Abstract
The cell nucleus harbors a variety of different bodies that vary in number, composition, and size. Although these bodies coordinate important nuclear processes, little is known about how they are formed. Among the most intensively studied bodies in recent years is the PML body. These bodies have been implicated in gene regulation and other cellular processes and are disrupted in cells from patients suffering from acute promyelocytic leukemia. Using live cell imaging microscopy and immunofluorescence, we show in several cell types that PML bodies are formed at telomeric DNA during interphase. Recent studies revealed that both SUMO modification sites and SUMO interaction motifs in the promyelocytic leukemia (PML) protein are required for PML body formation. We show that SMC5, a component of the SUMO ligase MMS21-containing SMC5/6 complex, localizes temporarily at telomeric DNA during PML body formation, suggesting a possible role for SUMO in the formation of PML bodies at telomeric DNA. Our data identify a novel role of telomeric DNA during PML body formation.
INTRODUCTION
The cell nucleus harbors a variety of distinct compartments and bodies, which are involved in a variety of nuclear activities, such as transcription and RNA processing. These bodies are not surrounded by membranes but accumulate specific sets of proteins by means of protein–protein interactions. Furthermore, most proteins that reside in bodies are in a dynamic equilibrium with their surroundings (Misteli, 2001), and a few of these proteins have been reported to shuttle between various bodies (Snaar et al., 2000). Typical examples of nuclear bodies are nucleoli, which are sites of rRNA synthesis and ribosome subunit assembly, speckles, sites involved in RNA-splicing metabolism, and Cajal bodies, which are processing sites for several ribonucleoproteins (Spector, 2001).
The promyelocytic leukemia (PML) body has been implicated in many different cellular pathways and is characterized by the presence of the PML protein, first identified in patients with acute promyelocytic leukemia (APL; de The et al., 1991). Virtually all APL patients carry the chromosomal translocation t(15,17), resulting in a fusion protein between the retinoic acid receptor-α (RAR) and the PML protein (de The et al., 1991; Melnick and Licht, 1999). The PML-RARα fusion protein fails to locate to PML bodies (Melnick and Licht, 1999) and is thought to block differentiation of bone marrow cells (Naeem et al., 2006). In addition, the leukemic blast cells of APL patients reveal fragmented PML bodies. Treatment of APL patients with all-transretinoic acid or arsenic trioxide results in the degradation of the PML-RARα fusion protein, restoration of PML bodies, and remission of the disease (Koken et al., 1994; Weis et al., 1994).
Each cell nucleus contains, depending on cell type and cell cycle stage, ∼10–30 PML bodies ranging in size from 0.2 to 1 μm. In addition to PML, more than 50 PML body–associated proteins have been characterized, including Sp100, SUMO-1, Daxx, pRB, p53, HAUSP, CBP, Hp1, and BLM, which function in transcription, DNA replication, DNA repair, antiviral defense, chromatin organization, cell cycle control, and apoptosis (Borden, 2002; Dellaire and Bazett-Jones, 2004; Bernardi and Pandolfi, 2007; Everett and Chelbi-Alix, 2007). Thus, PML bodies play active roles in a broad variety of nuclear processes, but they also apparently function as nuclear storage depots regulating the availability of nucleoplasmic proteins in response to external stimuli (Negorev and Maul, 2001).
Most significantly, PML nuclear bodies apparently coordinate DNA repair and cell cycle checkpoint activities, as these bodies were shown to temporarily associate with sites of double-strand breaks and to recruit p53 and the hMre11/Rad50/NBS1 DNA repair complex after ionizing radiation (Carbone et al., 2002). Also, PML nuclear bodies likely regulate and/or coordinate the expression of a variety of genes. Recently, it was shown that the expression of genes within the major histocompatibility class I genomic locus, which have a high degree of association with PML bodies (Shiels et al., 2001), is coordinated by the formation of higher-order chromatin-loop structures mediated by PML and SATB1 (Kumar et al., 2007). Also, it has been reported that a selection of gene-rich and transcriptional active genomic loci, present on several chromosomes, reveal a nonrandom association with PML bodies (Wang et al., 2004). Furthermore, the observation that a number of DNA viruses transcribe their genomes at PML bodies underscores a role of the PML body in transcription (Maul, 1998). Finally, consistent with the idea that PML bodies associate with transcriptionally active regions, newly synthesized mRNA transcripts have been found associated with the periphery of PML bodies (Boisvert et al., 2000; Kießlich et al., 2002).
Although the mechanism by which PML bodies move to and associate with specific genomic loci is not known yet, the frequency with which these associations are observed suggests that PML bodies are nonrandomly organized in the cell nucleus. To address this issue, we visualized the de novo formation of PML bodies after the disassembly of all PML bodies in the cell nucleus and identified the sites at which they form.
MATERIALS AND METHODS
Cell Culture
U2OS, immortalized mouse embryonic fibroblast (MEF) W8 cells (gift from T. Jenuwein, Vienna, Austria), immortalized mouse embryonic PML−/− fibroblasts (gift from P. P. Pandolfi, Harvard Medical School, Boston, MA), and HeLa and hematopoietic leukemia NB4 cells (gift from M. Lanotte, Hospital Saint-Louis, Paris, France) were cultured at 37°C on 3.5-cm glass-bottom culture dishes (MatTek, Ashland, MA) in DMEM without phenol red and containing 1.0 mg/ml glucose, 4% FBS, 2 mM glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin, buffered with 25 mM HEPES, pH 7.2 (all from Invitrogen). To induce alkylating DNA damage, cells were incubated with 0.02% methylmethane sulfonate (MMS; Sigma, St. Louis, MO) for 45 min or 1.5 h.
Plasmids and Cell Transfection
The construction of vectors EYFP-PML I and ECFP-Sp100 is previously described (Wiesmeijer et al., 2002). The coding sequence for PML III has been cloned in the pEYFP-C1 vector (Clontech, Palo Alto, CA) and the coding sequences for telomeric repeat binding factor (TRF) 1, TRF2, and ASF have been cloned into the DsRedExpress vector (Clontech) according to standard procedures. The SUMOylation-deficient yellow fluorescent protein (YFP)-tagged K65/160/490R PML mutant protein (verified by sequencing) was a gift from O. A. Vaughan (University of Durham, Durham, NC). Cells were transiently transfected with 0.5 μg vector DNA using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). For the bimolecular fluorescence complementation assay, the DNA sequences for PML III and TRF1 were cloned into vectors containing YN173, corresponding to residues 1–173 of EYFP or CC155, corresponding to residues 155–238 of ECFP (Hu et al., 2002). TEL-YN was a gift from D. Baker (LUMC, Leiden, The Netherlands). All protein coding sequences used in this study were of human origin. The correct localization of all expressed proteins was verified and confirmed in both U2OS and W8 MEF cells. PML-YFP–expressing cells were stained with antibodies specific for endogenous Sp100, Daxx, and Hausp to confirm the localization of PML-YFP in PML bodies. The localization of DsRed-TRF1 and DsRed-TRF2 at telomeric DNA was confirmed by peptide nucleic acid (PNA)-fluorescence in situ hybridization (FISH; Molenaar et al., 2003). Furthermore, it has been demonstrated before that the TTAGGG repeat binding sites in mouse and human TRF1 and TRF2 show a high level of sequence homology (Broccoli et al., 1997a) and bind to TTAGGG repeat DNA with the same preference (Broccoli et al., 1997b).
Analysis of Fluorescence Complementation
Cells were cotransfected with combinations of the plasmids encoding PML-CC155 and YN173-TRF1 or YN173-TEL as well as YN173-PML and TRF2-CC155. The complementation assay was essentially performed as described (Hu et al., 2002). Transfected cells were first incubated for 3 h at 37°C and then for 16 h at 30°C to promote fluorophore maturation. Cells were monitored either alive or after fixation in 2% formaldehyde.
Live Cell Imaging
Wide-field fluorescence microscopy was performed on a multidimensional workstation for live cell imaging (model DMIRE2; Leica Microsystems, Mannheim, Germany) equipped with a metal halide bulb and a 63× NA 1.4 PlanApo objective lens. Four-dimensional (4D) image stacks were collected using an automated motorized piezo Z-stage. The Z-stacks were collected with 0.4-μm steps and contained generally 20 Z-slides. During imaging, the microscope was heated to 37°C in a CO2 perfused and moisturized chamber. Image stacks were collected every 10 min for 1–4 h and deconvolved by using the Leica software. Deconvolution is a computational algorithm that restores out-of-focus fluorescent signals resulting in a decrease of blur and an improved contrast. For each experiment and cell type at least 10 movies were analyzed.
Immunofluorescence
The following antibodies were used for immunofluorescence staining: mouse mAb 5E10 against PML (gift from R. van Driel, Amsterdam, The Netherlands), rabbit polyclonal antibody against PML (1130 directed against sequence: MEPAPARSPRPQQDP), rabbit polyclonal antibody against SP100 (ab1380, Chemicon, Temecula, CA), rabbit polyclonal antibody against Daxx (sc-7152, Santa Cruz Biotechnology, Santa Cruz, CA), rabbit polyclonal antibody against Hausp (A300–033A, Bethyl Laboratories, Montgomery, TX), mouse mAb against TRF1 (ab10579–50, Abcam, Cambridge, MA), mouse mAb against TRF2 (IMG-124, Imgenex, San Diego, CA), human autoimmune serum against centromeres (Antibodies Incorporated, Davis, CA), rabbit polyclonal antibody against γH2AX (A300–081A, Bethyl Laboratories), rabbit polyclonal antibody against 53BP1 (NB100–304, Novus Biologicals, Littleton, CO), and rabbit polyclonal antibody against SMC5 (A300–236A, Bethyl Laboratories). Cells were grown on coverslips, washed three times in PBS, and then fixed in 2% formaldehyde in PBS for 10 min at room temperature. After fixation, cells were washed three times in PBS, permeabilized in PBS containing 0.2% Triton X-100 for 15 min, and washed once in TBS containing 0.1% Tween 20. Then, cells were incubated with primary antibody for 45 min at 37°C, followed by three washes in TBS containing 0.1% Tween 20. Finally, cells were incubated with an appropriate Alexa-Fluor 488, Alexa-Fluor 594 (both Invitrogen), or Cy3 secondary antibody conjugate for 45 min at 37°C, washed in TBS containing 0.1% Tween 20, and mounted in Citifluor (Agar Scientific, Stansted, Essex, United Kingdom) containing 400 μg/ml DAPI (Sigma-Aldrich).
Fluorescence In Situ Hybridization
NB4 cells were grown on coverslips, fixed in 4% formaldehyde in PBS for 10 min, and permeabilized in PBS containing 1% Triton X-100 for 10 min. Then, cells were washed twice with distilled water, dehydrated in a graded series of ethanol, and dried. For combined PML-immunostaining and PNA-FISH, cells were incubated with 1 ng/μl telomere PNA probe (DAKO, Carpinteria, CA) in 40% formamide/2× SSC. Cells and probe were denatured at 80°C for 3 min. After hybridization for 1 h at 37°C, the cells were washed three times for 5 min in TBS containing 0.5% Triton X-100. Then, cells were incubated with the respective primary and secondary antibodies as described above.
Protein Blot Analysis
Cells were lysed in NuPAGE LDS sample preparation buffer (Invitrogen). Protein samples were then size-fractionated on Novex 4–12% BisTris gradient gels using a MOPS buffer (Invitrogen) and were subsequently transferred onto Hybond-C extra membranes (Amersham Biosciences, Piscataway, NJ) using a submarine system (Invitrogen). Blots were stained for total protein using Ponceau S (Sigma-Aldrich). After blocking with PBS containing 0.1% Tween 20 and 5% milk powder, the membranes were incubated with antibody 5E10 against PML or a rabbit antibody against small ubiquitin-like modifier (SUMO; directed against sequence: MEDEDTIDVFQQQTG) and with a mouse polyclonal antibody against tubulin (1:2000; Sigma-Aldrich). The secondary antibodies used were HRP-conjugated anti-mouse (1:5000; Pierce, Rockford, IL) and HRP-conjugated anti-rabbit (1:2000; Pierce). Bound antibodies were detected by chemiluminescence using ECL Plus (Amersham Biosciences).
RESULTS
PML Bodies Do Not Necessarily Form at Previously Used Sites after Recovery from Stress
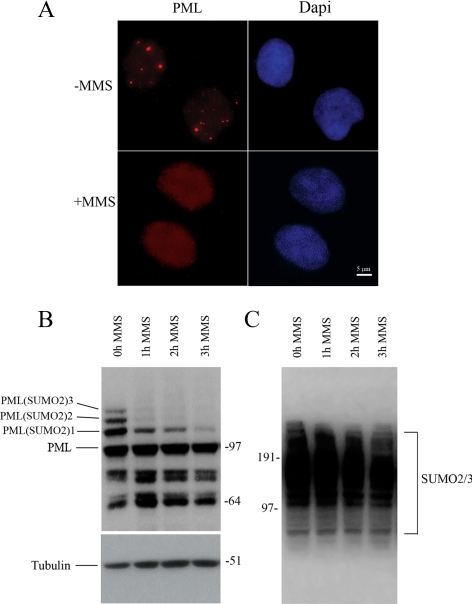
A number of different cellular stresses have been described that induce the disassembly of PML bodies and result in the formation of PML residual bodies, PML microstructures, or even in a complete disappearance of the PML body (Maul et al., 1995; Eskiw et al., 2003; Nefkens et al., 2003; Conlan et al., 2004). To examine whether PML bodies have predetermined spatial positions in the nucleus, we treated U2OS human osteosarcoma cells with the DNA-methylating agent MMS. This agent has previously been shown to cause a complete dispersal of PML bodies (Conlan et al., 2004). By immunofluorescence, we confirmed that PML bodies indeed disassemble completely in response to MMS treatment and that PML proteins redistribute throughout the nucleus in a diffuse manner (Figure 1A). However, these results do not exclude the possibility that the level of PML in PML bodies is greatly reduced below the detection level or that some of the other PML body components may organize into body structures. To confirm that PML bodies indeed disassemble in response to MMS treatment, we evaluated the levels of SUMO-modified PML by immunoblot analysis. Previous studies showed that the assembly and integrity of PML bodies is dependent on posttranslational modification of PML by SUMO (Zhong et al., 2000). Furthermore, it has been shown that PML is a preferential SUMO2-specific target protein (Vertegaal et al., 2006). As illustrated in Figure 1B, the amount of SUMOylated PML was markedly reduced by MMS treatment, whereas the total amount of SUMO2/3 did not decrease (Figure 1C). Also shown in Figure 1B is that the total amount of PML protein did not significantly change by MMS treatment, which is consistent with the observed subnuclear redistribution of PML and with previous data (Conlan et al., 2004).
Figure 1.
PML nuclear bodies disperse by MMS treatment. (A) U2OS cells were stained with an anti-PML antibody to detect endogenous PML and with DAPI (blue). In untreated cells, the PML protein is localized in PML nuclear bodies (top panel) as shown by fluorescence microscopy. In cells treated with the DNA-alkylating agent MMS, the PML bodies are dispersed, and the PML protein is present throughout the nucleus in a diffuse and punctate pattern (bottom panel). Images were not deconvolved. (B) Immunoblot analysis of PML in total lysates of U2OS cells that are treated with MMS for increasing time periods. (C) Total cell lysates from U2OS cells that are treated with MMS for increasing time periods were subjected to immunoblotting with anti-PML and anti-SUMO2/3 antibodies, respectively.
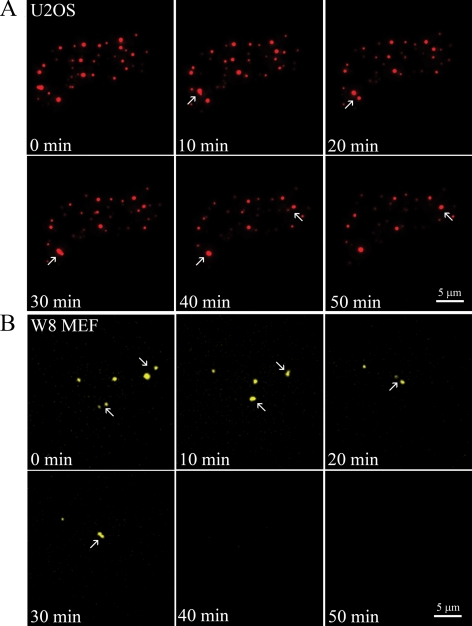
To image the disassembly of PML bodies in live cells, we collected 3D image stacks of U2OS cells and of W8 MEFs, both expressing EYFP-PML III, at 10-min time intervals. Image collection was started immediately after MMS treatment and continued throughout the dispersal process of the PML bodies, which took on average 1 h. Among cells there appeared to be quite some variability in the duration of the dispersal process, which could possibly due to a cell cycle dependent action of MMS. PML body dispersal was accompanied by a gradual loss of PML protein from these bodies. Some PML bodies, however, first fused into larger bodies which then rapidly dispersed (Figure 2 and Supplemental Movie 1). In a few cells some remnant PML bodies remained present after MMS treatment. Such bodies may have dispersed when cells were incubated with MMS for a longer time period. As a control, we analyzed the movement of PML bodies in untreated U2OS and W8 MEF interphase cells (10 cells each). 3D image stacks were collected every 10 min for 50 min (Supplemental Figure S1). The 4D stacks show that fusions of PML bodies occur but at low frequencies and that PML bodies show constrained as well as dynamic movements consistent with previous observations about PML body dynamics (Muratani et al., 2002; Wiesmeijer et al., 2002; Jegou et al., 2009). In a subfraction of cells, we noticed that the number of PML bodies increased during the imaging period, which is most likely related to cell cycle phase (Dellaire et al., 2006a,b).
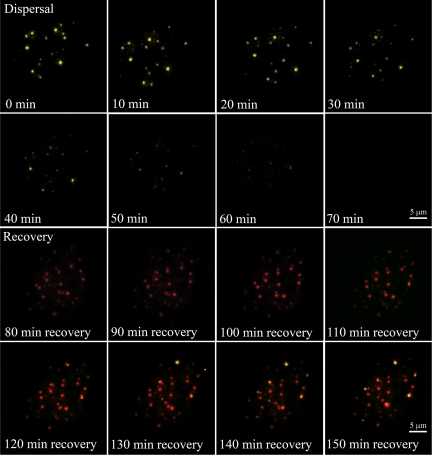
Figure 2.
PML body dispersal is preceded by a fusion of PML bodies in MMS-treated cells. (A) U2OS cells were transiently transfected with an expression vector encoding EYFP-PML. Image acquisition of live cells was started immediately after MMS addition to the culture medium, and a 3D image stack was taken every 10 min. Image stacks were captured using 50-ms exposure times and deconvolved to reduce out of focus information. Each picture shows a maximum projection of a deconvolved 3D image stack. Arrows indicate PML bodies that fuse and then disperse shortly after. (B) A similar procedure as described in A was used to image the disassembly of PML nuclear bodies in live W8 MEFs.
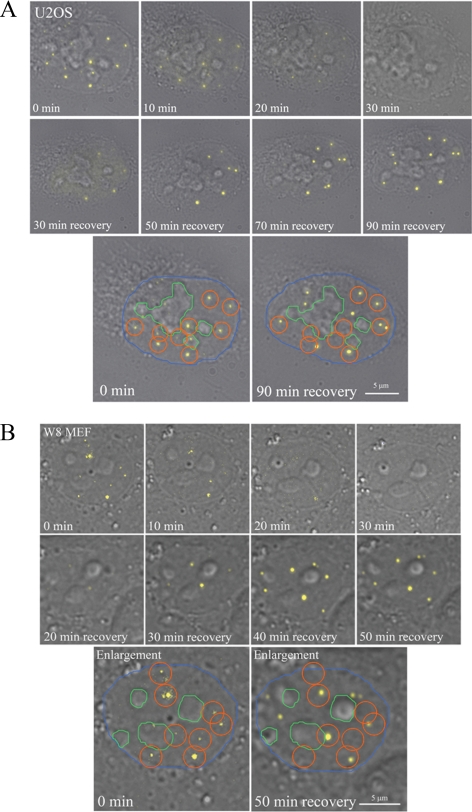
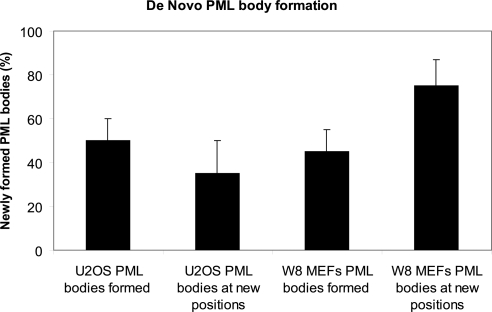
Interestingly, the dispersal of PML bodies proved reversible as we observed that PML bodies recover when the cells were incubated in fresh culture medium lacking MMS. Consistent with this observation, we found that the amount of SUMOylated PML increased when cells recovered from MMS treatment (Supplemental Figure S2). All live cell experiments were performed by expressing PML isoform III and were confirmed by expressing PML isoform I. We examined whether after MMS treatment PML bodies would recover at the same spatial position in the nucleus, possibly at the same chromatin site, as they were positioned before treatment. Both, U2OS and W8 MEF cells, were transiently transfected with EYFP-PML. The 3D image stacks were collected every 10 min before, during, and after MMS treatment using both the YFP channel and the differential interference contrast (DIC) channel and converted to maximum projections (Figure 3). These projections were used to denote the spatial areas occupied by PML bodies before MMS treatment by drawing -μm circles around them. After analyzing the maximum projections of 10 interphase U2OS cells, collected before and 90 min after recovery from MMS treatment, it appeared that only 50% (±10%) of the original number of PML bodies has been formed de novo after treatment. Of these bodies, 35% (± 15%) formed outside the circled areas. Because the total nuclear area occupied by circles is relatively large, it is expected that a significant number of PML bodies are formed in these areas just by chance. Hence, at least 35% of the de novo formed PML bodies are positioned at new locations (Figures 3A and 4). The analysis of the maximum projections of 4D image stacks of 10 interphase W8 MEFs taken before and 50 min after recovery of MMS treatment revealed that 45% (±10%) of the original number of PML bodies has been formed de novo after treatment and that a median of 75% of these PML bodies formed outside the circled areas (Figures 3B and 4). Whether previously used chromatin sites have moved in the intervening time to other positions in the nucleus should be addressed by additional experiments using photoactivatable GFP-tagged histone H4 (Wiesmeijer et al., 2008). Our observations suggest that a subset of PML bodies form at new locations in the cell nucleus and that this is particularly true for W8 MEFs.
Figure 3.
Not all PML bodies form at previously used sites in the cell nucleus. (A) Double projections of DIC and fluorescence images show the dispersal and formation of PML bodies in a U2OS cell. U2OS cells expressing EYFP-PML were treated with MMS for 40 min and then incubated in fresh medium without MMS. During the course of the experiment image stacks were taken every 10 min. To determine whether PML bodies recover at the same position, 2-μm orange circles were placed around the PML bodies of a cell before treatment, and the nucleoli were marked by a green line (enlarged image). The orange circles were grouped together with the most nearby nucleolus (green line), and the grouped items were placed on top of the same cell after recovery. The PML bodies outside the orange circles indicate those that are formed at a spatial position different from that which they occupied before MMS treatment. The contour of the nucleus is indicated with a blue line. (B) Double projections of DIC and fluorescence images show the dispersal and formation of PML bodies in a W8 MEF cell. Treatment and imaging conditions were the same as for U2OS cells. The image taken after 50 min of recovery time shows PML bodies that are positioned both inside and outside the orange circles. The contour of the nucleus is indicated with a blue line.
Figure 4.
De novo PML body formation in U2OS and W8 cells. Quantitative determination of PML bodies that form de novo in cells that recovered from MMS treatment compared with the original numbers and positions of PML bodies present in the same cells before MMS treatment. The data are averages of 10 U2OS and 10 W8 cells. Error bars, SD.
PML Bodies Form De Novo on Telomeric DNA
U2OS cells are ALT cells that contain extrachromosomal telomeric DNA and a subset of these cells contain variable numbers of PML bodies that are associated with telomeric DNA sequences (Yeager et al., 1999). Because we found in U2OS cells that ∼65% of the new PML bodies formed at approximately the same positions as they were found before MMS treatment, we speculated that these positions might contain telomeric DNA. Thus, PML bodies may form de novo at telomeric DNA after recovery from MMS treatment. We therefore monitored PML body dispersal in interphase U2OS cells, expressing EYFP-PML together with DsRed-TRF1 or EYFP-PML together with DsRed-TRF2, during MMS treatment and then visualized the formation of PML bodies during the recovery period by capturing 3D image stacks at 10-min time intervals. The collected 3D image data sets show that PML bodies were indeed formed at telomeric DNA foci labeled by DsRed-TRF1 or DsRed-TRF2 in U2OS cells. Approximately 1 h after incubating the cells in fresh medium without MMS, EYFP-PML was shown to accumulate first at a few sites that are labeled by DsRed-TRF1 or DsRed-TRF2. As time proceeded, EYFP-PML was accumulating at an increasing number of telomeric sites during the time course of the experiment (Figure 5 and Supplemental Movie 2). It should be noted, however, that we cannot discriminate between telomeres and extrachromosomal telomeric material, which are both present in ALT cells (Ogino et al., 1998). The amount of extrachromosomal telomeric DNA may even be increased by MMS treatment. It has been reported previously that DNA damage can lead to a twofold increase in extrachromosomal telomeric DNA (Fasching et al., 2007). Hence, PML bodies may form at both telomeres and extrachromosomal telomeric material in U2OS cells. The specificity of DsRed-TRF1 and DsRed-TRF2 localization at telomeric DNA was confirmed by their localization at Cy3-peptide nucleic acid–labeled telomere sequences as described previously (Molenaar et al., 2003). Both, MMS-treated and nontreated U2OS cells were stained for telomeric DNA using a PNA probe and for endogenous or exogenous TRF1 using antibodies. Double-labeled cells, both treated and nontreated, revealed 1–4 TRF1 foci that did not colocalize with telomeric DNA (Supplemental Figure S3). Thus, a few TRF1 or TRF2 foci may label other sites in the nucleus, which may represent protein aggregates formed by over expression. The presence of TRF1 or TRF2 foci that do not colocalize with telomeric DNA staining may also be explained by a poor hybridization and detection efficiency of PNA probes at short telomeres.
Figure 5.
New PML bodies form on telomeric sites when U2OS cells recover from MMS treatment. U2OS cells were cotransfected with expression vectors encoding EYFP-PML and DsRed-TRF1, and 3D stacks were acquired every 10 min. 3D image stacks were captured using 50-ms exposure times and deconvolved. The images taken during the recovery phase show the spatial position of telomeric DNA in red and the accumulation of PML protein in yellow.
W8 MEFs, expressing EYFP-PML and DsRed-TRF1, were included as non-ALT control cells in live cell experiments and subjected to the same treatment as U2OS cells. W8 MEF cells contain relatively long telomeres but do not contain extrachromosomal telomeric DNA and ALT associated PML bodies. Similar to what we observed for U2OS cells, a median of 60% of EYFP-PML bodies was shown to accumulate at fluorescently labeled telomere sequences in W8 MEFs (Supplemental Movie 3). These data suggest that after dispersal, new PML bodies form and that a substantial set of these bodies use telomeric DNA as initial assembly sites.
To confirm our findings using nontransfected U2OS cells, we analyzed the formation of PML bodies in 10 U2OS cells that were allowed to recover from MMS treatment and were fixed and incubated with antibodies against PML and TRF2. Consistent with the live cell experiments, we observed that 70% (±15%) of the newly formed PML bodies were in association with telomeric DNA (Figure 6, A and B). As a non-ALT human control, we studied the de novo formation of PML bodies in telomerase-expressing HeLa cells. These cells were first treated with MMS and then allowed to recover in fresh medium before they were fixed and incubated with antibodies specific for PML and TRF2. Similar to what we observed in U2OS cells, a significant number of PML bodies colocalized with telomeres (Figure 6C).
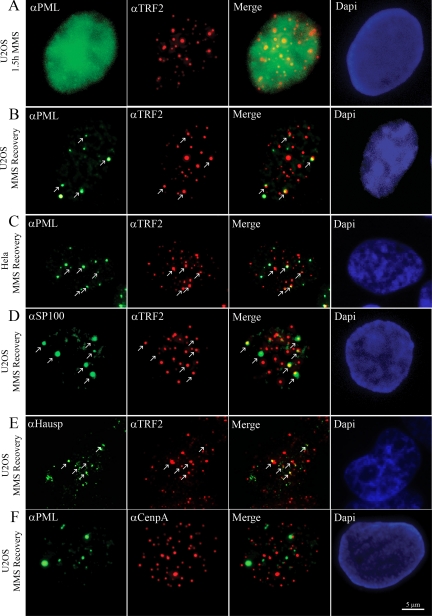
Figure 6.
Endogenous PML, Sp100 and Hausp accumulate at telomeric sites but not at centromeres in U2OS cells recovering from MMS treatment. (A) Immunofluorescence image of a U2OS cell treated with MMS, fixed and stained with anti-PML (green) and anti-TRF2 (red) antibodies. (B) Image of a U2OS cell that recovers from MMS treatment and is stained with antibodies against PML (green) and TRF2 (red). (C) Image of a HeLa cell that recovers from MMS treatment and is stained with antibodies against PML (green) and TRF2 (red). Arrows in B and C indicate the positions where PML colocalize or associate with TRF2 foci. (D) Localization of Sp100 at telomeric sites in a U2OS cell that recovers from MMS treatment. During recovery from MMS treatment, U2OS cells were fixed and stained with anti-Sp100 and anti-TRF2 antibodies. (E) Immunofluorescence image of a U2OS cell that recovers from MMS treatment. Sites where Hausp colocalize with telomeric DNA are indicated by arrows. (F) PML does not colocalize with centromeres in a U2OS cell that recovers from MMS treatment. After MMS treatment, U2OS cells were incubated in fresh medium, fixed and stained with anti-PML (green) and anti-CENPA (red) antibodies. All cell nuclei are counterstained with DAPI (blue).
To examine whether together with PML other typical PML body proteins are recruited to telomere sequences when PML bodies are formed, we stained cells recovering from MMS treatment for TRF2 and Sp100, Daxx or Hausp using specific antibodies. As shown in Figure 6D, Sp100 is recruited to telomeric sites when PML bodies are formed. Before MMS treatment, no colocalization of Sp100 with TRF2 foci was observed and during MMS treatment Sp100 appeared diffuse (Supplemental Figure S4). In addition to Sp100, both Hausp (Figure 6E) and Daxx (not shown) were also observed to be recruited by telomeric sites in cells recovering from MMS treatment. These results suggest that together with PML all other major PML-body components assemble in de novo formed PML bodies at telomeric sites.
Because MMS is a DNA-damaging agent, it could be possible that PML bodies form preferentially at damaged telomeric DNA. Therefore, we investigated the presence of DNA damage at sites of telomeric DNA before, during, and after MMS treatment by staining DsRed TRF1-expressing U2OS cells for PML and γH2AX. The latter is a marker for double-stranded DNA breaks. Before MMS treatment there is no significant amount of γH2AX staining observed in nucleus. Small γH2AX foci were observed in the nucleus of MMS-treated cells showing dispersal of PML bodies. In the recovery phase when PML bodies are formed γH2AX was not necessarily found present at telomeric DNA that colocalized with de novo–formed PML bodies (Supplemental Figure S5, A–C). Similar results were obtained when cells were stained for 53BP1, a component of DNA damage repair foci (Supplemental Figure S5, D and E).
Next, we examined the possibility that PML bodies may form at other nuclear compartments as well. We therefore expressed the nuclear body markers EYFP-ASF, EYFP-coilin, or EYFP-S5, which localize respectively at speckles, Cajal bodies, and nucleoli together with DsRed-PML in U2OS cells and W8 MEFs. After MMS treatment and PML body recovery, we observed that only 2–4% of PML bodies colocalized with these other nuclear bodies (data not shown). Because a previous study reported a dynamic association of PML with centromeres (Everett et al., 1999a), we also wanted to examine whether PML bodies could form at these chromosomal sites consisting of tandem repetitive elements. Using antibodies to the centromere protein CENPA and PML, no significant (<1%) colocalization of PML with centromeres was observed in both U2OS and W8 MEF cells that recovered from MMS treatment (Figure 6F).
Together, these data suggest that at least a subset of PML bodies are formed at telomeric DNA sequences and not at any other specific nuclear structure, in both ALT and non-ALT cells. However, we cannot rule out the possibility that PML bodies may be formed at other nuclear sites as well.
Ectopical Expression of PML in PML−/− MEFs Leads to De Novo PML Body Formation at Telomeric DNA
As PML bodies may assemble at telomeric DNA because of some effect of MMS treatment other than inducing DNA damage at telomeres, we wanted to investigate whether PML bodies may also form at telomeric DNA in untreated cells. To this end we examined the formation of PML bodies in PML−/− MEFs, which lack intact PML bodies (Wang et al., 1998). Both, EYFP-PML and DsRed-TRF1 were transiently expressed in PML−/− MEFs, and 3D image stacks were captured at 3 h after transfection. Analysis of 20 deconvolved image stacks showed a consistent colocalization of assembled PML bodies with fluorescently tagged telomeric DNA (Figure 7A). Similar results were obtained when cells were transfected with a vector encoding EYFP-PML, incubated for 3 h in culture medium, fixed, and then stained with a TRF2 antibody or hybridized with a telomere-specific PNA probe (data not shown).
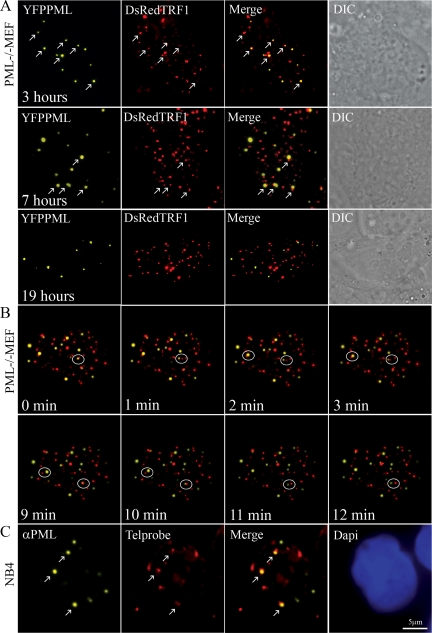
Figure 7.
New PML bodies form on telomeric DNA in a DNA damage–independent manner. (A) Ectopically expressed EYFP-PML form de novo PML bodies at telomeric DNA in PML−/− MEFs. PML−/− MEFs are cotransfected with EYFP-PML and DsRed-TRF1, and 3D image stacks are collected after 3, 7, and 19 h. Arrows indicate the positions where PML colocalize with TRF1. Note that the number of PML bodies that are associated with telomeric sites decreased at 7 and 19 h after transfection. (B) Newly formed PML bodies dissociate from telomeric sites. PML−/− MEFs were cotransfected with EYFP-PML and DsRed-TRF1, and at 3 h after transfection one 3D image stack was taken each minute (0–3 and 9–12). The area in the circle points to a PML body separating from a telomere. (C) NB4 cells were treated with arsenic trioxide for 8 h to initiate PML body assembly. Cells were fixed, subjected to FISH using a telomere-specific PNA probe, and stained with anti-PML antibody. Arrows indicate positions where PML localize at telomeric DNA. Finally, cells were counterstained with DAPI.
Because we observed a variable number of PML bodies that were not associated with telomeric DNA at 3 h after transfection, we also followed the fate of PML bodies at later time points after a cotransfection of PML−/− MEFs with EYFP-PML together with DsRed-TRF1. Image stacks collected at 7 and 19 h after transfection demonstrated that at 7 h fewer PML bodies were associated with telomeric DNA compared with 3 h after transfection and that at 19 h none or only very few PML bodies were associated with telomeric DNA (Figure 7A). Together, these observations suggest that PML bodies dissociate from telomeric DNA after they are formed at these sites. To monitor the dissociation of de novo–formed PML bodies from telomeric DNA sites, PML−/− MEF cells were cotransfected with EYFP-PML together with DsRed-TRF1, and at 3 h after transfection 4D image stacks were collected every 1 min for 30 min. Analysis of 10 image stacks suggests that PML bodies dissociate from telomeric DNA once they are formed at these sites (Figure 7B and Supplemental Movie 4). By analyzing single Z planes in the 3D stacks, we confirmed that PML bodies are indeed associated with telomeric DNA sites before they released from these sites.
Recovery of PML Bodies in the APL-Patient–derived Cell Line NB4
A characteristic morphological feature of APL is that patient-derived cells lack any intact PML bodies; however, treatment of patients with retinoic acid or arsenic trioxide results in the restoration of PML bodies. As expected, when NB4 cells (Lanotte et al., 1991) were treated with arsenic trioxide, fixed, and stained with an antibody against PML, we observed the formation of PML bodies, whereas untreated cells revealed a diffuse and punctate staining pattern of nuclear PML (Supplemental Figure S6). This punctate pattern of PML staining is consistent with the observation that PML is localized in nuclear microspeckles in APL cells (Koken et al., 1994). To address whether the formation of PML bodies in APL patient–derived cells also involve telomeric DNA, we examined the accumulation of PML protein at telomeric DNA after arsenic trioxide treatment for 2–8 h. Combined immunofluorescence and telomere PNA-FISH revealed a high extent of colocalization between newly formed PML bodies and telomeric DNA (Figure 7C; Supplemental Figure S6), further validating our hypothesis that non-ALT PML bodies form de novo at telomeric DNA. Maximum projections of image stacks collected from untreated cells revealed many PML microspeckles, some of which colocalize with telomeric DNA. Notably, this colocalization was only sporadically observed when viewing single optical sections.
PML Directly Interacts with TRF1 and TRF2
To further confirm that PML bodies assemble at telomeric DNA, we made use of a bimolecular fluorescence complementation assay (Hu et al., 2002). This assay is based on the reconstitution of a fluorescent protein by the close proximity of two fragments of fluorescent proteins, which are nonfluorescent themselves, when proteins fused to the fragments interact. To visualize the possible interaction of PML with TRF1 and/or TRF2, we engineered the expression constructs PML-CC155, YN173-PML, YN173-TRF1, and -TRF2-CC155, cotransfected PML−/− MEFs with the combinations PML-CC155 and YN173-TRF1 or YN173-PML and TRF2-CC155 and monitored the appearance of fluorescence signals in living cells using fluorescence microscopy. As shown for the combination PML-CC155 and YN173-TRF1, ∼16 h after transfection, we observed the appearance of fluorescent spots in the nucleus (Figure 8A). Similar results were obtained with the combination YN173-PML and TRF2-CC155 (data not shown). The occurrence of fluorescent spots indicates complementation and thus suggests that PML directly interacts with TRF1 as well as with TRF2. As a negative control we cotransfected PML−/− MEFs with the constructs YN173-TRF1 and TEL-CC155. TEL is a transcription repressor protein and is not expected to have an interaction with TRF1. Indeed, we observed no fluorescence appearing at 16 h after transfection, indicating that complementation did not take place (Figure 78B). In addition, expression of the single complementation halves did not result in any fluorescent signal (data not shown). Finally, coexpression of PML-CC155 and YN173-TRF1 in MEF W8 cells that were not treated with MMS did also not result in fluorescence complementation (data not shown). This experiment suggests that overexpressed TRF1 does not localize to already existing PML bodies. Thus, the observed complementation between PML-CC155 and YN173-TRF1 and between YN173-PML and TRF2-CC155 suggest that PML bodies form at telomeric DNA, at least in part through protein–protein interactions.
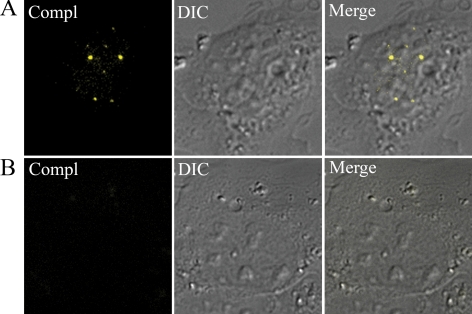
Figure 8.
PML directly interacts with TRF at telomeric DNA. (A) Ectopically expressed PML interacts with TRF1 in the nucleus of PML−/− MEFs as shown by the appearance of yellow fluorescent signals in a combined fluorescence-DIC image. PML−/− MEFs were cotransfected with the fluorescent complementation expression vectors PML-CC155 and YN173-TRF1 and incubated at 30°C for 16 h to allow for correct folding of fluorescent YFP. (B) Ectopically expressed YN173-TRF1 and TEL-CC155 does not interact as shown by the absence of fluorescence complementation.
PML Body Formation at Telomeric DNA: a Role for SUMO?
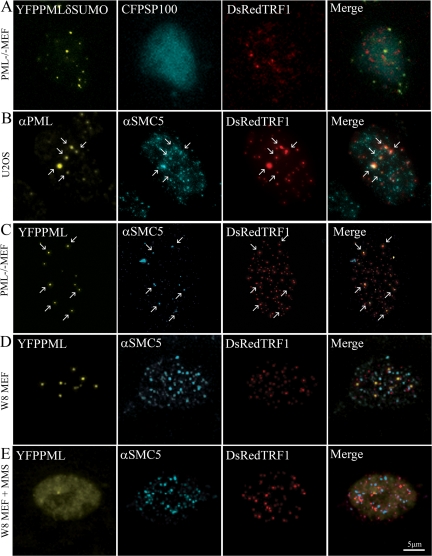
It has recently been reported that PML requires SUMOylation sites and SUMO interaction motifs for the nucleation and formation of PML bodies (Shen et al., 2006). The SUMO interaction motifs are possibly important for the interaction of PML with other SUMOylated proteins. We therefore investigated whether SUMOylation of PML and telomere-associated proteins was involved in the formation of PML bodies at telomeric DNA. Transient expression of DsRed-TRF1, ECFP-Sp100, and an YFP-tagged PML mutant that cannot be SUMOylated in PML−/− MEFs resulted in the formation of PML aggregates, which did not accumulate Sp100. Furthermore, the newly formed PML aggregates did not colocalize with telomeric DNA (Figure 9A), suggesting that SUMOylation of PML is required for PML to localize at telomeric DNA. Similar observations were made in U2OS cells that were cotransfected with EYFP-tagged SUMOylation-deficient PML and DsRed-TRF1, treated with MMS and then incubated in fresh medium (Supplemental Figure S7).
Figure 9.
A role for SUMO in the formation of new PML bodies. (A) SUMOylation-deficient PML does not localize at telomeric DNA in PML−/− MEFs. PML−/− MEFs were cotransfected with DsRed-TRF1, ECFP-Sp100 and YFP-tagged SUMOylation-deficient PML. The image shows the formation of PML aggregates (yellow), which do not colocalize with Sp100 (cyan) and do not localize at telomeric DNA that are stained by DsRed-TRF1 (red). (B) The SMC5/6 complex is present at sites where PML bodies form at telomeric DNA. U2OS cells were transfected with DsRed-TRF1, treated with MMS, and allowed to recover in fresh medium. Cells were fixed and stained with anti-PML and anti-SMC5 antibodies. The arrows point to positions in the nucleus where PML (yellow) and SMC5 (cyan) localize at telomeric sites (red). (C) Ectopically expressed PML localizes together with SMC5 at telomeric DNA in PML−/− MEFs. PML−/− MEFs were cotransfected with EYFP-PML, to induce PML body formation, and DsRed-TRF1, fixed, and stained with anti-SMC5 antibody. The arrows indicate the positions where PML (yellow) and SMC5 (cyan) localize at telomeric DNA (red). (D) PML and SMC5 do not localize at telomeric DNA in untreated and (E) MMS-treated W8 MEFs. W8 MEFs were cotransfected with YFP-PML and DsRed-TRF1 and either not treated or treated with MMS. Cells were fixed (without recovery) and stained with anti-SMC5 antibody. Both untreated and MMS-treated cells show that the SMC5/6 complex (cyan) localize in a punctate/diffuse pattern, which does not colocalize with telomeric sites (red) or PML (yellow).
Because the SMC5/6 complex, containing the SUMO ligase MMS21, has recently been shown to stimulate SUMOylation of telomere-binding proteins, including TRF1 and TRF2, and to be required for the recruitment of telomeres to PML bodies in ALT cells (Potts and Yu, 2007), we investigated whether the SMC5/6 complex colocalized with telomeric DNA at PML bodies. U2OS cells, recovering from MMS treatment, and PML−/− MEFs expressing EYFP-PML showed a specific staining of the SMC5/6 complex at sites of newly formed PML bodies (Figure 9, B and C). Similar results were obtained in MMS-treated W8 MEFs (data not shown). In cells that were not treated with MMS to elicit PML body formation, the SMC5/6 complex showed both a diffuse and dot-like staining in the nucleus. As shown for nontreated and MMS-treated W8 MEFs, this pattern did clearly not colocalize with telomeric DNA (Figure 9, D and E). Together, these results suggest a role for the SMC5/6 complex in the assembly of PML bodies at telomeric DNA, possibly by mediating SUMOylation of telomere-binding proteins.
DISCUSSION
To date, little is known about the molecular mechanisms that drive and regulate the formation of specific bodies in the cell nucleus. Only recently it was shown that PML SUMOylation and binding of PML to SUMOylated PML through a SUMO-binding motif are instrumental for PML body formation and for the recruitment of other proteins that localize at PML bodies (Shen et al., 2006). PML bodies are formed when cells progress from G1 to G2 during the cell cycle, with the highest number of PML bodies found in G2 (Everett et al., 1999b; Dellaire et al., 2006a). It remained, however, unclear how PML body formation is initiated. Our results indicate for the first time an unexpected role for telomeric DNA in the formation of PML bodies.
To investigate whether PML nuclear bodies occupy preferred positions in the interphase cell nucleus, we first treated cells with the DNA-demethylating agent MMS to disrupt PML bodies and then followed the reassembly of PML bodies during a recovery phase.
We found that all PML bodies disassemble in response to the DNA-demethylating agent MMS and that new PML bodies form during a recovery phase. Generally, the number of PML bodies formed is less than the original number of PML bodies in cells. This difference might be explained by the short time window of recovery in which we analyze the cells. Also, cells may first have to complete a full cell cycle to obtain a full set of PML bodies. Interestingly, our data indicate that new PML bodies do not necessarily form at their original positions in the cell nucleus. Previous data, however, indicated that PML bodies are formed at predetermined positions in the nucleus (Eskiw et al., 2003). PML bodies were shown to largely disassemble in response to heat shock, leaving behind residual PML bodies that maintain their spatial position. When cells were allowed to recover from the stress, these residual bodies were shown to recruit PML-containing microbodies to form intact PML bodies. Because PML bodies were not found at new locations, it was concluded that PML bodies are formed at predetermined positions only. Our results clearly indicate that PML bodies, after disassembly, do not necessarily recover at their original positions. Instead, we provide evidence that PML bodies nucleate at new sites, which we identified being telomeric DNA. It is probably in ALT cells only that newly formed PML bodies remain associated with telomeric DNA. In contrast to most tumor cells, ALT cells lack the enzyme telomerase for telomere maintenance and use an alternative lengthening of telomeres mechanism that is thought to involve homologous recombination (Dunham et al., 2000). A characteristic feature of ALT cells is that they contain, in addition to telomeres, extrachromosomal telomeric material and a subset of PML bodies that is in a complex with telomeric DNA. The possible function of these complexes is still not completely known (Henson et al., 2002). PML bodies that are in complex with telomeric DNA are referred to as ALT-associated PML bodies. It should be noted that the formation of ALT-associated PML bodies might be different from that of regular PML bodies that are present in both ALT and non-ALT cells. Recently, it has been described that ALT-associated PML bodies form by the binding of an existing PML body to a telomere and the subsequent recruitment of free PML protein (Jegou et al., 2009).
Although G2 cells contain the highest number of PML bodies, mitotic cells contain the fewest number. When cells enter mitosis PML bodies lose SUMO1 and Sp100, which is accompanied by a strong reduction in PML body number. However, accumulations of PML protein remained present during mitosis, some of which stably associated with chromatin (Dellaire et al., 2006). Although we analyzed the formation of new PML bodies in U2OS cells after mitosis, we noticed that many of the mitotic PML accumulations were associated with telomeric DNA, which hampered our analysis of PML body formation at late telophase/early G1. Our analysis of the formation of new PML bodies is therefore limited to interphase cells.
Although we observed that the majority of new PML bodies are initially positioned at telomeric sites, we do not exclude the possibility that some PML bodies are formed at other loci in the nucleus. Indeed, we observed in our time-lapse recordings that a few new PML bodies did not colocalize with a telomeric site. Because PML bodies have been found in association with some specific chromatin domains, it is conceivable that these domains may function as nucleation sites as well. It may, however, also be true that some new PML bodies dissociate rapidly from telomeric sites and that some PML bodies were already dissociated before we started capturing images. We frequently observed the dissociation of new PML bodies from telomeric DNA, suggesting that most PML bodies dissociate from telomeric sites shortly after their formation. Although we observed quite some variability in the time period PML bodies remain associated with telomeric DNA, the fact that they dissociate is possibly the reason why PML-telomere associations remained thus far unnoticed except for ALT cells containing ALT associated PML bodies. As we analyzed the formation of PML bodies in a limited number of cell types, we do not exclude the possibility that other cell types support a different mechanism of PML body formation.
In addition to PML protein, we showed that newly formed PML bodies also contained Sp100, Daxx, and Hausp, indicating that PML protein is not just temporarily aggregating at telomeric DNA. Whether all PML body components are recruited at the same time to the newly formed PML bodies has yet to be determined. Recent data suggest a stepwise recruitment of PML body constituents to PML bodies in early G1 cells, thereby preventing the early maturation of PML bodies at this stage (Chen et al., 2008). It can be envisaged that new, immature PML bodies still have the ability to move to and attach to specific nuclear sites, before they recruit factors that determine their function or strengthen their interaction with chromatin. Once settled, most PML bodies reveal a constrained movement, which is consistent with their association with chromatin (Chen et al., 2008).
We propose that SUMOylation of telomere-binding proteins may play an important role in the formation of PML bodies at telomeric DNA. Recent data demonstrated a role for the SMC5/6 complex in the formation of ALT-associated PML bodies by SUMOylation of six telomere-binding proteins, including TRF1 and TRF2 (Potts and Yu, 2007). Our data suggest that the SMC5/6 complex, containing the SUMO ligase MMS21, may not only fulfill a role in the telomere lengthening mechanism in ALT cells, but also in a mechanism supporting the formation of PML bodies at telomeric DNA. At present, it is unclear how and when the SMC5/6 complex is recruited to telomeric DNA and whether this complex is essential for PML body formation. A potential mechanism could be that PML protein is recruited to SUMOylated telomere-binding proteins by the SUMO-binding sites present in PML. SUMOylation of PML at these sites may then lead to the recruitment of more PML protein and other PML nuclear body proteins including Sp100. Consistent with this idea is that a PML mutant that cannot be SUMOylated does not accumulate at telomeric DNA, whereas a wild-type PML protein does. However, it remains unclear whether SUMOylation of telomere-associated proteins is indeed essential for PML body formation. Our initial attempts to detect SUMOylated forms of endogenous TRF1 or TRF2 in cells showing PML body formation by immunoblot analysis failed, possibly because of the rapid turnover of SUMOylated TRF1 and TRF2.
The suggestion that the telomere-binding proteins TRF1 and TRF2 are likely to play a role in the formation of PML bodies is supported by our observation that both TRF1 and TRF2 interact with PML in the fluorescence complementation assay. Whether other telomere-associated proteins may also be involved in PML body formation has to be investigated. Experiments aimed at reducing telomere-binding protein levels by RNA interference will perhaps shed more light on the function of these proteins in PML body formation. Furthermore, it will be interesting to investigate whether each telomere has the capacity to initiate PML body formation. We cannot exclude the possibilities that the size or activity of telomeric DNA play a role in the recruitment of PML protein. This could, for example, result in the impairment of PML body formation in aged cells, which generally have short telomeres, adding another potential mechanism as to why aged cells are less responsive to stress. Studies to test these hypotheses will enrich our understanding of the biological functions of PML bodies.
In conclusion, this study provides new insights in the assembly of new PML bodies in the cell nucleus and establishes a role for telomeric foci in the recruitment of PML protein.
Supplementary Material
ACKNOWLEDGMENTS
We thank P. Pandolfi for providing PML−/− MEFs, T. Jenuwein for W8 MEFs, A. I. Lamond (University of Dundee, Dundee, United Kingdom) for the NB4 cells, R. van Driel for the 5E10 anti-PML antibody, T. Kerppola (University of Michigan Medical School, Ann Arbor, MI) for bimolecular fluorescence complementation constructs, D. Baker for YN173-TEL, and O. A. Vaughan for the SUMOylation-deficient YFP-tagged K65/160/490R PML mutant protein. Furthermore, we are grateful to L. Fradkin and D. Baker for comments on the manuscript and to M. van Hagen for his help with the immunoblots. This work was partially supported by Cyttron Grant BSIK03036.
Abbreviations used:
- FISH
fluorescence in situ hybridization
- MEF
mouse embryonic fibroblast
- MMS
methylmethane sulfonate
- PML
promyelocytic leukemia
- PNA
peptide nucleic acid
- SUMO
small ubiquitin-like modifier
- TRF
telomeric repeat binding factor.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-04-0309) on September 30, 2009.
REFERENCES
- Bernardi R., Pandolfi P. P. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat. Rev. Mol. Cell. Biol. 2007;8:1006–1016. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- Boisvert F. M., Hendzel M. J., Bazett-Jones D. P. Promyelocytic leukemia (PML) nuclear bodies are protein structures that do not accumulate RNA. J. Cell Biol. 2000;148:283–292. doi: 10.1083/jcb.148.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden K.L.B. Pondering the promyelocytic leukemia protein (PML) puzzle: possible functions for PML nuclear bodies. Mol. Cell. Biol. 2002;22:5259–5269. doi: 10.1128/MCB.22.15.5259-5269.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broccoli D., Smogorzewska A., Chong L., de Lange T. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet. 1997a;17:231–235. doi: 10.1038/ng1097-231. [DOI] [PubMed] [Google Scholar]
- Broccoli D., Chong L., Oelmann S., Fernald A. A., Marziliano N., van Steensel B., Kipling D., Le Beau M. M., de Lange T. Comparison of the human and mouse genes encoding the telomeric protein, TRF1, chromosomal localization, expression and conserved protein domains. Hum. Mol. Genet. 1997b;6:69–76. doi: 10.1093/hmg/6.1.69. [DOI] [PubMed] [Google Scholar]
- Carbone R., Pearson M., Minucci S., Pelicci P. G. PML NBs associate with the hMre11 complex at sites of irradiation induced DNA damage. Oncogene. 2002;21:1633–1640. doi: 10.1038/sj.onc.1205227. [DOI] [PubMed] [Google Scholar]
- Chen Y. C., Kappel C., Beaudouin J., Eils R., Spector D. L. Live cell dynamics of promyelocytic leukemia nuclear bodies upon entry into and exit from mitosis. Mol. Biol. Cell. 2008;19:3147–3162. doi: 10.1091/mbc.E08-01-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlan A. L., McNees C. J., Heierhorst J. Proteasome-dependent dispersal of PML nuclear bodies in response to alkylating DNA damage. Oncogene. 2004;23:307–310. doi: 10.1038/sj.onc.1207119. [DOI] [PubMed] [Google Scholar]
- de The H., Lavau C., Marchio A., Chomienne C., Degos L., Dejean A. The PML-RARα fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell. 1991;66:675–684. doi: 10.1016/0092-8674(91)90113-d. [DOI] [PubMed] [Google Scholar]
- Dellaire G., Bazett-Jones D. P. PML nuclear bodies: dynamic sensors of DNA damage and cellular stress. BioEssays. 2004;26:963–977. doi: 10.1002/bies.20089. [DOI] [PubMed] [Google Scholar]
- Dellaire G., Eskiw C. H., Dehghani H., Ching R. W., Bazett-Jones D. P. Mitotic accumulations of PML protein contribute to the re-establishment of PML nuclear bodies in G1. J. Cell Sci. 2006a;119:1034–1042. doi: 10.1242/jcs.02817. [DOI] [PubMed] [Google Scholar]
- Dellaire G., Ching R. W., Dehghani H., Ren Y., Bazett-Jones D. P. The number of PML nuclear bodies increases in early S phase by a fission mechanism. J. Cell Sci. 2006b;119:1026–1033. doi: 10.1242/jcs.02816. [DOI] [PubMed] [Google Scholar]
- Dunham M. A., Neumann A. A., Fasching C. L., Reddel R. R. Telomere maintenance by recombination in human cells. Nat. Genet. 2000;26:447–450. doi: 10.1038/82586. [DOI] [PubMed] [Google Scholar]
- Eskiw C. H., Dellaire G., Mymryk J. S., Bazett-Jones D. P. Size, position and dynamic behavior of PML nuclear bodies following cell stress as a paradigm for supramolecular trafficking and assembly. J. Cell Sci. 2003;116:4455–4466. doi: 10.1242/jcs.00758. [DOI] [PubMed] [Google Scholar]
- Everett R. D., Earnshaw W. C., Pluta A. F., Sternsdorf T., Ainsztein A. M., Carmena M., Ruchaud S., Hsu W. L., Orr A. A dynamic connection between centromeres and ND10 proteins. J. Cell Sci. 1999a;112:3443–3454. doi: 10.1242/jcs.112.20.3443. [DOI] [PubMed] [Google Scholar]
- Everett R. D., Lomonte P., Sternsdorf T., van Driel R., Orr A. Cell cycle regulation of PML modification and ND10 composition. J. Cell Sci. 1999b;112:4581–4588. doi: 10.1242/jcs.112.24.4581. [DOI] [PubMed] [Google Scholar]
- Everett R. D., Chelbi-Alix M. K. PML and PML nuclear bodies: implications in antiviral defence. Biochimie. 2007;89:819–830. doi: 10.1016/j.biochi.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Fasching C. L., Neumann A. A., Muntoni A., Yeager T. R., Reddel R. R. DNA damage induces alternative lengthening of telomeres (ALT)-associated promyelocytic leukemia bodies that preferentially associate with linear telomeric DNA. Cancer Res. 2007;67:7072–7077. doi: 10.1158/0008-5472.CAN-07-1556. [DOI] [PubMed] [Google Scholar]
- Henson J. D., Neumann A. A., Yeager T. R., Reddel R. R. Alternative lengthening of telomeres in mammalian cells. Oncogene. 2002;21:598–610. doi: 10.1038/sj.onc.1205058. [DOI] [PubMed] [Google Scholar]
- Hu C.-D., Chinenov Y., Kerppola T. K. Visualization of interactions among bZIP and Rel proteins in living cells using bimolecular fluorescence complementation. Mol. Cell. 2002;9:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- Jegou T., Chung I., Heuvelman G., Wachsmuth M., Görisch S. M., Greulich-Bode K. M., Boukamp P., Lichter P., Rippe K. Dynamics of telomeres and promyelocytic leukemia nuclear bodies in a telomerase-negative human cell line. Mol. Biol. Cell. 2009;20:2070–2082. doi: 10.1091/mbc.E08-02-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kießlich A., von Mikecz A., Hemmerich P. Cell cycle-dependent association of PML bodies with sites of active transcription in nuclei of mammalian cells. J. Struct. Biol. 2002;140:167–179. doi: 10.1016/s1047-8477(02)00571-3. [DOI] [PubMed] [Google Scholar]
- Koken M.H.M., et al. The t(15;17) translocation alters a nuclear body in a retinoic acid-reversible fashion. EMBO J. 1994;13:1073–1079. doi: 10.1002/j.1460-2075.1994.tb06356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P. P., Bischof O., Purbey P. K., Notani D., Urlaub H., Dejean A., Galande S. Functional interaction between PML and SATB1 regulates chromatin loop architecture and transcription of the MHC class I locus. Nat. Cell Biol. 2007;9:45–56. doi: 10.1038/ncb1516. [DOI] [PubMed] [Google Scholar]
- Lanotte M., Martin-Thouvenin V., Najman S., Balerini P., Valensi F., Berger R. NB4, a maturation inducible cell line with t(15;17) marker isolated from a human acute promyelocytic leukemia (M3) Blood. 1991;77:1080–1086. [PubMed] [Google Scholar]
- Maul G. G. Nuclear domain 10, the site of DNA virus transcription and replication. BioEssays. 1998;20:660–667. doi: 10.1002/(SICI)1521-1878(199808)20:8<660::AID-BIES9>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Maul G. G., Yu E., Alexander M., Ishov A. M., Epstein A. L. Nuclear domain 10 (ND10) associated proteins are also present in nuclear bodies and redistribute to hundreds of nuclear sites after stress. J. Cell. Biochem. 1995;59:498–513. doi: 10.1002/jcb.240590410. [DOI] [PubMed] [Google Scholar]
- Melnick A., Licht J. D. Deconstructing a disease: RARα, its fusion partners, and their roles in the pathogenesis of acute promyelocytic leukemia. Blood. 1999;93:3167–3215. [PubMed] [Google Scholar]
- Misteli T. Protein dynamics: Implications for nuclear architecture and gene expression. Science. 2001;291:843–847. doi: 10.1126/science.291.5505.843. [DOI] [PubMed] [Google Scholar]
- Molenaar C., Wiesmeijer K., Verwoerd N. P., Khazen S., Eils R., Tanke H. J., Dirks R. W. Visualizing telomere dynamics in living mammalian cells using PNA probes. EMBO J. 2003;22:6631–6641. doi: 10.1093/emboj/cdg633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratani M., Gerlich D., Janicki S. M., Gebhard M., Eils R., Spector D. L. Metabolic-energy-dependent movement of PML bodies within the mammalian cell nucleus. Nat. Cell Biol. 2002;4:106–110. doi: 10.1038/ncb740. [DOI] [PubMed] [Google Scholar]
- Naeem M., Harrison K., Barton K., Nand K. S., Alkan S. A unique case of acute promyelocytic leukemia showing monocytic differentiation after ATRA (all-trans retinoic acid) therapy. Eur. J. Haematol. 2006;76:164–166. doi: 10.1111/j.0902-4441.2005.00583.x. [DOI] [PubMed] [Google Scholar]
- Nefkens I., Negorev D. G., Ishov A. M., Michaelson J. S., Yeh E. T., Tanquay R. M., Muller W. E., Maul G. G. Heat shock and CD(2+) exposure regulate PML and Daxx release from ND10 by independent mechanisms that modify the induction of heat-shock proteins 70 and 25 differently. J. Cell Sci. 2003;116:513–524. doi: 10.1242/jcs.00253. [DOI] [PubMed] [Google Scholar]
- Negorev D., Maul G. G. Cellular proteins localized at and interacting within ND10/PML nuclear bodies/PODs suggest functions of a nuclear depot. Oncogene. 2001;29:7234–7242. doi: 10.1038/sj.onc.1204764. [DOI] [PubMed] [Google Scholar]
- Ogino H., Nakabayashi K., Suzuki M., Takahashi E., Fujii M., Suzuki T., Ayusawa D. Release of telomeric DNA from chromosomes in immortal human cells lacking telomerase activity. Biochem. Biophys. Res. Comm. 1998;248:223–227. doi: 10.1006/bbrc.1998.8875. [DOI] [PubMed] [Google Scholar]
- Potts P. R., Yu H. The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere binding proteins. Nat. Struct. Mol. Biol. 2007;14:581–590. doi: 10.1038/nsmb1259. [DOI] [PubMed] [Google Scholar]
- Shen T. H., Lin -K. H., Scaglioni P. P., Yung T. M., Pandolfi P. P. The mechanisms of PML-nuclear body formation. Mol. Cell. 2006;24:331–339. doi: 10.1016/j.molcel.2006.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiels C., Islam S. A., Vatcheva R., Sasieni P., Sternberg M. J., Freemont P. S., Sheer D. PML bodies associate specifically with the MHC gene cluster in interphase nuclei. J. Cell Sci. 2001;114:3705–3716. doi: 10.1242/jcs.114.20.3705. [DOI] [PubMed] [Google Scholar]
- Snaar S., Wiesmeijer K., Jochemsen A. G., Tanke H. J., Dirks R. W. Mutational analysis of fibrillarin and its mobility in living human cells. J. Cell Biol. 2000;151:653–662. doi: 10.1083/jcb.151.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector D. L. Nuclear domains. J. Cell Sci. 2001;114:2891–2893. doi: 10.1242/jcs.114.16.2891. [DOI] [PubMed] [Google Scholar]
- Vertegaal A. C., Andersen J. S., Ogg S. C., Hay R. T., Mann M., Lamond A. I. Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol. Cell Proteonom. 2006;5:2298–2310. doi: 10.1074/mcp.M600212-MCP200. [DOI] [PubMed] [Google Scholar]
- Wang Z. G., Delva L., Gaboli M., Rivi R., Giorgio M., Cordon-Cardo C., Grosveld F., Pandolfi P. P. Role of PML in cell growth and the retinoic acid pathway. Science. 1998;279:1547–1551. doi: 10.1126/science.279.5356.1547. [DOI] [PubMed] [Google Scholar]
- Wang J., Shiels C., Sasieni P., Wu P. J., Islam S. A., Freemont P. S., Scheer D. Promyelocytic leukemia nuclear bodies associate with transcriptionally active genomic regions. J. Cell Biol. 2004;164:515–526. doi: 10.1083/jcb.200305142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis K., Ramboud S., Lavau C., Jansen J., Carvalho T., Carmo-Fonseca M., Lamond A., Dejean A. Retinoic acid regulates aberrant nuclear localization of PML-RARα in acute promyelocytic leukemia. Cell. 1994;75:347–386. doi: 10.1016/0092-8674(94)90341-7. [DOI] [PubMed] [Google Scholar]
- Wiesmeijer K., Krouwels I. K., Tanke H. J., Dirks R. W. Chromatin movement visualized with photoactivable GFP-labeled histone H4. Differentiation. 2008;76:83–90. doi: 10.1111/j.1432-0436.2007.00234.x. [DOI] [PubMed] [Google Scholar]
- Wiesmeijer K., Molenaar C., Bekeer C. I. M. L. A., Tanke H. J., Dirks R. W. Mobile foci of SP100 do not contain PML: PML bodies are immobile but PML and SP100 proteins are not. J. Struct. Biol. 2002;140:180–188. doi: 10.1016/s1047-8477(02)00529-4. [DOI] [PubMed] [Google Scholar]
- Yeager T. R., Neumann A. A., Englezou A., Huschtscha L. I., Noble J. R., Reddel R. R. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 1999;59:4175–4179. [PubMed] [Google Scholar]
- Zhong S., Muller S., Ronchetti S., Freemont P. S., Dejean A., Pandolfi P. P. Role of SUMO-1-modified PML in nuclear body formation. Blood. 2000;95:2748–2752. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.