Abstract
The microtubule (MT) network is essential in a broad spectrum of cellular functions. Many studies have linked CENP-F to MT-based activities as disruption of this protein leads to major changes in MT structure and function. Still, the basis of CENP-F regulation of the MT network remains elusive. Here, our studies reveal a novel and critical localization and role for CENP-F at the centrosome, the major MT organizing center (MTOC) of the cell. Using a yeast two-hybrid screen, we identify Hook2, a linker protein that is essential for regulation of the MT network at the centrosome, as a binding partner of CENP-F. With recently developed immunochemical reagents, we confirm this interaction and reveal the novel localization of CENP-F at the centrosome. Importantly, in this first report of CENP-F−/− cells, we demonstrate that ablation of CENP-F protein function eliminates MT repolymerization after standard nocodazole treatment. This inhibition of MT regrowth is centrosome specific because MT repolymerization is readily observed from the Golgi in CENP-F−/− cells. The centrosome-specific function of CENP-F in the regulation of MT growth is confirmed by expression of truncated CENP-F containing only the Hook2-binding domain. Furthermore, analysis of partially reconstituted MTOC asters in cells that escape complete repolymerization block shows that disruption of CENP-F function impacts MT nucleation and anchoring rather than promoting catastrophe. Our study reveals a major new localization and function of CENP-F at the centrosome that is likely to impact a broad array of MT-based actions in the cell.
INTRODUCTION
Characterization of CENP-F has revealed many different domains, binding partners, and functions. The large size of this protein lends itself to a multifaceted role within the cell and the orthologues studied in different species show significant variation in overall function and localization. Initially, CENP-F was visualized at the kinetochore (KT), the attachment point for the microtubule (MT) network at the centromere (Rattner et al., 1993). KT function of CENP-F has not yet been completely elucidated, but knockdown of CENP-F leads to mitotic delay, decreased KT tension, and MT instability (Bomont et al., 2005; Holt et al., 2005; Laoukili et al., 2005; Yang et al., 2005; Feng et al., 2006). Furthermore, the farnesylation site located at the extreme carboxy terminus is necessary for KT targeting of CENP-F (Ashar et al., 2000; Hussein and Taylor, 2002), and KT localization of CENP-F is also vital to normal KT recruitment of CENP-E and p150glued(Yang et al., 2005). The diversity of function within the C terminus of CENP-F is further illustrated by the recent characterization of its binding partner nucleoporin protein Nup133, an integral component of the nuclear pore complex (Zuccolo et al., 2007). Moreover, within the carboxy terminus resides an E2F1-like retinoblastoma protein (Rb)-binding domain that interacts with not only Rb but also other pocket proteins p107 and p130 (Ashe et al., 2004). Recently, analysis of the avian ortholog demonstrated that interaction with Rb is important for myogenic differentiation (Robertson et al., 2008). CENP-F was initially identified as an Rb-binding protein (Zhu et al., 1995b), and mutation of this site results in impaired differentiation of myotubes and reduced contractile protein expression (Robertson et al., 2008). Finally, the C terminus of CENP-F, the focus of the majority of studies on this protein, has also been established as a negative regulator of the transcription factor ATF-4 (Zhou et al., 2005). All of these functions are nuclear and are attributed to domains of CENP-F adjacent to the C-terminal nuclear localization signal characterized by Zhu et al. (Zhu et al., 1995a). However, given the substantial size of CENP-F and other predicted structural and interactive domains throughout the molecule, this nuclear region governs only a subset of functions of this protein.
Previous studies have also characterized cytoplasmic localization and function of CENP-F (Soukoulis et al., 2005; Pooley et al., 2006; Vergnolle and Taylor, 2007; Pooley et al., 2008). CENP-F contains a spectrin repeat region central to the C terminus and adjacent to these repeats is the Nde1/Ndel1-binding domain originally identified by Soukoulis et al. (2005) and confirmed by Vergnolle and Taylor (2007). This interaction domain regulates MT network organization through Nde1/Ndel1 interaction with the LIS1 pathway. Additionally, both termini of CENP-F have tubulin-binding capabilities and the C-terminal domain is capable of tubulin polymerization in vitro (Feng et al., 2006). Moreover, the N terminus of CENP-F has been shown to bind soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins soluble N-ethylmaleimide-sensitive factor attachment protein 25 (SNAP25) and syntaxin 4, linking MTs to vesicles in the regulation of protein trafficking (Pooley et al., 2006, 2008). These interactions with tubulin and the MT network further support additional functions of CENP-F in the cytoplasm.
Many studies have linked CENP-F to MT-based activities as disruption of this protein leads to major changes in MT structure and function. Still, the basis of CENP-F regulation of the MT network remains elusive. Here, our study reveals a novel role for CENP-F at the centrosome, the major MT organizing center (MTOC) of the cell. We demonstrate that Hook homologue 2 (Hook2), a newly identified centrosomal protein that is essential for centrosomal regulation of the MT network, as a binding partner of CENP-F. Using novel immunochemical reagents, we confirm this interaction and establish the localization of CENP-F at the centrosome. Importantly, we demonstrate that ablation of CENP-F in newly developed CENP-F−/− cells dramatically attenuates MT repolymerization after standard nocodazole treatment. Furthermore, this effect is centrosome specific because MT repolymerization is readily observed from the Golgi in CENP-F−/− cells. This centrosome-specific function of CENP-F is confirmed by expression of the Hook2-binding domain in CENP-F (NT-CENP-F). From analysis of NT-CENP-F–expressing cells where partially reconstituted MTOC asters are observed, we show that disruption of CENP-F function impacts MT nucleation and anchoring rather than promoting catastrophe. Thus, our studies reveal a major new function of CENP-F at the centrosome that is likely to impact a broad array of MT-based actions in the cell.
MATERIALS AND METHODS
Yeast Two-Hybrid (Y2H) Screen
The large N-terminal region of cytoplasmic murine CENP-F (amino acids 1-689, termed LCR), was used as bait in Y2H screen performed using our published methods with the Matchmaker Y2H System 3 (Clontech, Mountain View, CA) (Pooley et al., 2008). The bait was mated with a yeast strain pretransformed with a whole mouse embryonic day 17.5 cDNA library. Library plasmids were isolated from yeast colonies that survived on Quadruple dropout (QDO) medium (SD/-Ade/-His/-Leu/-Trp/X-a-Gal) and exhibited lacZ expression. The inserts were then sequenced by the Vanderbilt Sequencing Core Facility (Nashville, TN) and identified using National Center for Biotechnology Information Blast (Altschul and Lipman, 1990). A series of truncations of each of protein were constructed by polymerase chain reaction (PCR) and transformed into appropriate yeast strains. Yeast were grown and plated on QDO medium; positive associations grew and exhibited blue color upon galactosidase (Gal) testing. Positive control growth was indicated by yeast transformed with pGBKT7-53 and pGADT7-T, and the negative control used yeast expressing pGBTK-53 and the empty vector pGADT7. False positive tests with empty vector and random protein matings were conducted to eliminate spurious interactions according to manufacturer's recommendations.
Antibodies
A novel polyclonal murine CENP-F antibody was generated in rabbits from the peptide NTNKHSMSATD (aa 1122-1132; Biosynthesis, Lewisville, TX). Antisera were affinity purified using the injected peptide and a SulfoLink kit (Pierce Chemical, Rockford, IL). The polyclonal Hook2 antibody (epitope aa 427-719) was a generous gift from Dr. H. Kramer (The University of Texas Southwestern Medical Center at Dallas, Dallas, TX). The γ- and β-tubulin antibodies were obtained from Sigma-Aldrich (St. Louis, MO), myc and green fluorescent protein (GFP) antibodies were obtained from BD Biosciences (San Jose, CA). The ninein, pericentrin, centrin1, and MT network marker YL1/2 antibodies were purchased from Abcam (Cambridge, MA). The PCM-1 antibody was from Novus Biologicals (Littleton, CO). Alexa Fluor 488- and 568-conjugated secondary antibodies were also used (Invitrogen, Carlsbad, CA). For primary-antibody direct labeling immunofluorescence studies, polyclonal anti-CENP-F was directly labeled with the Zenon Alexa-488 labeling kit (Invitrogen). Alkaline phosphatase-conjugated secondary antibodies for western blot were also purchased from Sigma-Aldrich.
Cell Culture, Transfection, and DNA Constructs
COS-7 (American Type Culture Collection, Manassas, VA), 3T3 (American Type Culture Collection), mouse embryonic fibroblasts (MEFs), retinal pigment epithelial (RPE) cells (Clontech), and C2C12 cells (American Type Culture Collection) were maintained in DMEM (HyClone Laboratories, Logan, UT) supplemented with 10, 10, 10, and 20% fetal bovine serum (FBS), respectively, 100 μg/ml penicillin/streptomycin, and l-glutamine, in a 5% CO2 atmosphere at 37°C. For transfection, cells were grown to 50–75% confluence and transfected with DNA by using FuGENE 6 (Roche Diagnostics, Indianapolis, IN) according to manufacturer's recommendations. Full-length Hook2 cDNA (Invitrogen) was cloned into the pEGFP-C3 vector. This construct was then used as the PCR template for the truncation constructs, which were cloned into the manufactured yeast vector for Y2H analysis. NT-CENP-F was constructed by cloning the N-terminal 474 amino acids of CENP-F in frame into the pCMV-myc, pCerulean, and the pEGFP-C1 vectors (Clontech). The 3GFP-Ensc microtubule-binding domain (EMTB) construct was a kind gift of Dr. J. C. Bulinski (Columbia University, New York, NY).
Immunostaining, Microtubule Assays, and Microscopy
For studies of transiently transfected protein expression and analysis of endogenous protein localization, cells were gently washed with 1× phosphate-buffered saline (PBS) and fixed for 20 min at room temperature with either 4% paraformaldehyde to visualize directly labeled antibodies or with 70% methanol at room temperature to visualize all other proteins. Cells used for γ-tubulin labeling were fixed at −20°C. Subsequently, cells were washed with 1× PBS, permeabilized with 0.25% Triton X-100 in 1× PBS for 10 min, and blocked for 1 h in 2% bovine serum albumin in 1× PBS at room temperature. Primary antibodies were incubated overnight at 4°C. Cells were then washed three times in 1× PBS and incubated with secondary antibodies for 1 h at room temperature. Cells were again washed three times with 1× PBS and coverslips mounted with Poly AquaMount (PolySciences, Warrington, PA).
For MT repolymerization assays, cells were incubated with nocodazole (Sigma-Aldrich) for 2 h (2.5 μg/ml in culture medium). Cells were then washed three times with fresh culture medium and held in fresh medium at 37°C to allow repolymerization for a specific time period, dependent on the experiment. Cells were then washed once with PHEM buffer [60 mM piperazine-N,N′-bis(2-ethanesulfonic acid), 25 mM HEPES, 10 mM EGTA, and 3 mM MgCl2 pH 6.8] and treated for 1 min with 0.5% Triton-X in PHEM buffer before fixations, as described above.
Cells were visualized by fluorescence microscopy with an AX70 (Olympus, Melville, NY) or an LSM510 microscope (Carl Zeiss, Thornwood, NY) microscope for confocal analysis. Widefield images were captured using Elements Basic Science software (Nikon, Tokyo, Japan), and confocal images were captured with a ZEN LE laser scanning microscope (Carl Zeiss). Additional widefield images were captured with an IX71 inverted microscope (Olympus) and a CoolSNAP-HQ2 charge-coupled device (CCD) camera (Photometrics, Tucson, AZ), in a temperature- and CO2-controlled WeatherStation (Precision Control, Sammamish, Washington) as part of the DeltaVision platform (Applied Precision, Issaquah, WA) and deconvolved with the SoftWorx software included in the DeltaVision core. Image processing with performed with Photoshop (Adobe Systems, Mountain View, CA), contrasting with histogram stretching and using gamma correction in MT images to ensure visualization of all MTs. All images of control and experimental cells were processed identically. Statistical analysis of MTOC reformation in COS-7 cells was done with the Student's t test and the incomplete MT repolymerization statistical analysis was performed with the G test.
In live imaging studies of MT regrowth, cells were initially positioned and recorded in nocodazole medium before they were washed with new culture medium on the microscope stage for repolymerization. Live imaging experiments used cells plated on glass-bottomed dishes (MatTek, Ashland, MA). A heated stage (Warner Instruments, Hamden, CT) maintained cultures at 37°C. IPLab software (BD Biosciences Bioimaging, Rockville, MD) was used to capture images on a TE2000E inverted microscope (Nikon) (CFI PLAN APO VC 100× oil lens, numerical aperture 1.4, with or without 1.5× intermediate magnification) with an automated focusing device (Perfect-Focus; Nikon), Yokogawa QLC-100/CSU-10 spinning disk head (Visitec assembled by Vashaw, Norcross, GA), and a back-illuminated EM-CCD camera Cascade 512B (Photometrics). A krypton-argon laser (75-mW 488/568; Melles Griot Albuquerque, NM) with acousto-optical tunable filters was used for two-color excitation. Custom double dichroic mirror and filters (Chroma Technology, Brattleboro, VT) in a filter wheel (Ludl Electronic Products, Hawthorne, NY) were used in the emission light path (Efimov et al., 2007).
Coimmunoprecipitation Using Transient Transfections and Endogenous Proteins
Transfected COS-7 cells were grown on 10-cm plates; lysates were harvested 48 h after transfection. The ProFound Mammalian c-Myc Tag coimmunoprecipitation kit (Pierce Chemical) was used according to previous methods established by our laboratory (Pooley et al., 2008). In brief, cells were washed once with ice-cold Tris-buffered saline (TBS), incubated with M-Per Extraction Reagent (Pierce Chemical) containing protease inhibitor (11 697 498 001; Roche Diagnostics), and centrifuged at 16,000 × g for 20 min at 4°C. Protein lysate concentration of the supernatant was determined using a bicinchoninic acid solution assay (Pierce Chemical); 100 μg total lysate was incubated for two hours with gentle shaking at 4°C with 10 μl of anti-c-myc agarose slurry. Columns were washed three times with 1× TBS-Tween. Protein was eluted with 2× nonreducing sample buffer (Pierce Chemical) at 95°C for 5 min. To reduce proteins for SDS-polyacrylamide gel electrophoresis (PAGE) analysis and Western blot analysis, 2 μl of 2-mercaptoethanol was added. Ten microliters of total lysate supernatant was used to confirm protein expression. Blots were developed using nitro blue tetrazolium-5-bromo-4-chloro-3-indolyl phosphate (Roche Diagnostics) and scanned into digital images (Hewlett Packard, Palo Alto, CA).
For analysis of endogenous protein-protein interaction, Dynabeads protein G (Invitrogen) were washed three times with citrate phosphate buffer, incubated with CENP-F antibody for 1 h at room temperature, and washed again. C2C12 cells were lysed with NP-40 buffer with gentle sonication. Whole cell lysates were recovered and incubated with antibody-Dynabead complexes for 1 h at 4°C. The Dynabead magnet apparatus was used to wash with ice cold 1× PBS three times, and proteins were eluted with Laemmli sample buffer at a boiling temperature for 5 min. Proteins were resolved on a 10% SDS-PAGE gel after addition of β-mercaptoethanol and analyzed by Western blot analysis. Twenty micrograms of protein lysate was loaded to visualize Hook2 in whole cell lysate.
Isolation of Mouse Embryonic Fibroblasts
Embryos from a CENP-F−/WT × CENP-F−/WT cross were isolated from the uteri of 13.5-d-pregnant females, washed with PBS, and the head and visceral organs were dissected out. The remainder of the embryos were washed with PBS and finely minced before placement into individual tubes with fresh PBS. This mixture was well-triturated and plated into 10-cm cell culture dishes with MEF medium (DMEM, 4.5 g/ml glucose, 10% FBS, 2 mM glutamine, and 1% Pen/Strep). These were maintained as described above and MEFs were used within three passages to avoid replicative senescence. Genotyping was done with two sets of primers. The first flank the 5-prime loxP site and the sequences are 5′:AATAATGAAGCTGACACCAAAAACT and 3′: GAACCTACCGTCTGAGAACCACTG. The same 5′-primer is used for the recombination test PCR, only with a 3′-primer outside of the sixth exon: 3′: GAGGAGCACAGGAGGGAAATG.
RESULTS
Identification of a Novel Interaction between Murine CENP-F and Hook2
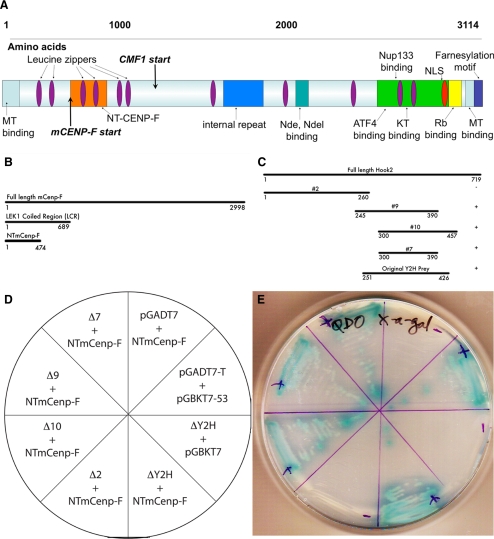
Molecular analysis of CENP-F function has been confined predominantly to the C-terminal sequences of the protein. Because the largely uncharacterized and extensive N terminus has several predicted protein–protein binding motifs, we used a Y2H approach to identify novel binding partners of CENP-F. The schematic in Figure 1A summarizes CENP-F domains studied previously as well as the N-terminal domains described here. The bait in this screen encompassed the first 689 amino acids of N-terminal CENP-F (base pairs 1-2067) and was screened against an embryonic whole mouse E17.5 cDNA library (Pooley et al., 2008). This screen identified specific proteins associated with the MT network and its various functions. Four independent hits matched the sequence of Hook2 and passed all false positive tests. Hook2 has been shown to regulate MT repolymerization at the centrosome and binds centrosomal protein CEP-110/centriolin (Szebenyi et al., 2007). Our laboratory has shown that a central region of CENP-F distant from the N terminus regulates the microtubule network via interaction with NudE (Soukoulis et al., 2005), and given this potential connection, we examined the interaction of Hook2 and CENP-F in greater detail.
Figure 1.
Identification of CENP-F/Hook2 interaction and characterization of binding domains. (A) Schematic of CENP-F protein domains. Our initial Y2H screen used the LEK coiled region (LCR) as bait and identified Hook2 directly interacting with CENP-F. (B) CENP-F truncation constructs. (C) Hook2 truncation constructs. In the column on the right, + indicates positive growth and − indicates no growth. Truncations were transformed into respective yeast strains to determine positive interactions. (D) Schematic of Hook2 truncation matings with NT-CENP-F and controls and plate of yeast growth. (E) Positive interactions grew and exhibited blue color upon Gal testing. Positive control growth was indicated by yeast transformed with pGBKT7-53 and pGADT7-T and our negative control used yeast expressing pGBTK-53 and the empty vector pGADT7.
We again used a Y2H approach to define the smallest effective binding domain of each protein with a series of truncation constructs. We determined that the first 474 amino acids of CENP-F are necessary to retain Hook2 binding, and amino acids 300-390 of Hook2 must be present to bind CENP-F (Figure 1, B–E). This N-terminal region of CENP-F contains a predicted leucine zipper, but no other predicted motifs. No other CENP-F sequences interact with Hook2 (Moynihan and Bader, unpublished data). The CENP-F–binding domain within Hook2 corresponds with a region between the microtubule-binding domain and organelle-binding domain identified by Szebenyi et al., 2007. Thus, the interaction of these two proteins reveals a third protein binding region within Hook2. These data also support the use of these interaction domains as endogenous function inhibitors in future experiments.
Transiently Expressed Hook2 and NT-CENP-F Interact and Colocalize at the Centrosome in Mammalian Cells
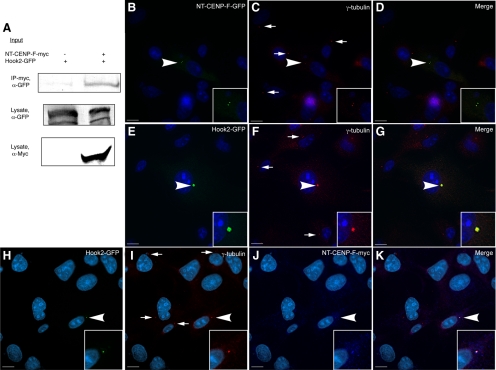
Given these initial data demonstrating CENP-F binding with Hook2, we sought to confirm this interaction in mammalian cell lines. Full-length Hook2 and NT-CENP-F were each tagged and expressed in COS-7 cells. Lysates from these cells were prepared, immunoprecipitated with an antibody to myc, and blotted for GFP (Figure 2A; Pooley et al., 2008). As seen in Figure 2A, effective immunoprecipitation of Hook2-GFP with NT-CENP-F-myc was readily observed, whereas no Hook2-GFP is pulled down in control lysates. This is consistent with the direct interaction found initially in yeast and led us to characterize the subcellular localization of these proteins. Again, the two tagged constructs were expressed in COS-7 cells and labeled with the respective antibodies to their epitope tags. NT-CENP-F was visualized at the centrosome by its colocalization with γ-tubulin (Figure 2, B–D), indicating the sufficiency of this peptide region for centrosomal targeting. Other expressed CENP-F constructs lacking amino acids 1-474 do not target to the centrosome, suggesting, along with our biochemical data, that this sequence contains information necessary to direct CENP-F to this subcellular domain (Feng et al., 2006; Evans et al., 2007). Full-length Hook2-GFP also localized to the centrosome when transfected in COS-7 cells (Figure 2, E–G). This result was not surprising, given the previously described role of Hook2 in MT dynamics at the centrosome (Szebenyi et al., 2007). As also demonstrated in Figure 2, transiently coexpressed NT-CENP-F-myc and Hook2-GFP colocalize at the centrosome, as identified by γ-tubulin (Figure 2, H–K). Therefore, these data with transiently expressed constructs support a novel localization of CENP-F to the centrosome and we next examined endogenous localization and interaction.
Figure 2.
Transiently expressed CENP-F and Hook2 proteins interact at the centrosome. COS-7 cells were cotransfected with NT-CENP-F-myc and Hook2-GFP. (A) Coimmunoprecipitation from transfected COS-7 cells. Lysate samples were pulled down with α-myc and blotted with α-GFP. (B–D) Singly transfected NT-CENP-F-GFP colocalizes with γ-tubulin centrosome marker (arrowhead). (E–G) Singly transfected Hook2-GFP colocalizes with γ-tubulin (arrowhead). (H–K) Coexpressed NT-CENP-F-myc and Hook2-GFP colocalized with γ-tubulin (arrowhead). Untransfected γ-tubulin at the centrosome marked with arrows. Bars 10 μm.
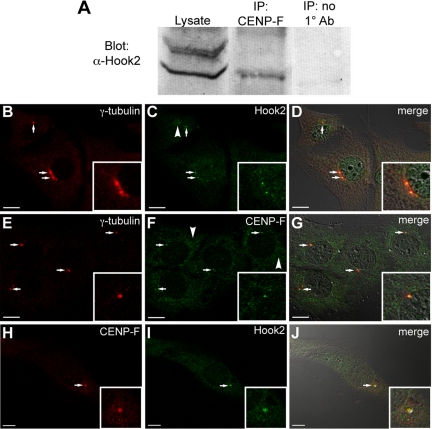
Endogenous Hook2 and CENP-F Interact and Colocalize at the Centrosome
The myoblast C2C12 cell line was used to coimmunoprecipitate CENP-F with binding partner Hook2 from murine cells. C2C12 cells were used in this experiment because the newly developed CENP-F antibody epitope is murine specific and does not recognize primate cells and tissue tested thus far. As seen in Figure 3A, Hook2 is identified in the CENP-F immunoprecipitant with an antibody to Hook2 (a generous gift of Dr. H. Kramer, The University of Texas Southwestern Medical Center at Dallas), further confirming the interaction of these two proteins. Although we do see an additional band in crude lysate, upon affinity purification with the CENP-F antibody, this nonspecific band is lost in Western blots. C2C12 murine myoblasts were also used to characterize the endogenous localization of these proteins. Cells were labeled with γ-tubulin centrosome marker and either Hook2 or CENP-F. Figure 3, B–G, shows the colocalization of each binding partner to the centrosome, whereas Figure 3, H–J, demonstrates colocalization of CENP-F and Hook2, with direct-labeled CENP-F antibody and Hook2. It should be noted that the centrosome is not the only localization for CENP-F or Hook2; these antibodies and previously reported antibodies show additional subcellular localizations (arrowheads, Figure 3). Because CENP-F plays roles within the cell separate from the centrosome, this result was expected. Together, these data demonstrate a novel localization and binding partner at the centrosome for CENP-F and predict another regulative capacity of this protein in MT network function.
Figure 3.
Endogenous CENP-F and Hook2 proteins interact and colocalize at the centrosome. (A) Endogenous coimmunoprecipitation with C2C12 cells. Lysate samples were pulled down with CENP-F antibody and blotted with Hook2 antibody. The third column shows no Hook2 pulled down when no CENP-F antibody was used in the pulldown. (B–J) Endogenous antibodies to CENP-F, Hook2, and γ-tubulin were used in C2C12 cells to show colocalization of all three proteins. (B) γ-tubulin centrosome marker. (C) Hook2. (D) Merge with differential interference contrast (DIC). (E) γ-tubulin. (F) CENP-F. (G) Merge. (H) CENP-F. (I) Hook2. (J) Merge with DIC. Arrows show colocalization and arrowheads show additional protein localizations. Bars 10 μm.
Ablated and Disrupted CENP-F Expression Attenuates MT Repolymerization after Nocodazole Challenge
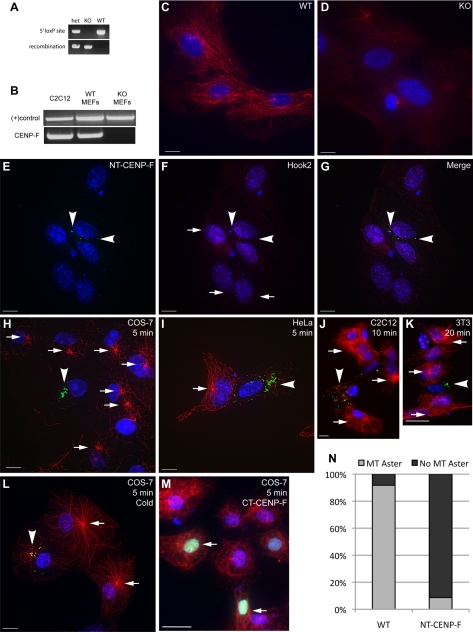
CENP-F function has been probed in a variety of ways: RNA interference, expression of truncation constructs, and morpholino knockdown (Holt et al., 2005; Soukoulis et al., 2005; Feng et al., 2006; Pooley et al., 2008). Here, we present the first report using CENP-F−/− mouse embryonic fibroblasts as a model to examine CENP-F function. To that effect, we used a genetic mouse model developed in the laboratory, in which the first five exons of CENP-F, containing the Hook2 binding domain, were flanked with loxP sites. This mouse line then crossed with the ubiquitously expressed cytomegalovirus (CMV)-Cre recombinase model (The Jackson Laboratory, Bar Harbor, ME). Figure 4A demonstrates effective excision and recombination of genomic DNA by PCR, and Figure 4B confirms the loss of CENP-F mRNA in the floxed region by reverse transcription (RT)-PCR. A complete characterization of the phenotype of this animal model will be presented elsewhere (Moynihan et al., unpublished data). Several preparations of MEFs were isolated from embryonic day 13.5 embryos and cultured for genotyping before experiments to analyze CENP-F function. Given the centrosomal localization of CENP-F described above and the role of the centrosome in MT initiation and organization, we used the wild-type (WT) and CENP-F−/− MEF populations in an assay of MT regulation: nocodazole washout and MT repolymerization.
Figure 4.
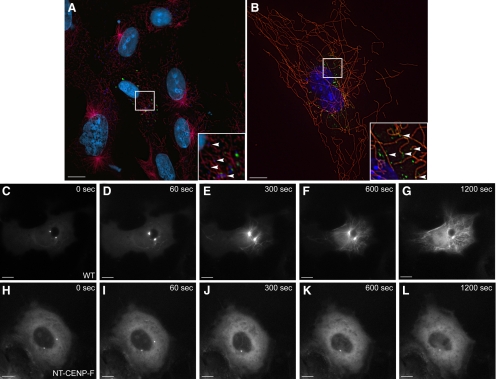
Disrupting the CENP-F/Hook2 interaction interferes with MT repolymerization after nocodazole challenge. (A) WT and CENP-F−/− MEFs were isolated and genotyped. The top bands use PCR primers flanking the 5′ loxP site of the floxed CENP-F allele, determining floxed versus WT. The bottom bands use PCR primers flanking the entire floxed sequence, allowing effective PCR only once the internal sequence has been excised, thus “recombination” has occurred. (B) RT-PCR confirmation of CENP-F−/− MEFs. NDRG4 primers and C2C12 lysate were used as positive controls. (C–D) WT and CENP-F−/− cells were treated with nocodazole, washed with fresh medium, and fixed after 5 min. (C) WT MEFs display reconstituted MT networks whereas (D) CENP-F−/− MEFs show absence of MT repolymerization. MTs are labeled in red. (E–G) Expression of NT-CENP-F (green) displaces endogenous Hook2 (red) expression from centrosome (arrowheads). Arrows indicate typical Hook2 localization to centrosome in untransfected cells. (H–M) Four cell lines were transfected with NT-CENP-F-GFP, treated with nocodazole for 2 h, washed with media, and allowed to recover. Transfected constructs are labeled green with an arrow, MTs are labeled red, and nuclei are labeled blue with 4,6-diamidino-2-phenylindole. (H–L) Arrows indicate MTOC aster formation in untransfected cells, whereas arrowheads point to NT-CENP-F–expressing cells without MTOC asters. (H) COS-7 cells at 5 min after washout (I) HeLa cells at 5 min. (J) C2C12 cells at 10 min. (K) 3T3 cells at 20 min. (L) Cold-treatment phenocopies nocodazole treatment in COS-7 cells. (M) Negative control C-terminal CENP-F construct (arrow) does not have the same effect, showing the specificity of this phenotype. (N) Quantification of MTOC aster formation in COS-7 cells after 5 min. Gray indicates percentage of counted cells with a MTOC aster, whereas black represents the percentage with no MTOC aster. WT cells reconstitute MTOC asters in >90% of cells, whereas NT-CENP-F–expressing cells repolymerize <10% MTOC asters. P < 0.0001. Bars, 10 μm.
Both WT and CENP-F−/− MEFs were plated and incubated with nocodazole, thoroughly washed with fresh medium, and allowed to recover for 5 min before fixation and labeling. WT MEFs display fully reconstituted MT networks and a substantive MTOC aster within this time period (Figure 4C). In contrast, Figure 4D demonstrates the markedly absent MT aster repolymerization in CENP-F−/− MEFs even at 5 min after washout. Some cytoplasmic MT repolymerization is seen in these null cells (see below; Figure 5B) but no clear MT aster forms. The lack of MT repolymerization from the centrosome in CENP-F−/− cells demonstrates a clear role for CENP-F in MTOC function. Specifically, these data signify a role for CENP-F in regulation of the MT network in correlation with MT repolymerization at the centrosome.
Figure 5.
CENP-F disruption of MT repolymerization is centrosome specific and inhibits MT aster nucleation. (A) RPE cells were transfected with NT-CENP-F (green), treated with nocodazole, and labeled for MTs (red), Golgi membrane (dark blue), and nuclei (4,6-diamidino-2-phenylindole, cyan). Transfected cells show attenuated MT aster repolymerization, but insets show MTs associated with Golgi membranes in the cytoplasm (arrowheads). (B) CENP-F−/− MEFs were treated with nocodazole and fixed after 5 min of recovery. Insets show MTs associated with Golgi membranes (arrowheads). MTs are labeled in red and Golgi marked with green. WT COS-7 cells (C–G) and NT-CENP-F transfected cells (H–L) were both transfected with EMTB-GFP, an MT marker, incubated with nocodazole, washed out, and immediately imaged for 20 min at 1 frame/20 s. (C–G) EMTB-marked MTs emanate from two MTOC asters very quickly, and MTs repopulate the entire cell within 10 min. (H–L) The EMTB-GFP pool remains cytoplasmic throughout the 20-min period in a cell expressing NT-CENP-F. After imaging, untransfected cells in the dish were checked and showed full MTOC recovery (data not shown). Bars, 10 μm.
Given that CENP-F−/− cells do not effectively repolymerize MT asters and the knowledge that centrosomal binding partner Hook2 affects MT organization, we next examined the effect of expression of the Hook2-binding domain of CENP-F on MT aster formation. Figure 2 shows that NT-CENP-F, the domain of CENP-F that binds Hook2, localizes to the centrosome. With expression of NT-CENP-F, cell viability was not adversely affected over the period of analysis (3–4 d), and in contrast to expression of the Nde-binding domain of CENP-F (Figure 1), the steady-state MT network is not overtly altered. However, the expression of NT-CENP-F does redistribute the localization of binding partner Hook2 away from the centrosome after nocodazole treatment, indicating a dominant disruption of endogenous CENP-F function (Figure 4, arrowheads, E–G). Endogenous Hook2 localization in untransfected cells (indicated by arrows in Figure 4F) marks the centrosome. Therefore, the NT-CENP-F truncation is a useful tool to study the function of CENP-F/Hook2 interaction at the centrosome. Previous studies of Hook2 function used protein truncations to elucidate function in MT repolymerization (Szebenyi et al., 2007), and we have used a similar strategy with NT-CENP-F as a analogous tool. To probe the effect NT-CENP-F on MTOC function and further validate CENP-F−/− data, nocodazole treatment and washout with MT repolymerization analysis was used. For nocodazole assays, COS-7 cells were used as standard single-MTOC aster cells (Clark and Meyer, 1999; Quintyne et al., 1999). Nonetheless, in total, four different mammalian cell lines were transfected with the tagged NT-CENP-F construct for nocodazole challenge and MT repolymerization experiments. MT repolymerization after washout varies slightly by cell type but generally, MTs begin to repopulate the cell within 180 s (Efimov et al., 2007). Similar to CENP-F-ablated MEFs, cells expressing NT-CENP-F exhibit a dramatic attenuation of MT repolymerization in COS-7 (Figure 4H), HeLa (Figure 4I), C2C12 (Figure 4J), and NIH-3T3 (Figure 4K) cell lines. Importantly, reduced MT repolymerization is not nocodazole specific, because depolymerization of MTs with cold treatment induces a similar phenotype in COS-7 cells (Figure 4L). Attenuation is specific to the expression of NT-CENP-F, as expression of C-terminal CENP-F does not result in any barrier to MT repolymerization (Figure 4M). Reformation of the MTOC aster was quantified in COS-7 cells and experimental cells showed fewer than 10% MTOC aster reformation, whereas WT cells show >90% MTOC aster reformation (Figure 4N; p < 0.0001). The impaired MT repolymerization can persist for a remarkably lengthy time period after washout (even at 20 min; Figure 4K), yet does come back after 2 h with the dominant-negative construct and some repolymerization in CENP-F−/− MEFs after 6 h (Supplemental Figure 4). This is a remarkable delay, considering the quick recovery seen in untransfected cells in under 5 min. Together, these data with CENP-F−/− cells and NT-CENP-F–expressing cells demonstrate the critical necessity of CENP-F in regulation of MT repolymerization.
CENP-F Regulation of MT Repolymerization Is Centrosome Specific
To delineate whether the attenuation of MT repolymerization is centrosome specific, we used two conditions to examine possible MT repolymerization from other subcellular localizations. Human RPE cells, a line recently shown to display abundant Golgi-nucleated MTs (Efimov et al., 2007), and WT and CENP-F−/− MEFs were treated with nocodazole treatment and washout. RPE cells were transfected with NT-CENP-F similar to experiments in Figure 4 with COS-7, HeLa, C2C12, and NIH-3T3 cells. RPE cells expressing NT-CENP-F phenocopy the attenuated MT repolymerization seen with other cell lines, with markedly decreased MTOC aster formation (Figure 5A). However, MTs can be seen emanating from disassociated Golgi membranes both in experimental and control cells (Figure 5A, inset). As noted above, some repolymerization was observed in CENP-F−/− cells, but not asters. In Figure 5B, CENP-F−/− MEFs that escape total block of MT repolymerization show a reduced and disorganized MT network and the MTs are associated with the dispersed Golgi membranes. WT MEFs do show a radial array but the images do not discount additional repolymerization from the Golgi membranes (Figure 4C). These data using CENP-F−/− MEFs and the RPE cell line with NT-CENP-F demonstrate the robust repolymerization of MTs from noncentrosomal sources, confirming that the role of CENP-F in regulating MT repolymerization is centrosome specific.
CENP-F Regulates MT Nucleation
To examine in detail the mechanism of the effect of CENP-F on MT repolymerization, a live-imaging approach was used to capture the entire repolymerization period after nocodazole washout. This strategy provided insight into the mechanism of the delay in MT repolymerization; visualization of the entire period shows concretely whether this delay is either due to lack of nucleation or instead, catastrophe in nascent MTs. Inhibited nucleation would obstruct any MT repolymerization, whereas MT catastrophe, depolymerization at the plus end of the growing MT, would allow initial growth from the centrosome before dispersal. Both control and experimental COS-7 cells were transfected with a MT-coating protein, EMTB-GFP, to visualize MT polymerization. Experimental samples were cotransfected with NT-CENP-F-Cerulean. After nocodazole treatment, both control and experimental cells show a typical cytoplasmic pool of EMTB-GFP before MT repolymerization begins (Figure 5, C and H). Post-nocodazole washout, control MTs clearly nucleate and polymerize from the centrosome into an organized network (Figure 5, C–G); asters form within 60 s and MTs reach the cell periphery in just over 6 min. In contrast, expression of NT-CENP-F alters this process considerably. As shown in Figure 5, H–L, NT-CENP-F–expressing COS-7 cells are unable to repolymerize the MT network after nocodazole washout, even up to 20 min after washout. This dramatic result is visualized in the full set of movies for both NT-CENP-F and WT cells shown in Supplemental Figures 1–3. No MT repolymerization is seen in the experimental condition, and this severe retardation of MT outgrowth with expression of NT-CENP-F demonstrates that CENP-F plays a role in nucleation of the MT network from the centrosome.
CENP-F Regulates Nucleation and Anchoring of MT Polymerization from the Centrosome
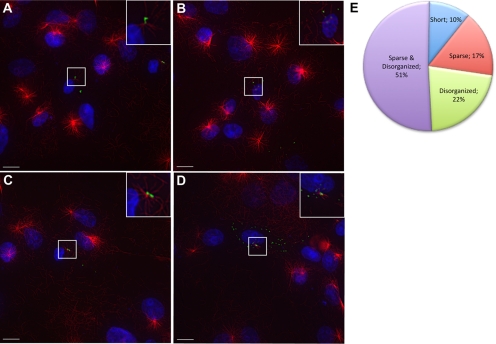
A minority of cells (<10%; Figure 4N) escape inhibited MT repolymerization induced by NT-CENP-F and thus provide an additional method of investigating the mechanism of CENP-F function. Live-imaging data demonstrated a clear nucleation defect with CENP-F disruption; therefore, we next examined experimental NT-CENP-F–expressing cells with an incomplete attenuation phenotype. This population of cells is characterized by significantly diminished MT repolymerization after nocodazole treatment and within this range of phenotypes, three phenotype categories would indicate the spectrum of CENP-F functional mechanism: sparse, disorganized, and short MT asters (Chakravarty et al., 2004; Zhapparova et al., 2007; Fumoto et al., 2009). The presence of full-length but fewer MTs emanating from the centrosome in expressing cells would further support our previously seen nucleation defects; this category is the sparse phenotype. Alternatively, if MTs are disorganized rather than arrayed as a centrosomal centered aster, there could be additional interference with MT anchoring; this category is termed disorganized. Whereas shorter, full MT asters with disrupted CENP-F function would suggest an elongation defect—the short phenotype. Using these categories, we classified NT-CENP-F–expressing cells that escape inhibition of MT repolymerization after nocodazole washout (<10% of all NT-CENP-F–expressing cells; Figure 4N). Figure 6 shows a representative example of each condition described. Figure 6A shows both a sparse and disorganized aster, which is most common in the cells with an incomplete phenotype (51.1 ± 11.4%). Figure 6B lacks a MTOC aster but disorganized MTs are present; in this case, both nucleation and anchoring could be affected and/or other parts of the cell could be acting as MTOCs (21.8 ± 5.3%). Figure 6C shows a representative image of a sparsely populated, yet organized radial array of MTs, indicative of inhibited nucleation (16.6 ± 5.8%). Figure 6D displays a characteristic example of a populated, yet disorganized MTOC with mostly short MTs, characteristic of a potential elongation defect (10.4 ± 2.1%). The relative populations of each of these phenotypes within the general category of “incomplete” is shown in Figure 6E and show a continuum of disrupted MT nucleation and anchoring, because nearly 90% of all cells counted fall into the sparse and disorganized characterization. Nevertheless, it should be noted that this range of incomplete phenotypes could also be attributed simply to a partial loss-of-funtion in nucleation with expression of NT-CENP-F. Thus, these results reveal a role in MT nucleation and possibly anchoring within CENP-F regulation of MT repolymerization.
Figure 6.
CENP-F functions in MTOC nucleation and anchoring. NT-CENP-F–expressing COS-7 cells were examined for incomplete inhibition of MT repolymerization. Various phenotypes are shown. (A) Sparse and disorganized MTs, most commonly seen with NT-CENP-F–expressing cells. (B) Sparse aster. (C) Disorganized MTs. (D) Short, disorganized aster. (E) Quantification of each phenotype along spectrum. Bars, 10 μm.
Disruption of CENP-F Function Redistributes Centrosomal Proteins
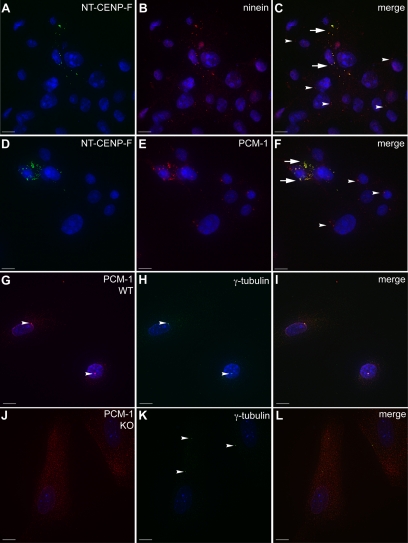
Interestingly, after application of nocodazole, the localization of NT-CENP-F scatters into cytoplasmic puncta and remains dispersed even after 10–20 min in fresh media. As seen in Figure 4, after nocodazole washout, NT-CENP-F is no longer confined to a centrosomal localization demonstrated in Figure 2. Instead, NT-CENP-F is spread out as a cloud of puncta in the cytoplasm. It is possible that these nocodazole-induced puncta harbor proteins normally present at the centrosome that function there in MT organization. As demonstrated above in Figure 4, binding partner Hook2 redistributed with NT-CENP-F after nocodazole treatment in this manner. We next examined other centrosomal proteins involved in MT organization. Ninein, a mother centriole MT-anchoring protein recently shown to also participate in MT nucleation (Bouckson-Castaing et al., 1996; Mogensen et al., 2000; Ou et al., 2002; Delgehyr et al., 2005), shows distinct redistribution to NT-CENP-F puncta (Figure 7, arrows, A–C). Disruption of ninein causes a delay in MT nucleation as well as anchoring defects, similar to the phenotypes seen in this study (Delgehyr et al., 2005). Overexpression of ninein has been shown to increase γ-tubulin recruitment to the centrosome (Stillwell et al., 2004), and knockdown of human ninein results in reduced expression of γ-tubulin at the centrosome (Lin et al., 2006); both of these studies indicate a link between ninein and the γ-tubulin ring complex. Intriguingly, NT-CENP-F does not redistribute γ-tubulin (Supplemental Figure 5, A–C), indicating the complex relationship between MT regulatory proteins at the centrosome. Also, pericentrin or centrin1 proteins are not redistributed with expression of NT-CENP-F either (Supplemental Figure 5, D–I). However, the anchoring of MTs at the centrosome generates the radial array of the MT aster and another specific centrosomal protein regulates the anchoring process: PCM-1, a protein seen in the centriolar satellites that are required for MT anchoring (Mack et al., 1998; Kubo et al., 1999; Balczon et al., 2002; Dammermann and Merdes, 2002). PCM-1 also affects the centrosomal proteins ninein, centrin, and pericentrin (Dammermann and Merdes, 2002). The characterization of incomplete attenuated MT repolymerization implicated an anchoring defect with expression of NT-CENP-F and it is possible that the disruption of CENP-F affects other centrosomal proteins in the anchoring process, such as PCM-1. To test whether PCM-1 is sequestered in NT-CENP-F puncta after nocodazole treatment, we used COS-7 cells fixed 5 min after nocodazole washout. Figure 7, D–F, clearly demonstrates that PCM-1 does disperse with NT-CENP-F (arrows), whereas untransfected cells show normal PCM-1 localization (arrowheads). In addition, in CENP-F−/− MEFs, proper recruitment and/or stabilization of centrosome proteins could be impaired due to lack of CENP-F. Accordingly, we examined PCM-1 localization in both MEF populations after nocodazole challenge and WT MEFs show clear centrosomal PCM-1 localization (Figure 7, arrowheads, G–I), whereas CENP-F−/− MEFs only exhibit diffuse cytoplasmic PCM-1 labeling (Figure 7, J–L). Interestingly, Hook2 and ninein localization remained centrosomal in CENP-F−/− MEFs (Supplemental Figure 5, J–R), indicating a potential shift in mechanism between expression of truncated CENP-F and ablation of endogenous CENP-F. However, the redistribution of the PCM-1–anchoring protein may explain the inability of CENP-F−/− cells and NT-CENP-F–expressing cells to coordinate MT growth from the centrosome, and together, these data provide a link to the mechanism of CENP-F in MT network regulation.
Figure 7.
Centrosomal proteins are redistributed by CENP-F disruption and deletion. (A–F) COS-7 cells expressing NT-CENP-F show colocalization of the CENP-F truncation puncta with ninein and PCM-1. An arrowhead indicates normal centrosomal protein expression and aberrant colocalization with NT-CENP-F is labeled with an arrow. (A) NT-CENP-F. (B) ninein. (C) Merge. (D) NT-CENP-F. (E) PCM-1. (F) Merge. (G–L) WT and CENP-F−/− MEFs were labeled for PCM-1 (red) and γ-tubulin (green). (I) PCM-1 clearly localizes with γ-tubulin in WT MEFs, whereas in (L) CENP-F−/− MEFs, PCM-1 is diffusely cytoplasmic. Bars, 10 μm.
DISCUSSION
In this study, we have shown a previously unrecognized centrosomal localization, binding partner, and function of CENP-F. Hook2, a centrosomal participant in MT regulation, was confirmed as a direct binding partner of CENP-F. Ablation of CENP-F function in newly developed CENP-F−/− cells dramatically attenuates MT repolymerization and overexpression of the N-terminal portion of CENP-F that binds Hook2 phenocopies this event. Our experiments demonstrate that this is due to lack of nucleation, not catastrophe events, and that the Golgi retains MT nucleation activity. Therefore, this phenotype is specific to microtubule repolymerization at the centrosome. In addition, incomplete phenotype analysis shows that experimentally altered cells that escape inhibited MT repolymerization have fewer and more disorganized MT emanating from the centrosome, indicating a defect in anchoring as well as nucleation. Expression of NT-CENP-F redistributes localization of centrosomal proteins PCM-1, ninein, and mislocalization of PCM-1 protein is also seen in CENP-F−/− MEFs, an important and additional indicator of CENP-F–anchoring function at the centrosome. Together, these data reveal a novel role for CENP-F at the centrosome in MT formation via its interaction with Hook2 and predict a broader role for this protein in regulation of the MT network.
CENP-F Displays a Novel Endogenous Localization, Binding Partner, and Function at the Centrosome
CENP-F (mitosin) was independently discovered as a cell cycle-dependent KT associated and Rb-binding protein (Rattner et al., 1993; Zhu et al., 1995b). Because those initial discoveries, homologous proteins have been found in other vertebrates, invertebrates, and prokaryotes (Litvin et al., 1993; Goodwin et al., 1999; Moore et al., 1999; Ortiz et al., 1999; Redkar et al., 2002). These are large proteins and have a predicted coiled-coil structure with multiple intervening protein/protein binding domains (Figure 1) (Zhu et al., 1995a; Wei et al., 1996; Goodwin et al., 1999) and varying localizations (Rattner et al., 1993; Liao et al., 1995; Zhu et al., 1995a,b; Ashar et al., 2000; Dees et al., 2000; Hussein and Taylor, 2002; Ashe et al., 2004; Holt et al., 2005; Soukoulis et al., 2005; Yang et al., 2005; Zhou et al., 2005; Pooley et al., 2006, 2008; Evans et al., 2007). Despite the various localizations seen in the analysis of CENP-F homologues, many well-conserved domains have been shown to function and/or bind similar partners in more than one family member. CENP-F has been widely described as a nuclear, KT-associated protein and the conserved region responsible for this localization is found in the C terminus (Zhu et al., 1995a). More recently, analysis of CENP-F function has moved beyond the carboxy terminus into the central and N-terminal regions of the protein (Soukoulis et al., 2005; Pooley et al., 2006, 2008). These more recently characterized domains show localization and function of CENP-F in the cytoplasm. In this study, we reveal that CENP-F localizes to the centrosome and regulates MT organizing activity from this organelle.
The interaction of CENP-F with Hook2 reveals additional cytoplasmic function and further substantiates centrosomal localization. Hook2 primarily localizes to the centrosome and binds another centrosomal protein, centriolin (Szebenyi et al., 2007). The Hook2 linker protein has been shown to participate in MT growth and organization at the centrosome, but the mechanism underlying this activity remains unknown. Our data identify a previously undetermined binding domain within Hook2 in the central coiled-coil region. The interaction with Hook2 is of particular interest as the Hook family has been described a linker between the MT and specific organelles (Walenta et al., 2001; Mendoza-Lujambio et al., 2002; Malone et al., 2003; Gönczy, 2004; Linstedt, 2004; Szebenyi et al., 2007). Given the established interaction of CENP-F with MT regulators such as Nude/Nudel (Soukoulis et al., 2005; Vergnolle and Taylor, 2007) and tubulin itself (Feng et al., 2006), the newly identified relationship with the Hook family potentially links CENP-F to a myriad of subcellular domains and organelle-specific activities.
CENP-F Regulates MT Nucleation and Anchoring at the Centrosome with Other Centrosomal Proteins
In this study, we see an even more dramatic attenuation of MT repolymerization with disruption of CENP-F, compared with disruption of binding partner Hook2 (Szebenyi et al., 2007). This effect is expansive, as CENP-F−/− MEFs shown a dramatic delay in MT repolymerization, providing essential supportive evidence of this new CENP-F function. It is significant to note that a highly similar phenotype is observed with several cell lines of different species when transfected with NT-CENP-F (Figure 4). Given that this role of CENP-F was heretofore unrecognized, we performed several experiments to investigate the mechanism of CENP-F function at the MTOC. First, live-imaging data showed that this effect was not due to premature catastrophe and subsequent depolymerization of nascent MT (Figure 5). In addition, this effect is centrosome specific because Golgi membranes, known to initiate MT nucleation and polymerization (Efimov et al., 2007), retain the capability to nucleate MTs in the presence of NT-CENP-F, whereas the centrosome does not (Figure 6). These data further corroborate centrosomal localization of CENP-F and led us to explore the specific function of CENP-F at the centrosome in MT repolymerization.
To determine this mechanism, we closely examined those NT-CENP-F–expressing cells that escaped complete inhibition of MT repolymerization. We reasoned that by characterizing these MTs, we would provide initial evidence of function in one or more of three processes controlling MT network emanation from the centrosome: nucleation, elongation, and/or anchoring. As seen in Figure 6, we saw substantially fewer MTs emanating from the centrosome and poor organization in the few that formed throughout the continuum of incomplete phenotypes. These data suggest a role for CENP-F in MT nucleation and possibly anchoring from the centrosome, and constitute an initial step in defining the mechanism of the attenuation phenotype.
Inhibition of MT regrowth through nucleation/anchoring defects at the centrosome is a constant observed with disruption of CENP-F function whether it be by ablation or expression of truncation mutants. With the loss of CENP-F function, it was plausible that specific centrosomal proteins be displaced from the centrosome where complex interactions are essential for protein targeting (Delgehyr et al., 2005; Hames et al., 2005; Srsen et al., 2006). Therefore, we examined the localization of several diverse centrosomal proteins concurrent with disruption of CENP-F. Our results show definitive dispersal of Hook2 along with endogenous ninein and PCM-1 to NT-CENP-F–positive puncta distal from the centrosome. Both ninein and PCM-1 have well-documented roles in nucleation and anchoring of MT from the centrosome (Dammermann and Merdes, 2002; Delgehyr et al., 2005; Azimzadeh and Bornens, 2007). Critically, dispersal of endogenous PCM-1 is observed in both CENP-F−/− MEFs and NT-CENP-F–expressing cells. PCM-1 has a well-documented role in MT anchoring from the centrosome (Mogensen et al., 2000; Dammermann and Merdes, 2002; Ou et al., 2002; Delgehyr et al., 2005), as both expression of PCM-1 truncations and knockdown results in disorganization of the MT array, both in steady state and nocodazole challenge (Dammermann and Merdes, 2002). In addition, with the expression of a C-terminal deletion PCM-1 construct, ectopic granules formed and redistributed endogenous ninein, centrin, and pericentrin, and PCM-1 (Dammermann and Merdes, 2002). Thus, the relocation of this key regulator may a central role in the centrosome-specific inhibition of MT regrowth in both experimental models presented here.
Studies by Delgehy and Stillwell suggest that localization of γ-tubulin to the centrosome is necessary for MT nucleation (Stillwell et al., 2004; Delgehyr et al., 2005). With this in mind, it is interesting to note that γ-tubulin does not seem to redistribute in either experimental model presented here. However, direct disruption of PCM-1 function does not alter γ-tubulin localization at the centrosome yet produces the same inhibition of MT regrowth observed in CENP-F−/− or NT-CENP-F–expressing cells (Figure 4; Dammermann and Merdes, 2002). Thus, it seems that γ-tubulin localization in the absence of CENP-F function is not adequate to support MT regrowth from the centrosome and that localization of PCM-1 is dependent on intact CENP-F. In addition, with expression of NT-CENP-F (the Hook2-binding domain), Hook2 redistributed to the NT-CENP-F–positive puncta and was not observed at the centrosome. However, when the same experiment was conducted with CENP-F−/− cells, Hook2 localized to the centrosome in the absence of its binding partner suggesting that other mechanisms direct its localization. Nevertheless, both NT-CENP-F expression and CENP-F ablation experiments lead to a common phenotype, attenuated MT repolymerization, and this difference in centrosomal protein localization could indicate different mechanisms in each scenario. Although the interactions and relationships of proteins are complex, our data reveal that CENP-F as an essential component critical for the assembly and/or stability of essential MT organizers at the centrosome and disruption of its function results in a loss of centrosomal regulation of the MT network.
CENP-F Is a Possible Master Regulator of MT Network Processes with Cellular Organelles
The novel role and localization of CENP-F presented here open a new perspective on CENP-F function as a whole. The understanding that CENP-F functions with MTs at the centrosome is reminiscent of the highly regulated nuclear MT association with KTs that is well studied with CENP-F. The KT is a complex organelle that captures plus ends of MTs and links them to the chromosome; the centrosome shows a similar level of complexity in regulating the minus ends of MTs and anchoring them in a radial array. The association between MTs and CENP-F is seen in another cellular process: vesicular transport with SNARE proteins SNAP25 and syntaxin 4 (Pooley et al., 2006, 2008). Vesicular trafficking is highly dependent on regulated association with MTs (Ishiki and Klip, 2005; Caviston and Holzbaur, 2006; Soldati and Schliwa, 2006; Vedrenne and Hauri, 2006; Hehnly and Stamnes, 2007). In this study, we have identified a third MT-related process regulated by CENP-F: centrosomal MT nucleation and anchoring. Together, these roles suggest CENP-F as a global linker protein to the MT network with many different organelles. Indeed, with so many unexplored motifs on this large protein, there are various other modalities possible within this context of CENP-F function.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. H. Kramer for the Hook2 reagents, Dr. Kilkenny Rocheleau for microscopy expertise, Samyukta Reddy for work with the yeast two-hybrid, Dr. Soukoulis for CENP-F antibody development, and members of the Bader and Dees laboratories for helpful discussions and critical reading of this manuscript. Some experiments and data analysis were performed in part through the use of the VUMC Cell Imaging Shared Resource (supported by National Institutes of Health grants CA-68485, DK-20593, DK-58404, HD-15052, DK-59637, and EY08126) and the Imaging Resource of the Epithelial Biology Center. We are thankful for the following support: K.L.M., American Heart Association Predoctoral grant 09PRE2170006 and NIH NRSA 5 T32HL07751; P.M.M., American Heart Association predoctoral grant 09PRE2260729; I. K., National Institutes of Health National Institute of General Medical Sciences grant 1R01 GM-078373-01; and D.M.B., National Institutes of Health grant R01 HL-037675.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-07-0560) on October 7, 2009.
REFERENCES
- Altschul S. F., Lipman D. J. Protein database searches for multiple alignments. Proc. Natl. Acad. Sci. USA. 1990;87:5509–5513. doi: 10.1073/pnas.87.14.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashar H. R., James L., Gray K., Carr D., Black S., Armstrong L., Bishop W. R., Kirschmeier P. Farnesyl transferase inhibitors block the farnesylation of CENP-E and CENP-F and alter the association of CENP-E with the microtubules. J. Biol. Chem. 2000;275:30451–30457. doi: 10.1074/jbc.M003469200. [DOI] [PubMed] [Google Scholar]
- Ashe M., Pabon-Peña L., Dees E., Price K. L., Bader D. LEK1 is a potential inhibitor of pocket protein-mediated cellular processes. J. Biol. Chem. 2004;279:664–676. doi: 10.1074/jbc.M308810200. [DOI] [PubMed] [Google Scholar]
- Azimzadeh J., Bornens M. Structure and duplication of the centrosome. J. Cell Sci. 2007;120:2139–2142. doi: 10.1242/jcs.005231. [DOI] [PubMed] [Google Scholar]
- Balczon R., Simerly C., Takahashi D., Schatten G. Arrest of cell cycle progression during first interphase in murine zygotes microinjected with anti-PCM-1 antibodies. Cell Motil. Cytoskeleton. 2002;52:183–192. doi: 10.1002/cm.10043. [DOI] [PubMed] [Google Scholar]
- Bomont P., Maddox P., Shah J. V., Desai A. B., Cleveland D. W. Unstable microtubule capture at kinetochores depleted of the centromere-associated protein CENP-F. EMBO J. 2005;24:3927–3939. doi: 10.1038/sj.emboj.7600848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckson-Castaing V., Moudjou M., Ferguson D. J., Mucklow S., Belkaid Y., Milon G., Crocker P. R. Molecular characterisation of ninein, a new coiled-coil protein of the centrosome. J. Cell Sci. 1996;109:179–190. doi: 10.1242/jcs.109.1.179. [DOI] [PubMed] [Google Scholar]
- Caviston J. P., Holzbaur E. L. Microtubule motors at the intersection of trafficking and transport. Trends Cell Biol. 2006;16:530–537. doi: 10.1016/j.tcb.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Chakravarty A., Howard L., Compton D. A. A mechanistic model for the organization of microtubule asters by motor and non-motor proteins in a mammalian mitotic extract. Mol. Biol. Cell. 2004;15:2116–2132. doi: 10.1091/mbc.E03-08-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark I. B., Meyer D. I. Overexpression of normal and mutant Arp1alpha (centractin) differentially affects microtubule organization during mitosis and interphase. J. Cell Sci. 1999;112:3507–3518. doi: 10.1242/jcs.112.20.3507. [DOI] [PubMed] [Google Scholar]
- Dammermann A., Merdes A. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J. Cell Biol. 2002;159:255–266. doi: 10.1083/jcb.200204023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dees E., Pabón-Peña L. M., Goodwin R. L., Bader D. Characterization of CMF1 in avian skeletal muscle. Dev. Dyn. 2000;219:169–181. doi: 10.1002/1097-0177(2000)9999:9999<::aid-dvdy1055>3.3.co;2-2. [DOI] [PubMed] [Google Scholar]
- Delgehyr N., Sillibourne J., Bornens M. Microtubule nucleation and anchoring at the centrosome are independent processes linked by ninein function. J. Cell Sci. 2005;118:1565–1575. doi: 10.1242/jcs.02302. [DOI] [PubMed] [Google Scholar]
- Efimov A., et al. Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev. Cell. 2007;12:917–930. doi: 10.1016/j.devcel.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans H. J., Edwards L., Goodwin R. L. Conserved C-terminal domains of mCenp-F (LEK1) regulate subcellular localization and mitotic checkpoint delay. Exp. Cell Res. 2007;313:2427–2437. doi: 10.1016/j.yexcr.2007.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Huang H., Yen T. J. CENP-F is a novel microtubule-binding protein that is essential for kinetochore attachments and affects the duration of the mitotic checkpoint delay. Chromosoma. 2006;115:320–329. doi: 10.1007/s00412-006-0049-5. [DOI] [PubMed] [Google Scholar]
- Fumoto K., Kadono M., Izumi N., Kikuchi A. Axin localizes to the centrosome and is involved in microtubule nucleation. EMBO Rep. 2009;10:606–613. doi: 10.1038/embor.2009.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P. Centrosomes: hooked on the nucleus. Curr. Biol. 2004;14:R268–R270. doi: 10.1016/j.cub.2004.03.020. [DOI] [PubMed] [Google Scholar]
- Goodwin R. L., Pabón-Peña L. M., Foster G. C., Bader D. The cloning and analysis of LEK1 identifies variations in the LEK/centromere protein F/mitosin gene family. J. Biol. Chem. 1999;274:18597–18604. doi: 10.1074/jbc.274.26.18597. [DOI] [PubMed] [Google Scholar]
- Hames R. S., Crookes R. E., Straatman K. R., Merdes A., Hayes M. J., Faragher A. J., Fry A. M. Dynamic recruitment of Nek2 kinase to the centrosome involves microtubules, PCM-1, and localized proteasomal degradation. Mol. Biol. Cell. 2005;16:1711–1724. doi: 10.1091/mbc.E04-08-0688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehnly H., Stamnes M. Regulating cytoskeleton-based vesicle motility. FEBS Lett. 2007;581:2112–2118. doi: 10.1016/j.febslet.2007.01.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt S. V., Vergnolle M. A., Hussein D., Wozniak M. J., Allan V. J., Taylor S. S. Silencing Cenp-F weakens centromeric cohesion, prevents chromosome alignment and activates the spindle checkpoint. J. Cell Sci. 2005;118:4889–4900. doi: 10.1242/jcs.02614. [DOI] [PubMed] [Google Scholar]
- Hussein D., Taylor S. S. Farnesylation of Cenp-F is required for G2/M progression and degradation after mitosis. J. Cell Sci. 2002;115:3403–3414. doi: 10.1242/jcs.115.17.3403. [DOI] [PubMed] [Google Scholar]
- Ishiki M., Klip A. Minireview: recent developments in the regulation of glucose transporter-4 traffic: new signals, locations, and partners. Endocrinology. 2005;146:5071–5078. doi: 10.1210/en.2005-0850. [DOI] [PubMed] [Google Scholar]
- Kubo A., Sasaki H., Yuba-Kubo A., Tsukita S., Shiina N. Centriolar satellites: molecular characterization, ATP-dependent movement toward centrioles and possible involvement in ciliogenesis. J. Cell Biol. 1999;147:969–980. doi: 10.1083/jcb.147.5.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laoukili J., Kooistra M. R., Brás A., Kauw J., Kerkhoven R. M., Morrison A., Clevers H., Medema R. H. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat. Cell Biol. 2005;7:126–136. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- Liao H., Winkfein R. J., Mack G., Rattner J. B., Yen T. J. CENP-F is a protein of the nuclear matrix that assembles onto kinetochores at late G2 and is rapidly degraded after mitosis. J. Cell Biol. 1995;130:507–518. doi: 10.1083/jcb.130.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. C., Cheng T. S., Hsu C. M., Wu C. H., Chang L. S., Shen Z. S., Yeh H. M., Chang L. K., Howng S. L., Hong Y. R. Characterization and functional aspects of human ninein isoforms that regulated by centrosomal targeting signals and evidence for docking sites to direct gamma-tubulin. Cell Cycle. 2006;5:2517–2527. doi: 10.4161/cc.5.21.3404. [DOI] [PubMed] [Google Scholar]
- Linstedt A. D. Positioning the Golgi apparatus. Cell. 2004;118:271–272. doi: 10.1016/j.cell.2004.07.015. [DOI] [PubMed] [Google Scholar]
- Litvin J., Montgomery M. O., Goldhamer D. J., Emerson C. P., Bader D. M. Identification of DNA-binding protein(s) in the developing heart. Dev. Biol. 1993;156:409–417. doi: 10.1006/dbio.1993.1088. [DOI] [PubMed] [Google Scholar]
- Mack G. J., Rees J., Sandblom O., Balczon R., Fritzler M. J., Rattner J. B. Autoantibodies to a group of centrosomal proteins in human autoimmune sera reactive with the centrosome. Arthritis Rheum. 1998;41:551–558. doi: 10.1002/1529-0131(199803)41:3<551::AID-ART22>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Malone C. J., Misner L., Le Bot N., Tsai M. C., Campbell J. M., Ahringer J., White J. G. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell. 2003;115:825–836. doi: 10.1016/s0092-8674(03)00985-1. [DOI] [PubMed] [Google Scholar]
- Mendoza-Lujambio I., Burfeind P., Dixkens C., Meinhardt A., Hoyer-Fender S., Engel W., Neesen J. The Hook1 gene is non-functional in the abnormal spermatozoon head shape (azh) mutant mouse. Hum. Mol. Genet. 2002;11:1647–1658. doi: 10.1093/hmg/11.14.1647. [DOI] [PubMed] [Google Scholar]
- Mogensen M. M., Malik A., Piel M., Bouckson-Castaing V., Bornens M. Microtubule minus-end anchorage at centrosomal and non-centrosomal sites: the role of ninein. J. Cell Sci. 2000;113:3013–3023. doi: 10.1242/jcs.113.17.3013. [DOI] [PubMed] [Google Scholar]
- Moore L. L., Morrison M., Roth M. B. HCP-1, a protein involved in chromosome segregation, is localized to the centromere of mitotic chromosomes in Caenorhabditis elegans. J. Cell Biol. 1999;147:471–480. doi: 10.1083/jcb.147.3.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz J., Stemmann O., Rank S., Lechner J. A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. 1999;13:1140–1155. doi: 10.1101/gad.13.9.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou Y. Y., Mack G. J., Zhang M., Rattner J. B. CEP110 and ninein are located in a specific domain of the centrosome associated with centrosome maturation. J. Cell Sci. 2002;115:1825–1835. doi: 10.1242/jcs.115.9.1825. [DOI] [PubMed] [Google Scholar]
- Pooley R. D., Moynihan K. L., Soukoulis V., Reddy S., Francis R., Lo C., Ma L. J., Bader D. M. Murine CENPF interacts with syntaxin 4 in the regulation of vesicular transport. J. Cell Sci. 2008;121:3413–3421. doi: 10.1242/jcs.032847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pooley R. D., Reddy S., Soukoulis V., Roland J. T., Goldenring J. R., Bader D. M. CytLEK1 is a regulator of plasma membrane recycling through its interaction with SNAP-25. Mol. Biol. Cell. 2006;17:3176–3186. doi: 10.1091/mbc.E05-12-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintyne N. J., Gill S. R., Eckley D. M., Crego C. L., Compton D. A., Schroer T. A. Dynactin is required for microtubule anchoring at centrosomes. J. Cell Biol. 1999;147:321–334. doi: 10.1083/jcb.147.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattner J. B., Rao A., Fritzler M. J., Valencia D. W., Yen T. J. CENP-F is a .ca 400 kDa kinetochore protein that exhibits a cell-cycle dependent localization. Cell Motil. Cytoskeleton. 1993;26:214–226. doi: 10.1002/cm.970260305. [DOI] [PubMed] [Google Scholar]
- Redkar A., deRiel J. K., Xu Y. S., Montgomery M., Patwardhan V., Litvin J. Characterization of cardiac muscle factor 1 sequence motifs: retinoblastoma protein binding and nuclear localization. Gene. 2002;282:53–64. doi: 10.1016/s0378-1119(01)00789-2. [DOI] [PubMed] [Google Scholar]
- Robertson J. B., Zhu T., Nasreen S., Kilkenny D., Bader D., Dees E. CMF1-Rb interaction promotes myogenesis in avian skeletal myoblasts. Dev. Dyn. 2008;237:1424–1433. doi: 10.1002/dvdy.21544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldati T., Schliwa M. Powering membrane traffic in endocytosis and recycling. Nat. Rev. Mol. Cell Biol. 2006;7:897–908. doi: 10.1038/nrm2060. [DOI] [PubMed] [Google Scholar]
- Soukoulis V., Reddy S., Pooley R. D., Feng Y., Walsh C. A., Bader D. M. Cytoplasmic LEK1 is a regulator of microtubule function through its interaction with the LIS1 pathway. Proc. Natl. Acad. Sci. USA. 2005;102:8549–8554. doi: 10.1073/pnas.0502303102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srsen V., Gnadt N., Dammermann A., Merdes A. Inhibition of centrosome protein assembly leads to p53-dependent exit from the cell cycle. J. Cell Biol. 2006;174:625–630. doi: 10.1083/jcb.200606051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillwell E. E., Zhou J., Joshi H. C. Human ninein is a centrosomal autoantigen recognized by CREST patient sera and plays a regulatory role in microtubule nucleation. Cell Cycle. 2004;3:923–930. [PubMed] [Google Scholar]
- Szebenyi G., Hall B., Yu R., Hashim A., Krämer H. Hook2 localizes to the centrosome, binds directly to Centriolin/CEP110 and contributes to centrosomal function. Traffic. 2007;8:32–46. doi: 10.1111/j.1600-0854.2006.00511.x. [DOI] [PubMed] [Google Scholar]
- Vedrenne C., Hauri H. P. Morphogenesis of the endoplasmic reticulum: beyond active membrane expansion. Traffic. 2006;7:639–646. doi: 10.1111/j.1600-0854.2006.00419.x. [DOI] [PubMed] [Google Scholar]
- Vergnolle M. A., Taylor S. S. Cenp-F links kinetochores to Ndel1/Nde1/Lis1/dynein microtubule motor complexes. Curr. Biol. 2007;17:1173–1179. doi: 10.1016/j.cub.2007.05.077. [DOI] [PubMed] [Google Scholar]
- Walenta J. H., Didier A. J., Liu X., Krämer H. The Golgi-associated hook3 protein is a member of a novel family of microtubule-binding proteins. J. Cell Biol. 2001;152:923–934. doi: 10.1083/jcb.152.5.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y., Bader D., Litvin J. Identification of a novel cardiac-specific transcript critical for cardiac myocyte differentiation. Development. 1996;122:2779–2789. doi: 10.1242/dev.122.9.2779. [DOI] [PubMed] [Google Scholar]
- Yang Z., Guo J., Chen Q., Ding C., Du J., Zhu X. Silencing mitosin induces misaligned chromosomes, premature chromosome decondensation before anaphase onset, and mitotic cell death. Mol. Cell Biol. 2005;25:4062–4074. doi: 10.1128/MCB.25.10.4062-4074.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhapparova O. N., Burakov A. V., Nadezhdina E. S. The centrosome keeps nucleating microtubules but looses the ability to anchor them after the inhibition of dynein-dynactin complex. Biochemistry. 2007;72:1233–1240. doi: 10.1134/s0006297907110090. [DOI] [PubMed] [Google Scholar]
- Zhou X., Wang R., Fan L., Li Y., Ma L., Yang Z., Yu W., Jing N., Zhu X. Mitosin/CENP-F as a negative regulator of activating transcription factor-4. J. Biol. Chem. 2005;280:13973–13977. doi: 10.1074/jbc.M414310200. [DOI] [PubMed] [Google Scholar]
- Zhu X., Chang K. H., He D., Mancini M. A., Brinkley W. R., Lee W. H. The C terminus of mitosin is essential for its nuclear localization, centromere/kinetochore targeting, and dimerization. J. Biol. Chem. 1995a;270:19545–19550. doi: 10.1074/jbc.270.33.19545. [DOI] [PubMed] [Google Scholar]
- Zhu X., Mancini M. A., Chang K. H., Liu C. Y., Chen C. F., Shan B., Jones D., Yang-Feng T. L., Lee W. H. Characterization of a novel 350-kilodalton nuclear phosphoprotein that is specifically involved in mitotic-phase progression. Mol. Cell Biol. 1995b;15:5017–5029. doi: 10.1128/mcb.15.9.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccolo M., et al. The human Nup107–160 nuclear pore subcomplex contributes to proper kinetochore functions. EMBO J. 2007;26:1853–1864. doi: 10.1038/sj.emboj.7601642. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.