3.2α: 5′-GATCCTCGGCCGTCATGGCGCTGCAGGAGGCA-3′ (Eliassen et al., 1998), adeno-associated virus (AAV) promoter (p5-60): 5′-GATCCGTTTTGCGACATTTTGCGACACA-3′ (Shi et al., 1991), and Cdc6 promoter region (Cdc6p): 5′-GATCTGTGGCCATTCGGATTTGGCGCGCGAG-3′ (Schlisio et al., 2002). To test the specificity of the YY1 shift, 1 μg of anti-YY1 (C-20), anti-green fluorescent protein (GFP), or anti-Sp1 antibodies (Santa Cruz Biotechnology) were added to the binding reactions.
Indirect Immunofluorescence
For indirect immunofluorescence, cells grown on coverslips were washed three times with PBS, fixed with 3.7% formaldehyde for 10 min RT, and then washed three times with PBS. Cells were permeabilized for 10 min at RT with PBS containing 0.2% Triton-X-100 and subsequently were washed three times with PBS. Immunostaining was performed by overlaying the coverslips with blocking solution (PBST, 1% IgG-free BSA) for 30 min at 37°C. Primary antibodies were then added to the coverslips, in blocking solution, and incubated for 1 h at 37°C. Anti-YY1 (H-414, Santa Cruz Biotechnology) and anti-Flag antibody (Sigma) were added at a final concentration of 1 μg/ml. Anti-lamin A/C (Santa Cruz Biotechnology) was added at 0.2 μg/ml. Coverslips were then washed three times with PBST, and anti-rabbit Alexa-Fluor 546 and anti-mouse Alexa-Fluor 647 (Molecular Probes, Eugene, OR) secondary antibodies (2 μg/ml in blocking solution) were then applied to the coverslips and incubated for 1 h at 37°C. After washing three times with PBST, cells were overlaid with DAPI solution (2 μg/ml in PBS) for 5 min, washed briefly, and mounted in Vectashield (Vector Laboratories, Burlingame, CA). Images were captured using a confocal microscope (Leica Microsystems, Exton, PA), taking 1-μm sections of the cells. The overlay images were generated using Leica LCS Lite Software.
In Vivo Labeling of YY1
An asynchronous population of HeLa cells was starved for phosphate for 30 min in phosphate-free medium (DMEM, Cellgro). 32P-Orthophosphate was added to a final concentration of 1 mCi/ml and incubated with the cells for 2 h. After washing three times in ice-cold PBS, WCE was prepared, and endogenous YY1 was immunoprecipitated with anti-YY1 (C-20) antibody. To label Flag-YY1 in vivo, HeLa cells were transiently transfected with pCS2(+)-Flag-YY1 in parallel with negative control cells that did not receive DNA. Eighteen hours after transfection, the 32P-orthophosphate labeling procedure described above was followed, except that the labeling time was extended to 4 h to enhance the signal. Flag-YY1 was immunoprecipitated using rabbit polyclonal anti-Flag antibody (Sigma), followed by IP with anti-YY1 (C-20) antibody. SDS-PAGE buffer was added to the beads and boiled for 5 min. The immunoprecipitated proteins were separated on a 10% SDS-PAGE gel and then transferred to a nitrocellulose membrane. The membrane was first exposed to a Phosphorimager screen and then probed with anti-YY1 (H-10) antibody. The screen was scanned using a Typhoon 9410 Imager (GE Healthcare).
Phosphospecific Protein Staining
Flag-YY1 was immunoprecipitated from asynchronous or nocodazole-arrested HeLa-Flag-YY1 cells. After boiling in SDS-PAGE buffer, the immunoprecipitated complex was loaded and separated on a 10% SDS-PAGE gel. The fluorescent phosphospecific stain ProQ Diamond (Molecular Probes) was applied according to the manufacturer's instructions. The fluorescent signal was captured on a Typhoon 9410 Imager (GE Healthcare).
Mass Spectrometric Analysis
Flag-YY1 was immunoprecipitated from nocodazole-arrested HeLa-Flag-YY1 cells. The immunoprecipitated proteins were separated on a 10% SDS-PAGE gel and stained with Coomassie blue. The Flag-YY1 band was excised from the gel and analyzed at the UNC-Duke Proteomics Center. Briefly, processing of Flag-YY1 was as follows: samples were submitted as in gel bands, which were tryptic-digested and extracted as previously described (Parker et al., 2005). The gel extracts were lyophilized and resuspended in water. The samples were enriched for phosphopeptides using Perkin Elmer-Cetus's PhosTrap beads and analyzed directly on the MALDI target (Raska et al., 2003), using a Bruker Reflex II MALDI TOF/TOF mass spectrometer (Marlborough, MA), operated in the positive ion, reflectron mode. The mass spectrometer was scanned over the mass range 739-4560 Da. The matrix used was DHB (2,5-dihydroxybenzoic acid). For comparison, an aliquot of the supernatant, depleted of phosphopeptides, was also analyzed in the same matrix.
Fluorescence-activated Cell Sorter Analysis
Asynchronous or synchronized HeLa cells were trypsinized, washed two times with PBS, and then fixed in 70% ethanol on ice for at least 2 h. After washing off the ethanol, cells were resuspended in propidium iodide (PI) solution (50 μg/ml PI, 200 μg/ml RNase A, 0.1% Triton-X 100 in PBS) and incubated for 30 min at 30°C. The cell suspension was then passed through a 50-μm nylon mesh to remove clumps. Cells were then analyzed based on DNA content on a fluorescence-activated cell sorter (FACS; FACS Canto; Becton Dickinson, San Jose, CA), and images were generated using BD FACS Diva software.
RESULTS
Changes in Subcellular Distribution of YY1 during Mitosis
YY1 is primarily a nuclear protein, in accordance with its function as a transcription factor. Previous studies have reported changes in the subcellular localization of YY1 during certain stages of development (Ficzycz et al., 2001), differentiation (Delehouzee et al., 2005), and G1/S transition of the cell cycle (Palko et al., 2004). The localization and distribution of YY1 during mitosis, in the absence of the nuclear envelope and relative to the condensed chromosomes, have not been reported. To examine the subcellular distribution of YY1 during mitosis, HeLa cells were synchronized in early S-phase and then released back into the cell cycle. Cells were fixed for immunostaining of YY1 at 7–9 h after release. The nuclear envelope was visualized by immunostaining with anti-lamin A/C antibody, and the DNA was stained with DAPI. The physical appearance of the DNA was used to define the different stages of mitosis (Mitchison and Salmon, 2001; Pines and Rieder, 2001; Watrin and Legagneux, 2003).
As shown in Figure 1, YY1 is primarily nuclear at the G2/M transition of the cell cycle, colocalizing with the DAPI signal of the uncondensed DNA. As cells proceed into prophase and DNA appears to condense into chromosomes, YY1 is dispersed into the cytoplasm. By the end of prophase, YY1 appears to be homogenously dispersed throughout the cell. This dispersed pattern of YY1 is maintained in prometaphase, metaphase, and anaphase, and YY1 appears to be excluded from the condensed DNA. In telophase, the YY1 signal colocalizes with the decondensing chromosomes very rapidly. The distribution of YY1 into the cytoplasm correlates with the breakdown of the nuclear envelope in prophase, as is observed in Figure 1. The return of YY1 back to the DNA during telophase appears to precede the complete formation of the nuclear envelope (Supplementary Figure S1).
Figure 1.
Subcellular distribution of YY1 changes during mitosis. Indirect immunofluorescence of HeLa cells during mitosis. Endogenous YY1 (red) was immunostained with anti-YY1 antibody (H-414). The nuclear envelope was visualized by immunostaining of lamin A/C (green), followed by DAPI staining of DNA (blue). The different phases of mitosis were determined based on DNA conformation and nuclear envelope breakdown and reassembly (Mitchison and Salmon, 2001; Pines and Rieder, 2001; Watrin and Legagneux, 2003). Bar, 5 μm. Further analysis of telophase is presented in Supplementary Figure S1.
Loss of YY1 DNA-binding Activity in Prometaphase HeLa Cells
To determine whether the change in subcellular distribution of YY1 in prophase correlates with loss of YY1's DNA-binding activity, WCEs were prepared from asynchronous, S-phase, and prometaphase HeLa cells. S-phase cells were obtained by thymidine arrest and prometaphase cells were obtained by nocodazole arrest. The DNA-binding activity of YY1 in these extracts was tested in an in vitro EMSA. A radioactively labeled double-stranded oligonucleotide, encompassing the YY1-binding site in histone H3.2 coding region (H3.2α) was used in this assay (Eliassen et al., 1998). The YY1 protein in the WCEs from asynchronously growing and thymidine-blocked HeLa cells was able to bind the H3.2α probe efficiently. However, the DNA-binding activity in extracts from nocodazole-arrested HeLa cells was greatly decreased (Figure 2A). To confirm that the observed shift is the result of the YY1 binding to H3.2α, anti-YY1 (C-20) antibody was incubated with the binding reaction using asynchronous WCE. This antibody specifically recognizes and binds to the DNA-binding domain of YY1, interfering with its DNA-binding activity (Eliassen et al., 1998). As shown in Figure 2A addition of Anti-YY1(C-20) antibody significantly decreased the shift. Addition of antibodies against GFP or Sp1 had no effect on the shift. This experiment was repeated several times, and decreased binding activity of YY1 was always observed in nocodazole-arrested cells. It has been previously reported that the YY1 expression and protein levels do not fluctuate significantly during the various stages of the mammalian cell cycle (Whitfield et al., 2002; Palko et al., 2004; Wilkinson et al., 2006). However, to exclude the possibility that YY1 is degraded in nocodazole-arrested cells, samples from the WCEs were analyzed on a Western blot, using monoclonal anti-YY1 specific antibody (H-10). As shown in Figure 2B, comparable levels of YY1 protein were detected in all three cell extracts. The levels of cyclin B were elevated in nocodazole-arrested cells, as expected (Sullivan and Morgan, 2007). GAPDH was used to show equal loading. In addition, the cell cycle distribution of asynchronous, thymidine-arrested, or nocodazole-arrested HeLa cells was confirmed by FACS analysis (Figure 2C).
Figure 2.
YY1 DNA-binding activity is lost in prometaphase. WCEs were prepared from asynchronous (Asy), nocodazole-arrested (Noc), or thymidine-arrested HeLa cells (Th), separated by SDS-PAGE, and analyzed by Western blot using anti-YY1 (H-10), anti-cyclin B, or anti-GAPDH antibodies (A) and electrophoretic mobility shift assay (EMSA) using 32P-labeled H3.2α as probe (B). The specificity of the YY1 shift was tested by addition of anti-YY1 (C-20), anti-GFP, or anti-Sp1 to the WCE from asynchronous cells as indicated. The cell cycle distribution of the cells was assessed by FACS analysis (C). Confocal images of nocodazole-arrested cells are presented in Supplementary Figure S2.
The inability of YY1 in extracts from nocodazole-arrested cells to bind DNA in vitro was correlated with in vivo dissociation of YY1 from the DNA. This was shown by immunostaining of YY1 in nocodazole-blocked HeLa cells (Supplementary Figure S2). The distribution of YY1 in nocodazole-arrested cells was identical to what was observed in prometaphase cells in Figure 1.
Increase in DNA-binding Activity of YY1 as Cells Progress through Mitosis
To examine the DNA-binding activity of YY1 in cells progressing from prometaphase through mitosis, HeLa cells were first blocked by nocodazole. After 18 h, cells were collected, washed, and then replated into fresh growth medium. Samples were harvested for WCE preparation at time points 0, 30, 60, 90, and 120 min after release. These extracts were tested in an in vitro EMSA with labeled H3.2α probe. A gradual increase in the binding activity of YY1 was observed as cells progressed toward telophase. Anti-YY1(C-20), anti-GFP, or anti-Sp1 antibodies were incubated with WCE from the 120-min time point to confirm the specificity of the YY1 shift. WCE from the different time points were also assessed by Western blot with anti-YY1 (H-10) and anti-cyclin B, and anti-GAPDH antibodies. The YY1 protein levels remained constant as the cells progressed through mitosis (Figure 3A). Progression through mitosis was verified by examining cyclin B protein levels. Expression of cyclin B increases during S-phase and accumulates at the G2/M boundary when it plays a key role in activating Cdk1 (cyclin-dependent kinase 1), which initiates entry into mitosis. In anaphase, cyclin B levels drop dramatically through rapid degradation by the anaphase-promoting complex (APC; Sullivan and Morgan, 2007). Thus, cyclin B can be utilized as a marker for the end of metaphase and progression through the end of mitosis. As observed in Figure 3B, declining levels of cyclin B indicate that the cells proceeded through mitosis, after release from the nocodazole block.
Figure 3.
YY1 DNA-binding activity increases after release from nocodazole arrest. HeLa cells were blocked with nocodazole for 18 h, then washed, and released into fresh medium. WCEs were prepared from blocked cells and from cells released for 30, 60, 90, and 120 min. (A) Electrophoretic mobility shift assay (EMSA) was performed using WCE from released cells using 32P-labeled H3.2α as a probe. The time points for WCEs are indicated. The specificity of the YY1 shift was assessed by addition of anti-YY1 (C-20), anti-GFP, or anti-Sp1 to the WCE from cells released for 120 min, as indicated. (B) Western blot of extracts prepared from the different time points after release from nocodazole arrest was performed. The blot was first probed with anti-YY1 (H-10) antibody, stripped, and reprobed with anti-cyclin B antibody and then anti-GAPDH.
The elimination of cyclin B after metaphase is usually very rapid and is required for the cell to proceed into anaphase. The gradual decrease observed in Figure 3B indicates that, as might be expected, the cells do not exit from the nocodazole block at the exact same rate. The released cells were observed under the microscope before WCE preparation. A large variety of cell shapes was detected at each time point. Some of the cells were already in telophase stage as early as 60 min after the release, while some were still in late metaphase (data not shown). Thus, each time point after the removal of the block contained cells that are in slightly different stages. From these results, it is clear that the inhibition of the DNA-binding activity of YY1 in prometaphase is reversed later in mitosis.
Loss of YY1 DNA-binding Activity Due to Phosphorylation during Mitosis
The YY1 protein levels do not fluctuate during the cell cycle, including mitosis, as previously reported (Palko et al., 2004) and shown in Figures 2B and 3B. Thus, it seems likely that the change in the DNA-binding activity is due to posttranslational modification. To investigate whether phosphorylation plays a role in the loss of DNA-binding activity of YY1 in mitosis, Flag-YY1 was purified from the HeLa-Flag-YY1 cell line blocked at prometaphase by nocodazole for 18 h. WCEs were prepared and purification of Flag-YY1 was performed by IP using an anti-Flag antibody and competitive elution with excess Flag peptide. Three different double-stranded DNA oligonucleotides were used as probes in the EMSA experiment, containing three different YY1-binding sites. The three probes were defined in Materials and Methods. The H3.2α-binding site was described earlier (Eliassen et al., 1998), AAV p5-60 is part of the promoter region of the adeno-associated virus (Shi et al., 1991), and Cdc6p is part of the Cdc6 promoter (Schlisio et al., 2002). The use of three different YY1-binding sites excluded the possibility of context-specific DNA-binding activities. Flag-YY1 purified from nocodazole-blocked HeLa cells showed almost no binding activity to H3.2α, shown in Figure 4A. EMSA using p5-60 and Cdc6p probes is shown in Supplementary Figure S3. When λ-phosphatase, a general protein phosphatase, was added to Flag-YY1 at 30°C, a significant increase in the binding activity was observed with all three probes (Figure 4A and Supplementary Figure S3). The addition of phosphatase inhibitors along with λ-phosphatase prevented this increase in binding activity, further demonstrating that dephosphorylation of Flag-YY1 is the cause of this recovery in binding. Equal levels of the Flag-YY1 protein in all three reactions (with or without λ-phosphatase and phosphatase inhibitors) was confirmed by Western blotting using anti-YY1 antibody (Figure 4B). The specificity of the YY1 shift was confirmed by incubation of anti-YY1 (C-20), anti-GFP, or anti-Sp1 antibodies to the binding reactions containing Flag-YY1 and λ-phosphatase. Only anti-YY1(C-20) was able to abolish the shift (Figure 4A and Supplementary Figure S3).
Figure 4.
Dephosphorylation of Flag-YY1 from nocodazole-arrested HeLa cells increases its DNA-binding activity. Flag-YY1, purified from nocodazole-arrested HeLa cells, stably transfected with pCS2(+)-Flag-YY1, was incubated 30°C, with or without λ-phosphatase for 30 min before use in the EMSA. The specificity of the YY1 shift was assessed by addition of anti-YY1 (C-20), anti-GFP, or anti-Sp1 to the reaction containing Flag-YY1 and λ-phosphatase, as indicated in A. Equal loading of Flag-YY1 in these reactions was assessed by Western blotting using anti-YY1 (H-10) antibody (B).
These results clearly indicate that specific phosphorylation events negatively regulate the DNA-binding activity of YY1 in mitosis.
Increase in the Phosphorylation Level of YY1 during Mitosis
If a phosphorylation event negatively regulates the binding activity of YY1 in mitosis, then we might expect the phosphorylation level of the protein to increase in prometaphase cells, relative to asynchronous cells.
Direct evidence of phosphorylation of a protein under physiological conditions is obtained by metabolic radioactive labeling of the protein in vivo. It has been previously reported that YY1 is phosphorylated, using 32P-orthophosphate labeling, but the results were not shown (Austen et al., 1997). To confirm this result, radioactive orthophosphate was added to the growth medium of asynchronously growing HeLa cells for 2 h under normal growth conditions. WCEs were then prepared, and endogenous YY1 was immunoprecipitated.
When analyzed on an SDS-PAGE gel and transferred to a nitrocellulose membrane, a radioactive band was observed at the molecular weight where YY1 is usually observed (Figure 5A, left panel). To confirm the identity of this radioactive band, the membrane was probed with mouse monoclonal YY1 antibody (H-10). Indeed, the immunoprecipitated radioactive band was confirmed to be YY1 (Figure 5A, right panel). The radioactive labeling of endogenous YY1 in a relatively short incubation time with orthophosphate indicates that phosphorylation of YY1 in vivo is a dynamic process.
Figure 5.
Phosphorylation of YY1 increases in nocodazole-arrested HeLa cells. (A) Endogenous YY1 was immunoprecipitated from HeLa cells incubated with radioactive orthophosphate for 2 h. Radioactive proteins were detected by Phosphorimager, and the identity of the protein was confirmed by Western blot using anti-YY1 (H-10). (B) HeLa cells transfected with pCS2(+)-Flag-YY1 or with no DNA as negative control were incubated with radioactive orthophosphate for 4 h, and WCEs were prepared. Two successive IPs were performed, a first with anti-Flag antibody (top) and the second with anti-YY1 (C-20) (bottom). Radioactive proteins were detected by Phosphorimager (left panels), and the identity of the proteins was confirmed by Western blot using anti-YY1 (H-10). (C) Flag-YY1 was immunoprecipitated from asynchronous (Asy) or nocodazole-arrested (Noc) HeLa cells, stably transfected with pCS2(+)-Flag-YY1, and separated on a 10% SDS-PAGE gel. The gel was stained with phosphospecific ProQ Diamond and subsequently with Coomassie blue. The graph represents results of quantification of five independent experiments. The synchrony of the cells was verified by FACS analysis.
The increase in DNA-binding activity of YY1 from nocodazole-arrested cells, upon in vitro dephosphorylation (Figure 4), was performed using overexpressed Flag-tagged form of YY1. To confirm that Flag-YY1 is phosphorylated as well as the endogenous YY1, the in vivo labeling experiment described above was repeated with HeLa cells transiently transfected with pCS2(+)-Flag-YY1. When anti-Flag antibody was used in the IP, only one radioactive band was detected, because the anti-Flag antibody would only detect the overexpressed Flag-tagged form of YY1. The nontransfected negative control lane did not show a radioactive band (Figure 5B, top left panel). The identity of the radioactive protein in the Flag-YY1 lane was further confirmed as YY1 in the Western blot analysis (Figure 5B, top right panel). Subsequently, IP by the YY1 C-terminal specific antibody (C-20) was performed. Two radioactive proteins were detected in the transfectant cell lane because this antibody recognizes both endogenous and overexpressed Flag-tagged YY1 (Figure 5B, bottom left panel). Only endogenous YY1 was detected in the negative control lane. The identity of these bands was confirmed by Western blot analysis (Figure 5B, bottom right panel).
To directly test if there is an increase in the phosphorylation status of YY1 in nocodazole-blocked cells, Flag-YY1 was immunoprecipitated from either nocodazole-blocked or asynchronous HeLa-Flag-YY1 cells. The immunoprecipitates were separated on an SDS-PAGE gel and stained with ProQ Diamond. ProQ Diamond is a fluorescent stain that binds to phosphorylated amino acid residues on proteins. An increase in phosphorylation of a protein is observed by increase in the fluorescent signal. In this case, Flag-YY1 immunoprecipitated from cells blocked by nocodazole in prometaphase showed an increase of fluorescent signal when compared with Flag-YY1 immunoprecipitated from asynchronous cells. Staining of the gel with the general protein stain, Coomassie blue, showed equal protein levels for both lanes (Figure 5C). The difference in ProQ Diamond signal was a threefold increase. This quantification is an average resulting from five independent experiments. The synchrony of the nocodazole-arrested cells was verified by FACS analysis (Figure 5C).
Mapping of Three Phosphorylation Sites on Flag-YY1 Purified from Nocodazole-blocked Cells
To identify potential phosphorylation sites on YY1, Flag-YY1 was immunoprecipitated from nocodazole-arrested HeLa-Flag-YY1 cells and loaded on an SDS-PAGE gel. After staining the gel with Coomassie blue, the Flag-YY1 protein band was excised and subjected to mass spectrometry analysis. Flag-YY1 was digested by trypsin, and YY1 phosphopeptides were enriched on phospho-specific binding beads (PhosphoTrap beads). The enriched peptides were analyzed by MALDI-TOF mass spectrometry. Three phosphopeptides were detected and further sequenced to identify the amino acid residues carrying the phosphorylations. The three amino residues identified were serine 247 and threonines 348 and 378. Figure 6 shows the location of the three sites relative to the functional domains of YY1. The diagram of YY1 used here is based on previous studies characterizing the various functional domains of YY1 (Bushmeyer et al., 1995; Austen et al., 1997). Serine 247 is located within what is known as the spacer region of YY1. The function of this region is not understood. Threonines 348 and 378 are located within the DNA-binding domain of YY1. The DNA-binding domain of YY1 is composed of four C2H2 zinc fingers. Threonine 348 is located between zinc fingers 2 and 3, and threonine 378 is located between zinc fingers 3 and 4. A closer examination of these two sites revealed that they are part of a consensus linker peptide sequence. Linker peptides, short amino acid sequences found in almost all zinc-finger proteins, play a critical role in joining and regulating the flexibility of the two adjacent fingers (Choo and Klug, 1993; Wuttke et al., 1997; Laity et al., 2000).
Figure 6.
Three phosphorylation sites are identified in YY1. Three phosphorylation sites were identified by mass spectrometry in tryptic digests of Flag-YY1 immunoprecipitated from nocodazole-arrested HeLa cells. The location of these sites is indicated relative to the functional domains of YY1 previously reported by Austen et al. (1997). Serine 247 is found in the spacer region. Threonines 348 and 378 are found in the DNA-binding domain, specifically in the linker peptides between fingers 2–3, and 3–4.
In a previous mass spectrometric analysis of Flag-YY1 purified from an asynchronous population of HeLa-Flag-YY1 cells, only the phosphorylation of serine 247 was detected (Hurt lab, unpublished results). The phosphorylation of threonines 348 and 378 was only detected on Flag-YY1 from nocodazole-arrested cells.
Effect of Mutation of Serine 247 of YY1 on Subcellular Localization and DNA-binding Activity
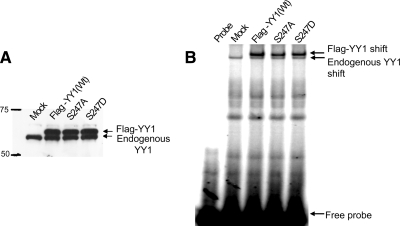
To test the possible roles that a modification of YY1 at serine 247 can play in its regulation, YY1 mutants with a serine 247 substitution to aspartic acid or to alanine (were generated). To examine effects of these substitutions on the DNA-binding activity of YY1, WCEs were prepared from HeLa cells transiently transfected with pCS2(+)-Flag-YY1(WT), (S247A), or (S247D). WCEs were also prepared from HeLa cells that received only the transfection reagent and served as negative control (Mock). Overexpression of these proteins was examined by Western blot, probed with anti-YY1 (H-10) antibody (Figure 7A). When the WCEs were tested in an in vitro EMSA with H3.2α-labeled probe, Flag-YY1 wild type generated a shift slightly higher than that of the endogenous YY1 shift, as expected. Alanine or aspartic acid substitutions at serine 247 had only minor effects on the DNA-binding activity of YY1 (Figure 7B). Because the loss of DNA-binding activity of YY1 in nocodazole-arrested cells was dramatic, it is improbable that phosphorylation of serine 247 is responsible for this loss.
Figure 7.
Mutation of S247 has little effect on YY1 DNA-binding activity. HeLa cells were transiently transfected with pCS2(+)-Flag-YY1 WT, S247A, or S247D mutants. Cells that did not receive DNA served as negative control (Mock). Eighteen hours after transfection, WCEs were analyzed by Western blot using anti-YY1 antibody (H-10) (A), and EMSA using 32P-labeled H3.2α as probe (B). Confocal imaging of Hela cells transfected with Flag-YY1 (WT) or S247 mutants is presented in Supplementary Figure S4.
To test the importance of serine 247 modification on the nuclear localization of YY1, HeLa cells were transfected with Flag-YY1 (WT), (S247A), or (S247D). The overexpressed proteins were visualized using anti-Flag antibody, and DNA was stained with DAPI to visualize the nucleus. No difference was detected between localization of the mutants and wild-type Flag-YY1, showing that phosphorylation at serine 247 does not regulate the localization of YY1 (Supplementary Figure S4).
Effect of Mutation of Threonines 348 and 378 of YY1 on Subcellular Localization and DNA-binding Activity
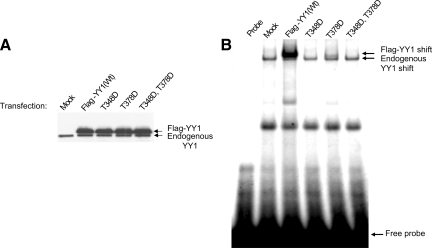
It has previously been shown that all four zinc fingers of YY1 are required for its full DNA-binding activity (Austen et al., 1997). The phosphorylation sites threonine 348 and 378 on YY1, identified in nocodazole-arrested cells, are not located in any of the four zinc fingers. Interestingly, they are located in the linker peptides (the “knuckles”) joining zinc fingers 2 and 3, and 3 and 4. To test the effect of phosphomimetic substitutions at these two sites, three YY1 mutants were generated. HeLa cells were transiently transfected with pCS2(+)-Flag-YY1 (T348D), pCS2(+)-Flag-YY1 (T378D), or pCS2(+)-Flag-YY1 (T348D, T378D), as well as pCS2(+)-Flag-YY1 (WT). HeLa cells receiving only the transfection reagent served as a negative control (Mock). WCEs prepared from the transfected cells were analyzed by Western blot with anti-YY1 (H-10) antibody. Equal levels of overexpression of Flag-YY1 wild type or Flag-YY1 mutants were detected in all transfections (Figure 8A). When tested in an EMSA experiment with labeled H3.2α as probe, equivalent shifts for the endogenous YY1 were detected in all lanes. Flag-YY1(WT)-bound DNA efficiently. However, no binding activity was detected for Flag-YY1 (T348D) and Flag-YY1(T348D, T378D). Only minimal binding activity was detected for Flag-YY1(T378D) (Figure 8B). This result indicates that modification of either of these two sites would have drastic effects on YY1's DNA-binding activity, as observed in prometaphase YY1. Interestingly, substitution of threonines 348 and 378 with alanine residues resulted in the same loss of binding activity (Supplementary Figure S5).
Figure 8.
T348D and T378D mutations abolish YY1 DNA-binding activity. HeLa cells were transiently transfected with pCS2(+)-Flag-YY1 wild type (WT), (T348D), (T378D), and the double mutant (T348D, T378D). Cells that did not receive DNA served as negative control (Mock). Eighteen hours after transfection, WCEs were analyzed by Western blot using anti-YY1 antibody (H-10) (A), and EMSA using 32P-labeled H3.2α as probe (B). Confocal imaging of Hela cells transfected with Flag-YY1 (WT) or T348, T378 mutants is presented in Supplementary Figure S6.
Threonines 348 and 378 are also found in the region of YY1 that has been shown to be essential for nuclear localization (Austen et al., 1997). Because YY1 also distributes to the cytoplasm during mitosis, we wanted to test whether substitutions of these two sites would have effects on the localization of YY1. pCS2(+)-Flag-YY1 (WT), (T348, T378D), or (T348, T378A) were transfected into HeLa cells growing on coverslips. Eighteen hours after transfection, cells were fixed, permeabilized, and immunostained with anti-Flag antibody for visualization of the overexpressed proteins. DNA was stained with DAPI to visualize the nucleus. Flag-YY1 wild type, and both mutants (to aspartic acid or alanine) localized equally to the nucleus (Supplementary Figure S6). This indicates that although modifications at these two sites abolish the DNA-binding activity of YY1, it is not likely involved in the export of YY1 to the cytoplasm.
DISCUSSION
The regulation of the transcription factor YY1 by phosphorylation has long been proposed, but how a specific phosphorylation event can regulate the function of YY1 has never been reported. The results presented in this study elucidate a mechanism for the inactivation of YY1, during cell division, through phosphorylation of its DNA-binding domain.
YY1 is a global regulator of vertebrate gene expression. The pivotal role of YY1 has become more evident as our knowledge of the genes regulated by this factor has grown since its identification. Genes shown to be repressed or activated by YY1 are involved in the regulation of most basic biological processes, including development, cell proliferation, and differentiation (Affar el et al., 2006; Gordon et al., 2006). Furthermore, YY1 has been shown to play a role in resistance to apoptosis in cancerous cells and down-regulation of YY1 led to increased sensitivity of HeLa cells to apoptotic stimuli (Affar el et al., 2006). Inhibition of the DNA-binding activity of YY1, through S-nitrosation of its DNA-binding domain upon nitric oxide treatment of some prostatic and ovarian cancer cell lines, resulted in sensitization of these cells to apoptotic stimuli (Garban and Bonavida, 2001; Hongo et al., 2005; Huerta-Yepez et al., 2009). Many of the cancer therapies in use or in development today are based on inhibition of certain phosphorylation pathways (Zhang et al., 2009). Previous studies have proposed that phosphorylation can affect the DNA-binding activity of YY1 (Becker et al., 1994; Kaludov et al., 1996; Shi et al., 1997; Eliassen et al., 1998). However, these studies did not provide direct evidence or a mechanism for this effect. In addition, they reported contradictory effects for the phosphorylation of YY1. For example, phosphorylation was proposed to be essential for the ability of YY1 to bind the upstream conserved region in the murine leukemia virus long terminal repeat (Becker et al., 1994), to have no effect on YY1-binding activity to the AAV P5 promoter (Shi et al., 1997), or to have a negative effect on the binding activity of YY1 to the alpha element of the replication-dependent histone gene H3.2 (Kaludov et al., 1996; Eliassen et al., 1998). These discrepancies could be due to the use of different experimental systems, including cell lines, DNA-binding sites, and phosphatases. But, more importantly, these studies have applied indirect in vitro approaches, using nuclear extracts and not purified YY1. The heterogeneous protein composition of nuclear or WCEs allows for the possibility of complicating effects.
YY1 has been shown to regulate specific genes at the entry into mitosis (Affar el et al., 2006) and exit from mitosis (Elkon et al., 2003). However, the activity of YY1 between these two stages has not been examined. In this study, immunofluorescence staining of HeLa cells during the different stages of mitosis revealed that YY1 leaves the DNA and distributes to the cytoplasm in prophase. This redistribution persists throughout metaphase and anaphase of mitosis and then YY1 is rapidly recruited back to the DNA at telophase, before complete decondensation of the chromosomes and nuclear envelope reassembly. Exclusion from DNA in vivo was correlated with loss of YY1 DNA-binding activity in vitro, as was shown by EMSA assays. Using WCEs of synchronized HeLa cells, we show that YY1 regains its DNA-binding activity as cells progress toward the exit from mitosis.
The YY1 protein levels do not fluctuate significantly throughout the different phases of the cell cycle, including mitosis. Therefore, we hypothesized that YY1 could be the substrate of a posttranslational modification that inactivates its DNA-binding activity, and consequently, its transcriptional activity, upon entry into mitosis. To test if phosphorylation is responsible for this inactivation, Flag-YY1 was purified from prometaphase HeLa cells using a mild purification approach, yielding a fully functional protein. When Flag-YY1 was dephosphorylated in vitro before an EMSA assay, a significant increase was observed in its DNA-binding activity. Three different DNA probes (H3.2α, p5-60, and Cdc6p) were used in this experiment and gave the same result. These probes represent three different genetic contexts for YY1 binding in vivo, which suggests that the negative effect of phosphorylation on the DNA-binding activity of YY1 is not context specific.
To identify the specific phosphorylation sites causing this loss of binding activity, mass spectrometric analysis was performed using Flag-YY1 purified from prometaphase HeLa cells. Three phosphorylation sites were identified: serine 247, threonine 348, and threonine 378. Consistent with this result, a recent large-scale study of the mitotic phosphoproteome identified three mitotic phosphopeptides that match these three sites in YY1 (Dephoure et al., 2008).
To examine if phosphorylation at these sites can cause the loss of YY1 DNA-binding activity, we generated phosphomimetic mutations at the identified sites. Phosphomimetic substitution at serine 247 did not have a substantial effect on the DNA-binding activity of YY1. On the other hand, phosphomimetic substitutions at threonines 348 and 378 abolished YY1 DNA-binding activity.
Threonines 348 and 378 are not located in any of YY1's four zinc finger domains, all of which are required for full DNA-binding activity (Austen et al., 1997). In fact, these critical threonines are located within linker peptides joining zinc fingers 2 and 3 (threonine 348), and fingers 3 and 4 (threonine 378). The presence of linker peptides is very well conserved among C2H2 zinc finger proteins (Wolfe et al., 2000), and a critical role has been attributed to these peptides in DNA-binding activity (Choo and Klug, 1993; Wuttke et al., 1997; Laity et al., 2000). It is noteworthy to mention that substitution of threonines 348 and 378 with alanine residues also abolished the DNA-binding activity of YY1. It appears that the hydroxyl group of threonines 348 and 378 is essential for the “capping” mechanism that occurs during binding of zinc finger protein to DNA. This group mediates interactions between the linker part and the preceding zinc finger (Laity et al., 2000). Addition of a phosphate molecule to this group would prevent this interaction and affects the ability of the protein to bind DNA properly. Thus, it is not unexpected that the addition of a phosphate group at threonines 348 and 378 of YY1 would drastically affect its DNA-binding activity.
Additional support of this conclusion was shown in an in vitro biochemical study that demonstrated that addition of a phosphate group to the linker peptides of zinc finger proteins results in a dramatic reduction in DNA-binding affinity (Jantz and Berg, 2004). This mode of inactivation is not a unique to YY1. Similar observations have been reported for the zinc finger transcription factors Ikaros and Sp1 (Dovat et al., 2002).
Silencing of gene expression during mitosis has long been known in mammalian cells (Prescott and Bender, 1962). It has also been shown that the loss of transcriptional activity in mitosis is not only due to condensation of chromatin, but is also due to inactivation of binding activities of some transcription factors (Gottesfeld and Forbes, 1997). Other studies have proved this model for several sequence-specific transcription factors like Oct-1 (Segil et al., 1991), GHF-1 (Caelles et al., 1995), and Sp1 (Martinez-Balbas et al., 1995; Dovat et al., 2002). Here we show that the DNA-binding activity of YY1 is also inactivated upon entry into mitosis, further enhancing our understanding of mitotic cessation of transcription.
YY1 has been shown to physically interact with and recruit several chromatin modifiers to sequence specific regions of the genome, including histone acetyltransferases (Lee et al., 1995, 1998), deacetylases (Yang et al., 1996, 1997; Ren et al., 2009), and methyltransferases (Rezai-Zadeh et al., 2003; Baumeister et al., 2005; Ko et al., 2008). In addition, YY1 has been shown to play a role in genomic imprinting (Kim et al., 2006, 2008). The rapid recruitment of YY1 to the decondensing chromosomes in telophase, before the formation of the nuclear envelope (Supplementary Figure S1), raises the possibility of a role for YY1 in marking active and repressed genes in newly formed daughter cells. Adding evidence to this hypothesis is that in our immunostaining studies, YY1 appears to be equally distributed between the two new cells possibly by an active mechanism as is the case for the transcription factor Sp1 (He and Davie, 2006).
In summary, herein we provide concrete and direct evidence for the regulation of the DNA-binding activity of YY1 by phosphorylation. We show that specific phosphorylation of YY1 at threonines 348 and 378 plays an essential role in the dynamic removal of YY1 from DNA during mitosis.
Supplementary Material
ACKNOWLEDGMENTS
We thank Karen Alexander for her outstanding technical support and assistance in this project and her critical reading of this manuscript. We thank Dr. Bernhard Lüscher for his generous gifts of various reagents and plasmids including pCMV-HA-YY1 and for critical reading of this manuscript. We also thank Dr. Yoichi Kato for the pCS2(+)-Flag vector, Ruth Didier for her technical assistance with confocal microscopy and FACS analysis, and Dr. Viorel Mocanu at UNC-Duke Michael Hooker Proteomic Center for mass spectrometric analyses. This work was supported by a grant from the Heart Fund of the Florida Order of Eastern Star.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-04-0264) on September 30, 2009.
REFERENCES
- Affar el B., Gay F., Shi Y., Liu H., Huarte M., Wu S., Collins T., Li E., Shi Y. Essential dosage-dependent functions of the transcription factor yin yang 1 in late embryonic development and cell cycle progression. Mol. Cell. Biol. 2006;26:3565–3581. doi: 10.1128/MCB.26.9.3565-3581.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austen M., Luscher B., Luscher-Firzlaff J. M. Characterization of the transcriptional regulator YY1. The bipartite transactivation domain is independent of interaction with the TATA box-binding protein, transcription factor IIB, TAFII55, or cAMP-responsive element-binding protein (CPB)-binding protein. J. Biol. Chem. 1997;272:1709–1717. doi: 10.1074/jbc.272.3.1709. [DOI] [PubMed] [Google Scholar]
- Baumeister P., Luo S., Skarnes W. C., Sui G., Seto E., Shi Y., Lee A. S. Endoplasmic reticulum stress induction of the Grp78/BiP promoter: activating mechanisms mediated by YY1 and its interactive chromatin modifiers. Mol. Cell. Biol. 2005;25:4529–4540. doi: 10.1128/MCB.25.11.4529-4540.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker K. G., Jedlicka P., Templeton N. S., Liotta L., Ozato K. Characterization of hUCRBP (YY1, NF-E1, delta): a transcription factor that binds the regulatory regions of many viral and cellular genes. Gene. 1994;150:259–266. doi: 10.1016/0378-1119(94)90435-9. [DOI] [PubMed] [Google Scholar]
- Bushmeyer S., Park K., Atchison M. L. Characterization of functional domains within the multifunctional transcription factor, YY1. J. Biol. Chem. 1995;270:30213–30220. doi: 10.1074/jbc.270.50.30213. [DOI] [PubMed] [Google Scholar]
- Caelles C., Hennemann H., Karin M. M-phase-specific phosphorylation of the POU transcription factor GHF-1 by a cell cycle-regulated protein kinase inhibits DNA binding. Mol. Cell. Biol. 1995;15:6694–6701. doi: 10.1128/mcb.15.12.6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo Y., Klug A. A role in DNA binding for the linker sequences of the first three zinc fingers of TFIIIA. Nucleic Acids Res. 1993;21:3341–3346. doi: 10.1093/nar/21.15.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nigris F., Botti C., de Chiara A., Rossiello R., Apice G., Fazioli F., Fiorito C., Sica V., Napoli C. Expression of transcription factor Yin Yang 1 in human osteosarcomas. Eur. J. Cancer. 2006;42:2420–2424. doi: 10.1016/j.ejca.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Delehouzee S., et al. GABP, HCF-1 and YY1 are involved in Rb gene expression during myogenesis. Genes Cells. 2005;10:717–731. doi: 10.1111/j.1365-2443.2005.00873.x. [DOI] [PubMed] [Google Scholar]
- Deng Z., Wan M., Sui G. PIASy-mediated sumoylation of Yin Yang 1 depends on their interaction but not the RING finger. Mol. Cell. Biol. 2007;27:3780–3792. doi: 10.1128/MCB.01761-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N., Zhou C., Villén J., Beausoleil S. A., Bakalarski C. E., Elledge S. J., Gygi S. P. A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. USA. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe M. E., Zhang X., McGinnis L., Biggers J., Li E., Shi Y. Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol. Cell. Biol. 1999;19:7237–7244. doi: 10.1128/mcb.19.10.7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovat S., Ronni T., Russell D., Ferrini R., Cobb B. S., Smale S. T. A common mechanism for mitotic inactivation of C2H2 zinc finger DNA-binding domains. Genes Dev. 2002;16:2985–2990. doi: 10.1101/gad.1040502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliassen K. A., Baldwin A., Sikorski E. M., Hurt M. M. Role for a YY1-binding element in replication-dependent mouse histone gene expression. Mol. Cell. Biol. 1998;18:7106–7118. doi: 10.1128/mcb.18.12.7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkon R., Linhart C., Sharan R., Shamir R., Shiloh Y. Genome-wide in silico identification of transcriptional regulators controlling the cell cycle in human cells. Genome Res. 2003;13:773–780. doi: 10.1101/gr.947203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkeland S. J., Valkhof M., Heijmans-Antonissen C., Delwel R., Valk P. J., Hermans M. H., Touw I. P. The gene encoding the transcriptional regulator Yin Yang 1 (YY1) is a myeloid transforming gene interfering with neutrophilic differentiation. Blood. 2003;101:1111–1117. doi: 10.1182/blood-2002-04-1207. [DOI] [PubMed] [Google Scholar]
- Ficzycz A., Eskiw C., Meyer D., Marley K. E., Hurt M., Ovsenek N. Expression, activity, and subcellular localization of the Yin Yang 1 transcription factor in Xenopus oocytes and embryos. J. Biol. Chem. 2001;276:22819–22825. doi: 10.1074/jbc.M011188200. [DOI] [PubMed] [Google Scholar]
- Garban H. J., Bonavida B. Nitric oxide inhibits the transcription repressor Yin-Yang 1 binding activity at the silencer region of the Fas promoter: a pivotal role for nitric oxide in the up-regulation of Fas gene expression in human tumor cells. J. Immunol. 2001;167:75–81. doi: 10.4049/jimmunol.167.1.75. [DOI] [PubMed] [Google Scholar]
- Gordon S., Akopyan G., Garban H., Bonavida B. Transcription factor YY1, structure, function, and therapeutic implications in cancer biology. Oncogene. 2006;25:1125–1142. doi: 10.1038/sj.onc.1209080. [DOI] [PubMed] [Google Scholar]
- Gottesfeld J. M., Forbes D. J. Mitotic repression of the transcriptional machinery. Trends Biochem. Sci. 1997;22:197–202. doi: 10.1016/s0968-0004(97)01045-1. [DOI] [PubMed] [Google Scholar]
- Gronroos E., Terentiev A. A., Punga T., Ericsson J. YY1 inhibits the activation of the p53 tumor suppressor in response to genotoxic stress. Proc. Natl. Acad. Sci. USA. 2004;101:12165–12170. doi: 10.1073/pnas.0402283101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S., Davie J. R. Sp1 and Sp3 foci distribution throughout mitosis. J. Cell Sci. 2006;119:1063–1070. doi: 10.1242/jcs.02829. [DOI] [PubMed] [Google Scholar]
- Hiromura M., Choi C. H., Sabourin N. A., Jones H., Bachvarov D., Usheva A. YY1 is regulated by O-linked N-acetylglucosaminylation (O-glcNAcylation) J. Biol. Chem. 2003;278:14046–14052. doi: 10.1074/jbc.M300789200. [DOI] [PubMed] [Google Scholar]
- Hongo F., Garban H., Huerta-Yepez S., Vega M., Jazirehi A. R., Mizutani Y., Miki T., Bonavida B. Inhibition of the transcription factor Yin Yang 1 activity by S-nitrosation. Biochem. Biophys. Res. Commun. 2005;336:692–701. doi: 10.1016/j.bbrc.2005.08.150. [DOI] [PubMed] [Google Scholar]
- Huerta-Yepez S., Vega M., Escoto-Chavez S. E., Murdock B., Sakai T., Baritaki S., Bonavida B. Nitric oxide sensitizes tumor cells to TRAIL-induced apoptosis via inhibition of the DR5 transcription repressor Yin Yang 1. Nitric Oxide. 2009;20:39–52. doi: 10.1016/j.niox.2008.08.001. [DOI] [PubMed] [Google Scholar]
- Hurt M. M., Bowman T. L., Marzluff W. F. A common transcriptional activator is located in the coding region of two replication-dependent mouse histone genes. Mol. Cell. Biol. 1991;11:2929–2936. doi: 10.1128/mcb.11.6.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantz D., Berg J. M. Reduction in DNA-binding affinity of Cys2His2 zinc finger proteins by linker phosphorylation. Proc. Natl. Acad. Sci. USA. 2004;101:7589–7593. doi: 10.1073/pnas.0402191101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaludov N. K., Bowman T. L., Sikorski E. M., Hurt M. M. Cell cycle-regulated binding of nuclear proteins to elements within a mouse H3.2 histone gene. Proc. Natl. Acad. Sci. USA. 1996;93:4465–4470. doi: 10.1073/pnas.93.9.4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Kim J. D. In vivo YY1 knockdown effects on genomic imprinting. Hum. Mol. Genet. 2008;17:391–401. doi: 10.1093/hmg/ddm316. [DOI] [PubMed] [Google Scholar]
- Kim J. D., Hinz A. K., Bergmann A., Huang J. M., Ovcharenko I., Stubbs L., Kim J. Identification of clustered YY1 binding sites in imprinting control regions. Genome Res. 2006;16:901–911. doi: 10.1101/gr.5091406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko C. Y., Hsu H. C., Shen M. R., Chang W. C., Wang J. M. Epigenetic silencing of CCAAT/enhancer-binding protein delta activity by YY1/polycomb group/DNA methyltransferase complex. J. Biol. Chem. 2008;283:30919–30932. doi: 10.1074/jbc.M804029200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krippner-Heidenreich A., Walsemann G., Beyrouthy M. J., Speckgens S., Kraft R., Thole H., Talanian R. V., Hurt M. M., Luscher B. Caspase-dependent regulation and subcellular redistribution of the transcriptional modulator YY1 during apoptosis. Mol. Cell. Biol. 2005;25:3704–3714. doi: 10.1128/MCB.25.9.3704-3714.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laity J. H., Dyson H. J., Wright P. E. DNA-induced alpha-helix capping in conserved linker sequences is a determinant of binding affinity in Cys(2)-His(2) zinc fingers. J Mol. Biol. 2000;295:719–727. doi: 10.1006/jmbi.1999.3406. [DOI] [PubMed] [Google Scholar]
- Lee H. Y., Chaudhary J., Walsh G. L., Hong W. K., Kurie J. M. Suppression of c-Fos gene transcription with malignant transformation of human bronchial epithelial cells. Oncogene. 1998;16:3039–3046. doi: 10.1038/sj.onc.1201843. [DOI] [PubMed] [Google Scholar]
- Lee J. S., Galvin K. M., See R. H., Eckner R., Livingston D., Moran E., Shi Y. Relief of YY1 transcriptional repression by adenovirus E1A is mediated by E1A-associated protein p300. Genes Dev. 1995;9:1188–1198. doi: 10.1101/gad.9.10.1188. [DOI] [PubMed] [Google Scholar]
- Martinez-Balbas M. A., Dey A., Rabindran S. K., Ozato K., Wu C. Displacement of sequence-specific transcription factors from mitotic chromatin. Cell. 1995;83:29–38. doi: 10.1016/0092-8674(95)90231-7. [DOI] [PubMed] [Google Scholar]
- Mitchison T. J., Salmon E. D. Mitosis: a history of division. Nat. Cell Biol. 2001;3:E17–E21. doi: 10.1038/35050656. [DOI] [PubMed] [Google Scholar]
- Oei S. L., Griesenbeck J., Schweiger M., Babich V., Kropotov A., Tomilin N. Interaction of the transcription factor YY1 with human poly(ADP-ribosyl) transferase. Biochem. Biophys. Res. Commun. 1997;240:108–111. doi: 10.1006/bbrc.1997.7621. [DOI] [PubMed] [Google Scholar]
- Oei S. L., Griesenbeck J., Schweiger M., Ziegler M. Regulation of RNA polymerase II-dependent transcription by poly(ADP-ribosyl)ation of transcription factors. J. Biol. Chem. 1998;273:31644–31647. doi: 10.1074/jbc.273.48.31644. [DOI] [PubMed] [Google Scholar]
- Palko L., Bass H. W., Beyrouthy M. J., Hurt M. M. The Yin Yang-1 (YY1) protein undergoes a DNA-replication-associated switch in localization from the cytoplasm to the nucleus at the onset of S phase. J. Cell Sci. 2004;117:465–476. doi: 10.1242/jcs.00870. [DOI] [PubMed] [Google Scholar]
- Parker C. E., Warren M. R., Loiselle D. R., Dicheva N. N., Scarlett C. O., Borchers C. H. Identification of components of protein complexes. Methods Mol. Biol. 2005;301:117–151. doi: 10.1385/1-59259-895-1:117. [DOI] [PubMed] [Google Scholar]
- Petkova V., Romanowski M. J., Sulijoadikusumo I., Rohne D., Kang P., Shenk T., Usheva A. Interaction between YY1 and the retinoblastoma protein. Regulation of cell cycle progression in differentiated cells. J. Biol. Chem. 2001;276:7932–7936. doi: 10.1074/jbc.M007411200. [DOI] [PubMed] [Google Scholar]
- Pines J., Rieder C. L. Re-staging mitosis: a contemporary view of mitotic progression. Nat. Cell Biol. 2001;3:E3–E6. doi: 10.1038/35050676. [DOI] [PubMed] [Google Scholar]
- Prescott D. M., Bender M. A. Synthesis of RNA and protein during mitosis in mammalian tissue culture cells. Exp. Cell Res. 1962;26:260–268. doi: 10.1016/0014-4827(62)90176-3. [DOI] [PubMed] [Google Scholar]
- Raska C. S., Parker C. E., Sunnarborg S. W., Pope R. M., Lee D. C., Glish G. L., Borchers C. H. Rapid and sensitive identification of epitope-containing peptides by direct matrix-assisted laser desorption/ionization tandem mass spectrometry of peptides affinity-bound to antibody beads. J. Am. Soc. Mass. Spectrom. 2003;14:1076–1085. doi: 10.1016/S1044-0305(03)00405-7. [DOI] [PubMed] [Google Scholar]
- Ren G., et al. Recruitment of HDAC4 by transcription factor YY1 represses HOXB13 to affect cell growth in AR-negative prostate cancers. Int. J. Biochem. Cell Biol. 2009;41:1094–1101. doi: 10.1016/j.biocel.2008.10.015. [DOI] [PubMed] [Google Scholar]
- Rezai-Zadeh N., Zhang X., Namour F., Fejer G., Wen Y. D., Yao Y. L., Gyory I., Wright K., Seto E. Targeted recruitment of a histone H4-specific methyltransferase by the transcription factor YY1. Genes Dev. 2003;17:1019–1029. doi: 10.1101/gad.1068003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs K. J., Saleque S., Wong K. K., Merrell K. T., Lee J. S., Shi Y., Calame K. Yin-yang 1 activates the c-myc promoter. Mol. Cell. Biol. 1993;13:7487–7495. doi: 10.1128/mcb.13.12.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlisio S., Halperin T., Vidal M., Nevins J. R. Interaction of YY1 with E2Fs, mediated by RYBP, provides a mechanism for specificity of E2F function. EMBO J. 2002;21:5775–5786. doi: 10.1093/emboj/cdf577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segil N., Roberts S. B., Heintz N. Mitotic phosphorylation of the Oct-1 homeodomain and regulation of Oct-1 DNA binding activity. Science. 1991;254:1814–1816. doi: 10.1126/science.1684878. [DOI] [PubMed] [Google Scholar]
- Seligson D., et al. Expression of transcription factor Yin Yang 1 in prostate cancer. Int. J. Oncol. 2005;27:131–141. [PubMed] [Google Scholar]
- Shi Y., Lee J. S, Galvin K. M. Everything you have ever wanted to know about Yin Yang 1. Biochim. Biophys. Acta. 1997;1332:F49–F66. doi: 10.1016/s0304-419x(96)00044-3. [DOI] [PubMed] [Google Scholar]
- Shi Y., Seto E., Chang L. S., Shenk T. Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- Shrivastava A., Saleque S., Kalpana G. V., Artandi S., Goff S. P., Calame K. Inhibition of transcriptional regulator Yin-Yang-1 by association with c-Myc. Science. 1993;262:1889–1892. doi: 10.1126/science.8266081. [DOI] [PubMed] [Google Scholar]
- Shrivastava A., Yu J., Artandi S., Calame K. YY1 and c-Myc associate in vivo in a manner that depends on c-Myc levels. Proc. Natl. Acad. Sci. USA. 1996;93:10638–10641. doi: 10.1073/pnas.93.20.10638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui G., et al. Yin Yang 1 is a negative regulator of p53. Cell. 2004;117:859–872. doi: 10.1016/j.cell.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Sullivan M., Morgan D. O. Finishing mitosis, one step at a time. Nat. Rev. Mol. Cell Biol. 2007;8:894–903. doi: 10.1038/nrm2276. [DOI] [PubMed] [Google Scholar]
- Takasaki N., Kurokawa D., Nakayama R., Nakayama J., Aizawa S. Acetylated YY1 regulates Otx2 expression in anterior neuroectoderm at two cis-sites 90 kb apart. EMBO J. 2007;26:1649–1659. doi: 10.1038/sj.emboj.7601619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner D. L., Weintraub H. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 1994;8:1434–1447. doi: 10.1101/gad.8.12.1434. [DOI] [PubMed] [Google Scholar]
- Walowitz J. L., Bradley M. E., Chen S., Lee T. Proteolytic regulation of the zinc finger transcription factor YY1, a repressor of muscle-restricted gene expression. J. Biol. Chem. 1998;273:6656–6661. doi: 10.1074/jbc.273.12.6656. [DOI] [PubMed] [Google Scholar]
- Watrin E., Legagneux V. Introduction to chromosome dynamics in mitosis. Biol. Cell. 2003;95:507–513. doi: 10.1016/j.biolcel.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Whitfield M. L., et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield M. L., Zheng L. X., Baldwin A., Ohta T., Hurt M. M., Marzluff W. F. Stem-loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol. Cell. Biol. 2000;20:4188–4198. doi: 10.1128/mcb.20.12.4188-4198.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson F. H., Park K., Atchison M. L. Polycomb recruitment to DNA in vivo by the YY1 REPO domain. Proc. Natl. Acad. Sci. USA. 2006;103:19296–19301. doi: 10.1073/pnas.0603564103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe S. A., Nekludova L., Pabo C. O. DNA recognition by Cys2His2 zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- Wuttke D. S., Foster M. P., Case D. A., Gottesfeld J. M., Wright P. E. Solution structure of the first three zinc fingers of TFIIIA bound to the cognate DNA sequence: determinants of affinity and sequence specificity. J. Mol. Biol. 1997;273:183–206. doi: 10.1006/jmbi.1997.1291. [DOI] [PubMed] [Google Scholar]
- Yakovleva T., et al. YY1 binding to a subset of p53 DNA-target sites regulates p53-dependent transcription. Biochem. Biophys. Res. Commun. 2004;318:615–624. doi: 10.1016/j.bbrc.2004.04.065. [DOI] [PubMed] [Google Scholar]
- Yang W. M., Inouye C., Zeng Y., Bearss D., Seto E. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc. Natl. Acad. Sci. USA. 1996;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W. M., Yao Y. L., Sun J. M., Davie J. R., Seto E. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J. Biol. Chem. 1997;272:28001–28007. doi: 10.1074/jbc.272.44.28001. [DOI] [PubMed] [Google Scholar]
- Yao Y. L., Dupont B. R., Ghosh S., Fang Y., Leach R. J., Seto E. Cloning, chromosomal localization and promoter analysis of the human transcription factor YY1. Nucleic Acids Res. 1998;26:3776–3783. doi: 10.1093/nar/26.16.3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y. L., Yang W. M., Seto E. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol. Cell. Biol. 2001;21:5979–5991. doi: 10.1128/MCB.21.17.5979-5991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Yang P. L., Gray N. S. Targeting cancer with small molecule kinase inhibitors. Nat. Rev. Cancer. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.