Abstract
Gas1p is a glucan-elongase that plays a crucial role in yeast morphogenesis. It is predominantly anchored to the plasma membrane through a glycosylphosphatidylinositol, but a fraction was also found covalently bound to the cell wall. We have used fusions with the green fluorescent protein or red fluorescent protein (RFP) to determine its localization. Gas1p was present in microdomains of the plasma membrane, at the mother-bud neck and in the bud scars. By exploiting the instability of RFP-Gas1p, we identified mobile and immobile pools of Gas1p. Moreover, in chs3Δ cells the chitin ring and the cross-linked Gas1p were missing, but this unveiled an additional unexpected localization of Gas1p along the septum line in cells at cytokinesis. Localization of Gas1p was also perturbed in a chs2Δ mutant where a remedial septum is produced. Phenotypic analysis of cells expressing a fusion of Gas1p to a transmembrane domain unmasked new roles of the cell wall-bound Gas1p in the maintenance of the bud neck size and in cell separation. We present evidence that Crh1p and Crh2p are required for tethering Gas1p to the chitin ring and bud scar. These results reveal a new mechanism of protein immobilization at specific sites of the cell envelope.
INTRODUCTION
In yeast, morphogenesis and cell wall biogenesis are two tightly linked and mutually dependent processes that are responsible for the changes of cell shape occurring at different stages of the life cycle. The cell wall results from the cross-linking of various polymers that forms a fabric strong enough to counteract the high turgor pressure of yeast cells. In the budding yeast Saccharomyces cerevisiae, the network consists of an ordered thread of chitin, β(1,3)/(1,6)-glucans and mannoproteins (constituting ∼1–2, 60, and 40–45% of the cell wall dry weight, respectively) (Klis et al., 2006; Lesage and Bussey, 2006). Cross-linking enzymes act on β(1,3)-glucan chains extruded by the plasma membrane β(1,3)-glucan synthases Fks1 and Fks2 and β(1,6)-glucan chains (the site of whose synthesis may be the cell surface or along the secretory pathway) (Montijn et al., 1999; Lesage et al., 2004; Lesage and Bussey, 2006). Moreover, surface mannoproteins and chitin molecules extruded by chitin synthases are also substrates of the cross-linking process.
Chitin synthesis is catalyzed by three distinct chitin synthases (CSI, II, and III), which use Chs1p, Chs2p, and Chs3p as the catalytic subunit, and is crucial for yeast morphogenesis. Most of the chitin in the cell is produced by Chs3p and is localized in the “chitin ring,” which forms in the cell wall shortly before bud emergence, whereas only a tiny amount is deposited in the lateral cell walls (Shaw et al., 1991). Chs2p is required for the synthesis of the primary septum (∼10% of total chitin), a thin disk of chitin that forms centripetally within the confines of the chitin ring at the onset of cytokinesis. Deposition of chitin by Chs2p is concomitant with the constriction of the actomyosin ring (for review, see Cabib, 2004). After cell separation the chitin ring remains on the mother cell as the crater-like “bud scar.” Finally, Chs1p has a repair function preventing lysis at the birth scar due to excessive hydrolysis of the septum by chitinase. The contribution of Chs1p to the chitin content of a cell is negligible (Cabib et al., 1989).
The chitin ring delimits the neck and its presence contributes to preventing growth at the mother-daughter neck region. In fact, a constriction of constant diameter is maintained during the budded phase of the cell cycle (Cabib, 2004). Both septins and chitin ring are known to cooperate in ensuring the integrity of the neck. Defects in either of these elements result in a dramatic widening of the neck diameter and lethality (Schmidt et al., 2003; Cabib, 2004).
To date, only a few classes of extracellular enzymes have been identified as cell wall cross-linking/remodeling enzymes. One of them is the Gas protein family that is composed by five members, Gas1 to Gas5 (Ragni et al., 2007b). Gas1p is expressed during vegetative growth together with Gas5p, which seems to play an auxiliary role. Gas2p and Gas4p are specifically expressed during meiosis and are essential for spore wall formation (Ragni et al., 2007a). Gas3p function is still unknown. Genes encoding Gas homologous proteins were found in all yeast and fungal species so far sequenced (Ragni et al., 2007b).
Gas proteins are modified by GPI, a glycolipid moiety bound to the C-terminal end that anchors the protein to the outer leaflet of the lipid bilayer in the plasma membrane. GPI confers lateral mobility in the lipid environment to the proteins (Malinska et al., 2004) and promotes association with specific membrane microdomains known as “lipid rafts.” These microdomains are enriched in sphingolipids and ergosterol. Gas1p was recovered in the detergent-resistant membranes (DRMs), also named detergent-insoluble glycolipid-enriched complexes, which is indicative of an association with lipid rafts (Bagnat et al., 2000; Simons and Vaz, 2004; Aronova et al., 2007).
In yeast, the protein modification by GPI attachment has an additional role compared with animals and plants. At the cell surface, GPI-containing proteins can undergo a transglycosylase reaction whereby they are cleaved inside the glycan moiety of the GPI and linked to the glucan network with consequent release of the remaining glucosamine-phosphatydylinositol (GlcN-PI) moiety of the GPI (Kollar et al., 1997). This class of proteins is called GPI-cell wall proteins (GPI-CWPs, where GPI indicates a GPI remnant) and is the most abundant in the cell wall. To date the molecular mechanism of this process is still obscure. The putative mannosidases Dfg5p and Dcw1p were proposed as candidate catalysts of the transglycosylase reaction (Kitagaki et al., 2002).
The nonreducing end of β(1-3)-glucan is an acceptor site for the attachment of chitin, PIR-proteins or β(1-6)-glucan to which the GPI-CWPs are linked (for a review see Klis et al., 2006). Short side branches of the β(1-6)-glucan provide a further attachment site for the reducing end of chitin. Thus, β(1-6)-glucan is a core component of the cell wall to which GPI-CWPs, chitin and β(1-3)-glucan can be linked (Kollar et al., 1997). The formation of the cross-links occurs by transglycosylation reactions because small energy-rich molecules are unavailable outside the cell.
Recently, it has been demonstrated that the GPI-containing proteins Crh1 and Crh2 are responsible of the linkage of chitin to the side branches of β(1-6)glucan (Cabib et al., 2008). These enzymes are localized to the bud neck, bud scar and in minor amount all over the cell periphery. Interestingly, the activity of transferring chitin on β(1,6)-glucan is high in the bud scars and may be involved in tethering mannoproteins at these sites (Rodriguez-Pena et al., 2002; Cabib et al., 2007, 2008).
Gas1p acts as a β(1,3)-glucan elongase in in vitro assays (Mouyna et al., 2000). Its presumable in vivo role is to incorporate new β(1,3)-glucan chains into the preexisting cell wall and also create anchoring sites for other cell wall components (Kollar et al., 1995; Mouyna et al., 2000). Gas1p is predominantly located in the plasma membrane, but it is also linked to cell wall β(1,3)-glucan (De Sampaio et al., 1999). A comprehensive mass spectrometry analysis of the cell wall proteome revealed that Gas1, Gas3, and Gas5 proteins are covalently cross-linked to the glycan network (Yin et al., 2005). Besides Gas proteins, other GPI-CWPs with putative cell wall cross-linking activity were recovered in the cell wall proteome, notably Crh1p and Crh2p. Because it is still unclear whether anchoring of GPI-mannoproteins to the cell wall is simply a consequence of some leakiness in the mechanism of plasma membrane retention or a specific destination of the protein (Yin et al., 2005), we have investigated the cellular location of Gas1p. We report novel and unexpected locations of Gas1p and show that anchoring of Gas1p to specific cell wall destinations contributes to yeast morphogenesis.
MATERIALS AND METHODS
Yeast Strains and Growth Conditions
The strains used are listed in Table 1. Cells were grown in batch at 30°C in YPD (1% yeast extract, 2% Bacto-peptone, and 2% glucose) or in synthetic dextrose minimal medium (SD) (Difco yeast nitrogen base without amino acids [Difco, Detroit, MI] at 6.7 g/l and 2% glucose) to which amino acids and uracil were added to a concentration of 50 and adenine to 100 mg/l. For solid media, 2% agar was added to YPD or SD media (YPDA and SDA). When required, YPD was buffered at pH 6.5 with 10 g/l 2-(N-morpholino)ethanesulfonic acid (MES). Growth was monitored as the increase in OD450. Duplication time (Td) was calculated using the equation Td = ln2/K, where K, the growth rate constant, is the slope of the line obtained by linear regression on a semilogarithmic plot of the OD values. The growth rate, μ (h−1) was calculated as 1/Td.
Table 1.
Strains used in this work
| Strain | Genotype | Relevant genotype | Source |
|---|---|---|---|
| W303-1A | Mata ade2-1 his3-11,15 trp1-1 ura3-1 leu2-3,112 can1-100 | Parental strain | CNRS, France |
| W303-1B | Matα ade2-1 his3-11,15 trp1-1 ura3-1 leu2-3,112 can1-100 | Parental strain | CNRS, France |
| chs3Δ | MATα chs3::LEU2 ade2-1 his3-11,15 trp1-1 ura3-1 leu2-3,112 can1-100 | chs3Δ | A. Duran, Universidad de Salamanca, Spain |
| WB2d | W303-1B gas1::LEU2 | gas1Δ | Vai et al. (1991) |
| WAH | W303-1A gas1::HIS3 | gas1Δ | Vai et al. (1991) |
| JC9 | WAH leu2::pMF608 mRFP-GAS1 LEU2 | gas1Δ mRFP-GAS1 | This work |
| JC10 | WAH leu2::pMF608TM mRFP-GAS1-MID2 LEU2 | gas1Δ mRFP-GAS1-MID2 | This work |
| W303-GAS1-GFP | W303-1B [pRS424-GAS1-GFP 2 μ TRP1] | GAS1-GFP | This work |
| WB2d-GAS1-GFP | WB2d [pRS424-GAS1-GFP 2 μ TRP1] | gas1Δ GAS1-GFP | This work |
| chs3Δ-GAS1-GFP | chs3Δ [pRS424-GAS1-GFP 2 μ TRP1] | chs3Δ GAS1-GFP | This work |
| AN117-4B | MATα ura3 leu2 his3 trp1 lys2 arg4 ho::LYS2 rme1::LEU2 | Parental strain | A. Neiman, Stony Brook University |
| AN117-16D | MATa ura3 leu2 his3 trp1 lys2 ho::LYS2 | Parental strain | A. Neiman |
| AN120 | (Cross AN117-4B x AN117-16D) MATa/α ura3/ura3 leu2/leu2trp1/trp1 his3/his3 lys2/lys2 arg4/ARG4 ho::LYS2/ho::LYS2rme1::LEU2/RME1 | Parental strain | Ragni et al. (2007a) |
| AN262 | AN120 and chs3::HIS5/chs3::HIS5 | chs3Δ | A. Neiman |
| ER310 | AN120 [YEp24 2m URA3] | Ragni et al. (2007a) | |
| ER320 | AN120 and gas1::KanMX2/gas1::KanMX2 | gas1Δ | This work |
| ER332 | ER320 [pRS416 CEN URA3] | gas1Δ | This work |
| ER333 | ER320 [YEp24] | gas1Δ | This work |
| ER334 | ER320 [pRS416-GAS1-URA3 CEN] | This work | |
| ER335 | ER320 [YEp24-GAS1] | This work | |
| ER336 | ER320 [pRS416-GAS1-GFP URA3 CEN] | gas1Δ GAS1-GFP | This work |
| ER337 | ER320 [YEp24-GAS1-GFP] | gas1Δ GAS1-GFP | This work |
| ER338 | AN120 [pRS416] | This work | |
| ER339 | AN120 [pRS416-GAS1] | This work | |
| ER340 | AN120 [YEp24-GAS1] | This work | |
| ER341 | AN120 [pRS416-GAS1-GFP] | GAS1-GFP | This work |
| ER342 | AN120 [YEp24-GAS1-GFP] | GAS1-GFP | This work |
| ER343 | AN262 [pRS416-GAS1-GFP] | chs3Δ GAS1-GFP | This work |
| ER344 | AN262 [YEp24-GAS1-GFP] | chs3Δ GAS1-GFP | This work |
| YPH499 | MATa ura3-52 leu2-Δ1 his3Δ200 trp1-Δ63 lys2-801 ade2-101 | Parental strain | E. Cabib, NIH |
| ECY46-1-8D | YPH499 chs1::HIS3 | chs1Δ | E. Cabib, NIH |
| YMS11 | YPH499 chs2::TRP1 | chs2Δ | E. Cabib, NIH |
| ECY46-4-1B | YPH499 chs3::LEU2 | chs3Δ | E. Cabib, NIH |
| CER1 | YPH499 gas1::LEU2 | gas1Δ | This work |
| CER2 | YPH499 chs1::HIS3 gas1::LEU2 | chs1Δ gas1Δ | This work |
| CER3 | CER1 [YEp24-GAS1-GFP] | gas1Δ GAS1-GFP | This work |
| CER4 | CER2 [YEp24-GAS1-GFP] | chs1Δ gas1Δ GAS1-GFP | This work |
| CER5 | YMS11 [YEp24-GAS1-GFP] | chs2Δ GAS1-GFP | This work |
| BY4742 | MATα his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 | Parental strain | Euroscarf |
| TG208 | BY4742 gas1::LEU2 | gas1Δ | This work |
| Y14895 | BY4742 dcw1::KanMX4 | dcw1Δ | Euroscraf |
| Y10824 | BY4742 dfg5::KanMX4 | dfg5Δ | Euroscarf |
| ER369 | Y14895 gas1::LEU2 | dcw1Δ gas1Δ | This work |
| ER370 | Y10824 gas1::LEU2 | dfg5Δ gas1Δ | This work |
| ER371 | TG208 [YEp24-GAS1-GFP] | gas1Δ GAS1-GFP | This work |
| ER372 | ER369 [YEp24-GAS1-GFP] | dcw1Δ gas1Δ GAS1-GFP | This work |
| ER373 | ER370 [YEp24-GAS1-GFP] | dfg5Δ gas1Δ GAS1-GFP | This work |
| BY4741 | MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 | Parental strain | Euroscarf |
| GRA006 | BY4741 crh2::HIS3 | crh2Δ | Cabib et al. (2007) |
| GRA007 | BY4741 crh1::hphMX4 crh2::HIS3 | crh1Δ crh2Δ | Cabib et al. (2007) |
| Y00897 | BY4741 gas1::KMX4 | gas1Δ | Cabib et al. (2007) |
| GR010 | BY4741 crh2::HIS3 gas1::KMX4 | crh2Δ gas1Δ | Cabib et al. (2007) |
| GR011 | BY4741 crh1::hphMX4 crh2::HIS3 gas1::KMX4 | crh1Δ crh2Δ gas1Δ | Cabib et al. (2007) |
| ER374 | Y00897 [YEp24-GAS1-GFP] | gas1Δ GAS1-GFP | This work |
| ER375 | GR010 [YEp24-GAS1-GFP] | crh2Δ gas1Δ GAS1-GFP | This work |
| ER376 | GR011 [YEp24-GAS1-GFP] | crh1Δ crh2Δ gas1Δ GAS1-GFP | This work |
Cloning of GAS1 Gene from SK1 Strain
The GAS1 gene, encompassing 400 base pairs upstream and 300 base pairs downstream the coding sequence, was amplified by polymerase chain reaction (PCR) from genomic DNA of AN120 strain using the primers (restriction sites underlined) GAS1-PROM-SmaI GCATATTCGACTGACCCGGGGCCAGCCCTGGCTATTCTTT and GAS1-TERM-BamHI ATCGTCGGGCTCGGATCCTATGGAGAAAGTACATAAATG and Expand HiFidelity DNA polymerase (Roche Diagnostics, Indianapolis, IN). The amplified fragment was cloned in pCRII TA-TOPO cloning vector (Invitrogen, Carlsbad, CA) to create plasmid pER-1. Sequence verification was carried out by sequencing both strands of plasmids extracted from three transformants. Single-nucleotide polymorphisms were found, in agreement with the reported polymorphism of the SK1 strain compared with S288c (Primig et al., 2000). In the GAS1 5′-flanking region, a T insertion was present at nucleotide −49 from the initiation codon (the A of ATG was defined as nucleotide 1). In the open reading frame, C-to-T and G-to-A transitions, T-to-A transversion, and A-to G and G-to-A transitions were found at nucleotides 51, 156, 822, 855, and 1374, respectively, but these changes were silent. A T-to-A transversion and an A-to-G transition, respectively, at nucleotides 822 and 932, cause the following amino acid substitutions: T211 to A and N311 to S. In the 3′-flanking region, a C-to-G transition was present at nucleotide 1691. Yeast plasmids pYER-1 and pRS-1 were obtained by cloning the SmaI-BamHI fragment from pER-1 into the like sites of the YEp24 and pRS416 vectors.
Construction of S. cerevisiae Strains and GAS1p-Green Fluorescent Protein (GFP) Fusion
The inactivation of GAS1 gene in AN120 strain was performed using short-homology polymerase chain reaction (PCR) gene targeting. Plasmid pFA6a-KanMX2, containing the module KanMX2 with the kanR gene, was used to amplify a PCR fragment used to inactivate GAS1 (GAS1KFOR, 5′-TGGTATTCCTCATACAGCCTGCGCGGTTTATTAGTAAAATACCCGATAATCCTCGAGGTTGATATCAAGCTTGCCTCG-3′; GAS1KREV, 5′-CTCCGTTTGGAGTTGGTGGTAATTCTGTTGCAGCGGACCAGTACTTACCAGTAGCTGGACGTCGACACTGGATGGCGG-3′—sequences annealing to kanR gene are underlined). The 1556-base pair PCR fragment, harboring ends complementary to the −248 to −189 and +1043 to +1102 from the AUG start codon of GAS1, was used to transform the haploid strains AN117-16D and AN117-4B (Table 1). S. cerevisiae cells were transformed using the S.C. EasyComp transformation kit (Invitrogen). Genomic DNA isolated from the transformant clones was subjected to five different PCR diagnostic tests to verify correct integration. The primers used were AEV25, 5′-AGTTTTCGTGCCGCAAACGT-3′; AEV26, 5′-AGTTCCGGAACCCAAATCAA-3′; GAS1ORF, 5′-AACACAGATGCATCTGCT-3′; GAS1OREV, 5′-AGCAGATGCATCTGTGTT-3′; KANMX2REV, 5′-GTAATGGCTGGCCTGTTG-3′; and KANMX2, 5′-CAACAGGCCAGCCATTAC-3′). The haploid strains carrying the gas1Δ mutation were crossed to generate gas1Δ/gas1Δ (ER320) null diploid. The lack of Gas1p was confirmed by immunoblotting.
Strain JC9 was constructed by transforming WAH cells (gas1Δ) with integrative plasmid pMF608, a kind gift of Prof. Y. Jigami (AIST, Tokyo, Japan). Plasmid pMF608 harbors an mRFP-GAS1 fusion under the control of the natural GAS1 promoter. The N-terminal sequence of the encoded hybrid protein is MLFKSLSKLATAAAFFAGVATA↓DTRASASSE—where the signal peptide of Gas1p is in italics, the amino acids of the linker are underlined, the first amino acids of monomeric red fluorescent protein (mRFP) are in bold, and the arrow indicates the cleavage site for leader peptidase. Plasmid pMF608 was linearized by digestion with BfrI for targeting into the LEU2 locus. Leu+ transformants were analyzed for the presence of the fluorescent protein and JC9 strain was analyzed in more detail (Table 1).
The GFP module was amplified by PCR from plasmid pFA6a-GFP(S65T)-KanMX6 with the following primers containing a BseAI restriction site (underlined): GAS1P-GFPup, ATATCCGACTGATCCGGAGGTGCCAGTAAAGGAGAAGAACTTTTCAC and GAS1P-GFPdown, ATCGTCCTCTATCCGGATTTGTATAGTTCATCCATG. The triplets encoding for Gly-Ala were introduced at the 5′ and 3′ flanking regions of the GFP coding sequence to act as spacers. The gel-purified PCR product was digested with BseAI and cloned in plasmid pER-1 (see above). Control digestions and sequencing were performed to verify the in-frame-fusion of the GFP module into the GAS1 cds. The SmaI-BamHI fragment, containing the GAS1-GFP fusion gene, was cloned into the corresponding site of the YEp24 (multicopy) and pRS416 (CEN) vectors, generating YEp24-GAS1-GFP and pRS416-GAS1-GFP. These plasmids were used to transform AN120 and AN262 strains. The SmaI-BamHI fragment from YEp24-GAS1-GFP was cloned into a similarly digested pRS424 vector to give pRS424-GAS1-GFP. This plasmid was used to transform W303 and derived strains. The strains and their genotypes are listed in Table 1. The GAS1 gene disruption in strains derived from YPH499 and BY4742 was performed as described previously (Vai et al., 1991).
Construction of the mRFP-GAS1-MID2 Fusion
The fusion gene was obtained by cloning in-frame the fragment of mRFP-GAS1 ending at codon 484 of the GAS1 open reading frame (ORF), with a DNA fragment of the cds (residues 213-376) of the MID2 gene encompassing the transmembrane segment (residues 225-247), the cytosolic tail (residues 248-376), and the downstream transcription termination sequence. In a first PCR step, the fragment of MID2 starting from nucleotide +637 to +1128 of the ORF and the 3′ downstream region from nucleotide +1129 to +1680 (being A of the starting ATG the nucleotide number +1) was amplified using the primers Mid2up (5′-ATATTCGACTGATCCGGATCCAAAAGTTCGGGTCTTTC-3′) and Mid2down2 (5′-ACTCTGCTCTACTCCGGACTCCTTATGCTTCTACAC-3′). In each primer, a sequence recognized by BseAI was introduced (in bold). The amplified fragment of ∼1.1 kbp was purified, BseAI digested, and ligated into the plasmid pMF608 previously linearized in the corresponding BseAI site at position +1453 of the GAS1 ORF. The DNA plasmid from 20 generated clones was digested with NdeI to check for correct orientation. Two independent positive plasmids were named pMF608TM-15 and -16. The plasmids were linearized with BfrI and 10 μg of digested DNA were used to transform the WAH strain via integration in the LEU2 locus to obtain JC10 strain (see Table 1).
α-Factor (α-F) Treatment
At a cell density of ∼5 × 106 cells/ml (∼0.3 OD450 nm), cells from strains JC9 and W303-1A, exponentially growing in YPD at 30°C, were collected by centrifugation, washed with fresh YPD, and suspended in YPD at the same cell density in the presence or absence of 20 μg/ml α-F (GenScript, Piscataway, NJ). At different time intervals from α-F addition, aliquots corresponding to 13, 20, or 50 OD450 were collected for the preparation of total extracts, for subcellular fractionation and cell wall purification, respectively.
Microscopy Analysis
Cells were routinely observed by phase-contrast microscopy and scored for budding by counting at least 200 cells after mild sonication. For fluorescence microscopy cells were either fixed or analyzed without fixation. In either case, cells were sonicated before being processed. Approximately 2 × 108 cells were fixed in 3.7% formaldehyde and 0.1 M K-phosphate, pH 6.5. After 30 min at room temperature, cells were filtered and resuspended in the same volume of a fixing buffer (3.7% formaldehyde and 0.1 M K-phosphate, pH 6.5). Cells were kept at 4°C for one or more days. Cells were collected by centrifugation at 6000 × g for 2 min, washed twice with phosphate-buffered saline (PBS), pH 7.4, at 4°C and left in PBS for 1 h in ice before examination at the microscope. For observation without fixation, cells were collected by centrifugation, washed twice with PBS, and incubated at least 15 min for Gas1p-GFP and 1 h for mRFP-Gas1p on ice before being examined under the microscope. If required, 8.3 μg/ml 4,6-diamidine-phenylindole (DAPI) was added to the cells. Samples were then incubated for further 15 min at room temperature in the dark. Staining of chitin on unfixed cells was performed by using 2 μg/ml calcofluor (CF) white (Sigma-Aldrich, St. Louis, MO). Cells were stained for 3 min and washed once with PBS. In the α-F experiments, chitin staining was performed by adding 0.2 μg CF/ml culture during the last 10 min of growth before viewing the cells (Warenda and Konopka, 2002).
The cells were examined as wet mounts using a BX60 microscope (Olympus, Melville, NY) and a DC290 digital photo camera (Eastman Kodak, Rochester, NY) or an Eclipse 90i microscope (Nikon, Tokyo, Japan) equipped with epifluorescence, Nomarski optics, and a Hamamatsu ORCA-ER device camera (Nuhsbaum, McHenry, IL). For stack acquisition analysis, the z-distance was set to 0.4 μm. For confocal microscopy cells were examined using a TCS SP2 AOBS confocal laser-scanning microscope (Leica Microsystems, Heidelberg, Germany), equipped with Ar/Kr and He/Ne lasers and a PLAPO 63 oil immersion objective. Gas1p-GFP was excited with a laser line of λ = 488 nm, and the fluorescence was collected between 493 and 539 nm. mRFP-Gas1p was excited with a laser line of λ = 561 nm, and the fluorescence collected between 555 and 620 nm. DAPI or calcofluor were excited in the UV (λ = 364 nm) and the fluorescence was collected in the range 410 and 470 nm. A focal series of horizontal planes of section were assessed by sequential scanning of sample with 1.0-μm step size. Neck diameters were measured in differential interference contrast images by measuring the distance between the inner parts of the cell wall.
Calcofluor Sensitivity Assay
Five microliters from a concentrated suspension of cells (8 OD450) and from 1:10 serial dilutions of it, were spotted on SDA or buffered SDA plates in the absence or presence of 2, 5, 10, and 20 μg of calcofluor white per milliliter. Growth was checked after 2 d at 30°C.
Flow Cytometry Analysis
Cells (1 OD450) were mildly sonicated, collected by centrifugation, washed, and suspended in ice-cold 70% ethanol. Fixed cells were centrifuged, suspended in 0.5 ml of RNase A (1 mg/ml in 50 mM Tris-HCl, pH 7.5) and incubated for 2 h at 37°C. Cells were centrifuged and suspended in 0.5 ml of proteinase K (1 mg/ml in 50 mM Tris-HCl, pH 7.5). After 2 h at 42°C cells were collected, resuspended in the same volume of fluorescence-activated cell sorting (FACS) buffer (200 mM Tris-HCl, pH 7.5, 200 mM NaCl, and 78 mM MgCl2), and then 100 μl was added to 1 ml Sytox Green 1× (1:5000 dilution in 50 mM Tris-HCl, pH 7.5; Invitrogen). The samples were sonicated for 10 s before analysis with a FACScan flow cytometer (BD Biosciences, San Jose, CA). We analyzed 104 cells for each sample, and the percentage of cells in G1, S, and G2 + M phases was determined. The percentage of cells with a DNA content >2c was measured starting from the end of the G2+M peak.
Extract Preparation, Precipitation of Protein from Medium, Electrophoresis, and Immunoblotting
For total extract, ∼2 × 108 cells were collected by filtration, washed, quickly frozen in dry ice-acetone and stored at −20°C. The procedure for extract preparation in the presence of protease inhibitors (1 mM phenylmethylsulfonyl fluoride, a protease inhibitors cocktail [Roche Diagnostics] and 1 μg/pepstatin A), and the determination of protein concentration was performed as described previously (Ragni et al., 2007a). For the analysis of Gas1p in the culture medium, cells were grown in YPD or in YPD-pH 6.5 and let grow to an OD450 of ∼1.5. Culture supernatants were obtained by filtration of 27 ml of culture on nitrocellulose filters and proteins were precipitated in 10% trichloroacetic acid (TCA) in ice for at least 1 h. After 10-min centrifugation at 13,000 × g, the pellet was washed with 1 ml of ice-cold acetone and allowed to evaporate. The pellet was denatured in SDS-sample buffer [0.0625 M Tris-HCl, pH 6.8, 2.3% (wt/vol) SDS, 5% (vol/vol) β-mercaptoethanol, and 10% glycerol] and neutralized by addition of 1 M Tris. SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblotting were performed as described previously (Gatti et al., 1994; Ragni et al., 2007a). Anti-Gas1p serum was obtained by immunizing rabbits with a soluble 6xHis-tagged form of Gas1p produced in Pichia pastoris (Ragni et al., 2007b). Immunization procedure was carried out by Areta International (Gerenzano, Varese, Italy). Anti-Gas1p serum was used at a dilution of 1:1000 in Tris-buffered saline (TBS)-bovine serum albumin (BSA) and 0.2% Tween 20. Monoclonal mouse anti-actin antibody, clone C4 (MP Biomedicals, Irvine, CA) was used at a dilution of 1:1000 in TBS-BSA, 0.1% Tween 20. Peroxidase-conjugated affinity purified F(ab′)2 fragment donkey anti-rabbit or anti-mouse immunoglobulin G were from The Jackson Laboratory (Bar Harbor, ME) and were diluted 1:10,000. Bound antibodies were revealed using the enhanced chemiluminescence Western blotting detection reagents (GE Healthcare, Chalfont St. Giles, Buckinghamshire, United Kingdom). Densitometric measurements of undersaturated films were performed using the program Scion Image (scion, Frederick, MD).
Subcellular Fractionation and Protein Extraction from Purified Cell Walls
For membrane separation, cells corresponding to 20 OD450 units were collected by centrifugation. The pellet was washed with distilled H2O (dH2O) and cells resuspended in 100 μl of 10 mM Tris-HCl, pH 7.5, supplemented with the protease inhibitors described above. Cells were mechanically broken in the presence of glass beads in a FastPrep 120 for three cycles or more at maximum speed until full cell disruption was confirmed by phase contrast microscopy. After removal of the glass beads and unbroken cells, the crude extract was centrifuged at 100,000 × g for 30 min at 4°C. The supernatant and the pellet, S100 and P100 fractions, were analyzed by immunoblot. For cell wall isolation, 50 OD450 units of cells were mechanically broken in 300 μl of SDS sample buffer supplemented with the protease inhibitors as described above. After removal of the glass beads and centrifugation of the crude extract for 1 min at 400 × g, the pellet contained the cell walls. The cell walls were processed essentially as described previously (de Groot et al., 2004). They were washed with 1 ml of 1 M NaCl at 4°C and boiled once at 100°C for 5 min in 1 ml of a buffer A (2% SDS, 40 mM β-mercaptoethanol, 100 mM EDTA, and Tris-HCl, pH 7.8), spun down, and then boiled again. After five washes with 1 ml of dH2O the pellet was suspended either in 50 μl of 50 mM Tris-HCl, pH 7.4 containing 20 U of Quantazyme, a recombinant β(1,3)-glucanase (Qbiogene Europe, Illkirch, France) or in 50 μl of 50 mM MES, pH 6.0, containing 30 mU of laminarinase, a β(1,3)-glucanase preparation (Sigma-Aldrich), or in 70 μl of 50 mM MES, pH 6.0, supplemented with 30 μl of pure exochitinase of Serratia marcescens (∼30 μg of the purified enzyme), kindly provided by Dr. Enrico Cabib (National Institutes of Health, Bethesda), and protease inhibitors. After a 16-h incubation at 37°C for Quantazyme and at 30°C for exochitinase or 2 h at 37°C for Laminarinase under gentle shaking, the samples were centrifuged at 11,000 × g for 5 min, and the supernatants were analyzed by immunoblot. In the sequential treatments, the pellet of the first digestion was boiled in buffer A for 10 min to inactivate the enzyme. After five washes with dH2O, the pellet was digested in 70 μl of 50 mM MES, pH 6.0, supplemented with 30 μl of pure exochitinase and the exochitinase pellet in 50 μl of 50 mM Tris-HCl, pH 7.4, containing 20 U of Quantazyme or in 50 μl of 50 mM MES, pH 6.0, containing 30 mU of laminarinase. After 2 h (laminarinase) or overnight digestion (Quantazyme and exochitinase) at 37 or 30°C (exochitinase), the samples were centrifuged and the supernatants were denatured. When the exochitinase treatment of the glucanase-released fraction was performed, the supernatant from the glucanase digestion was supplemented with 30 μl of exochitinase and incubated overnight (30°C). After centrifugation, the supernatant was analyzed. The samples were denatured in SDS-sample buffer for SDS-PAGE and immunoblot. SDS-PAGE was performed using 7% polyacrylamide slab gels (14 × 16 cm).
DRM Isolation
Isolation of the DRMs was performed essentially as described previously (Bagnat et al., 2000). Yeast cells (25 OD450) were mechanically broken in a FastPrep 120 in 500 μl of TNE buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 5 mM EDTA) supplemented with protease inhibitors and glass beads as described above. The lysates were cleared by centrifugation at 500 × g for 5 min at 4°C. Two cleared lysates were pooled and incubated with 1% Triton X-100 for 30 min at 4°C. The lysate (1.5 ml) was adjusted to 40% Opti-Prep (Sigma-Aldrich), and the resulting mixture (4.2 ml) was sequentially overlaid with 6.7 ml of 30% Opti-Prep in TNEX buffer (TNE buffer including 0.1% Triton X-100) supplemented with protease inhibitors and 1.1 ml of TNEX buffer. The samples were centrifuged at 100,000 × g in SW41Ti rotor for 2.5 h. At the end, 0.9-ml fractions were collected from the top of the gradient. Proteins were precipitated with 10% TCA on ice and analyzed by Western blot.
RESULTS
Construction of Fluorescent Versions of Gas1p
Gas1p is processed and transported to the plasma membrane through the secretory pathway (Popolo and Vai, 1999). Besides an N-terminal signal peptide required for targeting to the endoplasmic reticulum (ER), Gas1p has also a C-terminal signal sequence that directs the GPI attachment to the newly synthesized precursor. Because both these sequences are required for the maturation of Gas1p, we created internally tagged versions of GAS1 gene. In the first construct an mRFP was fused, in-frame, to the GAS1 coding sequence following the signal peptide (Figure 1A). This construct was previously described by others and used for a different purpose (Fujita et al., 2006). In the second construct, we inserted a bright version of GFP between the residues G486 and T487, within the Ser-rich region (Figure 1C). We chose this site on the basis of two criteria: 1) the Ser-rich region functions as a spacer and is dispensable for in vivo functionality and in vitro activity of Gas1p (Gatti et al., 1994, Carotti et al., 2004) and 2) the insertion site is downstream the two functional domains, GH72 and Cys-box, that are essential for Gas1p functionality (Popolo et al., 2008). The expression of both mRFP-Gas1p and Gas1p-GFP was driven by the natural GAS1 promoter. The fusion genes were either integrated into the genome (mRFP-GAS1) or placed on episomic plasmids (GAS1-GFP).
Figure 1.
Fusion to fluorescent proteins results in a functional Gas1 protein. (A and C) Schematic diagrams of the domain organization and site of insertion of the fluorescent proteins. The gray box represents the signal peptide. GH72, signature domain of GH72 glycosylhydrolases; Cys, cysteine-enriched module; Ser, serine-rich region and GPI, signal for GPI attachment at N528. For the CF sensitivity assays, cells were grown to log phase in YPD at 30°C. Ten-fold dilutions, from 2 to 4, of the concentrated suspension 1 of W303-1A, a derived gas1Δ mutant (WAH) and JC9, a gas1Δ mutant harboring an integrated copy of mRFP-GAS1, were prepared and spotted on YPDA plates. After 48 h at 30°C the plates were photographed. The CF assay for the Gas1-GFP was performed as described in A by using cells grown in YPD at 30°C of the diploid AN120 strain, a derived homozygous gas1Δ mutant carrying YEp24-GAS1 (ER335), YEp24 (ER333), YEp24-GAS1-GFP (ER337), or pRS416-GAS1-GFP (ER336). (B and D) Morphology of Formalin-fixed cells.
To establish whether the fusion proteins were functional, we tested their capability to suppress the defective phenotype of gas1Δ cells (Popolo et al., 1997; Valdivieso et al., 2000). As shown in Figure 1, A and B, mRFP-Gas1p fully complemented the CF hypersensitivity of gas1Δ cells and also suppressed other phenotypic traits such as the round morphology, the larger cell size and defective bud maturation and cell separation, these last two traits being responsible for the appearance of “mickey mouse” cells and the clumpy phenotype of gas1Δ cells (Popolo et al., 1993b). Moreover the reduced growth rate (μ) of gas1Δ cells (0.24 ± 0.02 h−1) reverted to the value of the parental strain (0.66 ± 0.05 h−1), as the growth rate of the strain expressing mRFP-Gas1p was 0.65 ± 0.04 h−1. Thus mRFP-Gas1p fully suppressed the defective gas1Δ phenotype.
Plasmids harboring the GAS1-GFP gene were introduced into a gas1 diploid null mutant derived from AN120 (see Table 1). Transformed cells were propagated in selective SD plates but inoculated in YPD because Gas1p-GFP did not mature into the fully glycosylated form and was not fluorescent in cells growing in liquid SD (our unpublished data). In YPD, GAS1-GFP complemented the CF hypersensitivity of gas1 cells when placed either on single or multicopy plasmids similarly to the plasmid carrying the wild type GAS1 (Figure 1C). GAS1-GFP placed on single or multicopy plasmids partially suppressed the slow growth rate of gas1Δ. The growth rates were 0.46 ± 0.04 and 0.44 ± 0.04 h−1, respectively, compared with a value of 0.32 ± 0.06 for the gas1 null mutant transformed with the empty vector and 0.55 ± 0.02 h−1 for the parental AN120 strain. The clumpy phenotype, the round shape and the large size were greatly attenuated in strains expressing Gas1p-GFP (Figure 1D). The construct restored the ellipsoidal morphology more completely when placed on a single copy plasmid as shown by the presence of more elongated cells in strain ER336. In contrast, the defect of cell separation was better complemented by the multicopy plasmid.
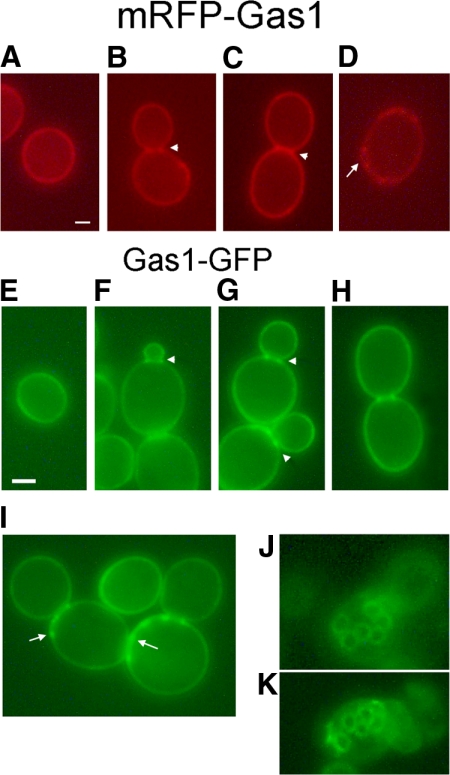
Gas1p Is Localized at the Cell Periphery, at the Mother-Daughter Neck Region, in the Septum and in Bud Scars
mRFP-Gas1p expressed in a gas1Δ mutant exhibited various locations. It decorated the cell periphery consistently, illustrating the plasma membrane localization of the protein (Figure 2). In addition, in medium-budded cells the fluorescence appeared as two bright dots at either side of the mother-daughter neck region and in large-budded cells clearly decorated the entire septum region (Figure 2, B and C). Interestingly, fluorescent rings or crater-like structures were observed, suggesting that mRFP-Gas1p was also localized in the bud scars (Figure 2D). In addition, we also detected bright crescent-shaped fluorescent segments at the edge of cells. These may represent lateral views of one or more adjacent bud scars (Figure 2D). When mRFP-Gas1p was expressed in the parental strain it was detected in perinuclear structures, in addition to the localization pattern previously described, suggesting a partial retention in the ER, probably due to competition in folding with endogenous Gas1p (data not shown).
Figure 2.
mRFP-Gas1 and Gas1p-GFP fluorescent proteins are associated to the plasma membrane, bud neck, septum and bud scars. (A–D) Cells of the haploid strain JC9, a gas1Δ expressing mRFP-Gas1p from a construct integrated into the genome, were grown in YPD at 30°C. Cells were fixed and washed with PBS, pH 7.4, before microscopy observation. Bar, 1 μm. (E–K) Cells of the diploid strain ER336 (a diploid gas1Δ mutant carrying the single copy plasmid pRS416-GAS1-GFP) were grown to log phase in YPD at 30°C. Cells were observed in PBS without fixation. Arrowheads, two dots aside the neck or septum. Arrows, bud scars. Bar, 3 μm.
Next, we analyzed the localization of Gas1p-GFP in a gas1Δ mutant. Gas1p-GFP localized to the plasma membrane (Figure 2E), in the mother-daughter neck (Figure 2, F–H), in the septum of large-budded cells (Figure 2H) and in bud scars (Figure 2, I–K). Similar results were also obtained for the Gas1p-GFP fusion expressed in the parental strain indicating that, for this hybrid protein, no competition with the endogenous protein occurs. The same pattern of localization was observed when the hybrid GAS1-GFP gene was placed either on single copy or multicopy plasmids. These data show that Gas1p maintains the same localization regardless of the type of fluorescent protein, site of fusion, genetic background or ploidy and gene dosage of the construct.
For a complete view of the various regions where Gas1p localizes, a stack acquisition was performed. Figure 3 shows surface and transversal sections of representative cells. In a small-budded cell, Gas1p-GFP is distributed in the cell periphery with a preferential location around the bud (Figure 3A). A bright fluorescent ring was visible in the surface sections supporting the inference that Gas1p-GFP was localized in a bud scar (Figure 3A, 1–3). In cross sections around the medial plane, the bright fluorescent structure present at the mother-daughter neck was visibly a ring (Figure 3A, 5–7). In the cross sections 6–8, a crater-like structure close to the neck was clearly visible. In the surface planes 1 and 2 and 8 and 9 of Figure 3A, a punctate pattern of very fine patches or dots was visible. Figure 3B shows a large-budded cell with a bright fluorescence in the septum area (Figure 3B, 2–4). In the medial plane Gas1p-GFP filled the inner part of the septum suggesting that the protein colocalizes with the chitin disk present at the primary septum (Figure 3B, 3 and 4). In surface sections the fluorescence was distributed in very fine dots all over the cell (image 1) and in discrete sites on the cell contour (images 1 and 2 and 7 and 8).
Figure 3.
Gas1-GFP protein has multiple locations in the single cell. Cells of strain ER337, a gas1Δ homozygous diploid mutant carrying the plasmid YEp24-GAS1-GFP were photographed through a fluorescence microscope equipped for stack acquisition. Optical sections of 400-nm intervals through individual cells are shown. (A) Small-budded cell, images 1–3: surface sections showing a ring and pattern of small dots all over the membrane. Images 4–7, in cross sections, the fluorescence forms a ring at the mother-daughter neck. Images 6–8, sections showing a bud scar next to the bud neck. (B) Large-budded cell, images 1 and 2 show the small dots pattern over the plasma membrane. Images 3–5, the fluorescence is inside the septum region. Images 6–8, surface sections showing a punctuate pattern in the membrane. (C) Magnification of surface sections of small-budded cells with a visible bud scar. Bar, 1 μm. (D and E) Gas1-GFP associates with the detergent-resistant membranes. Cell extracts were treated as described under Materials and Methods. Fractions (1–13) of the density gradient were collected from the top. (D) Immunoblot of fractions from ER366 strain, a gas1Δ mutant harboring pRS416-GAS1-GFP, obtained using anti-GFP monoclonal antibody. (E) Immunoblot from AN120 parental strain obtained using anti-Gas1p polyclonal serum.
Figure 3C shows two magnifications of representative images of surface sections. A punctate pattern of fine fluorescent dots is clearly visible. This pattern is consistent with the localization of the plasma membrane Gas1p-GFP in lipid rafts as indicated in previous biochemical reports (see Introduction). In Figure 3C, a fluorescent ring of Gas1p-GFP is also clearly visible. The brightness of the ring suggests that Gas1p is concentrated in these structures. In conclusion, these analyses demonstrate that Gas1p localizes in three districts: at discrete sites of the plasma membrane, in a ring at the mother-daughter neck and in the bud scars. In addition, these experiments suggest a fourth unexpected localization of Gas1p, in the primary septum.
To determine whether Gas1p-GFP associates with the lipid rafts, we used a biochemical approach to isolate the DRMs. A hallmark behavior of DRMs is their low-buoyant density after detergent extraction and centrifugation in iodixanol (e.g., Opti-Prep) discontinuous step gradients (Bagnat et al., 2000). Gas1p-GFP was recovered in fraction 2 at the top of an Opti-Prep gradient showing the same behavior of wild type Gas1p (Figure 3, D and E). The apparent molecular mass of Gas1p-GFP was ∼155 kDa in accordance with the size expected for a polypeptide constituted by Gas1p (∼125 kDa) and GFP (∼30 kDa). Another form of Gas1p-GFP (∼120–130 kDa) was abundant in fractions 7–9 of the gradient, suggesting an intracellular accumulation of immature forms (Figure 3D). These results extend the microscopy analysis and provide further support for the localization of Gas1p-GFP in the lipid rafts.
mRFP-Gas1p Is in Mobile or Immobile Pools Depending on Its Localization
In experiments originally designed to study Gas1p localization in conditions of α-F–induced polarized growth, we noticed that the mRFP-Gas1p fluorescence around the cell rapidly decreased after addition of α-F to the culture. In these studies MATa gas1Δ cells expressing mRFP-Gas1p were treated with α-F, and the effect on cell cycle progression was monitored by measuring the decrease of budding index and the appearance of shmoos (Figure 4A). 3.25 h after the addition of α-F, the red fluorescence signal at the cell contour was greatly reduced except at the bud scars and in the rare septa that were still fluorescent (Figure 4B). This suggested that mRFP-Gas1p in the membrane could be labile, whereas the association to the bud scar could protect it from degradation.
Figure 4.
mRFP-Gas1p is produced as an unstable 155-kDa polypeptide that is rapidly removed from cell contour in α-F–treated cells. (A) Cells from an exponentially growing culture were treated with 20 μg/ml α-F. A representative plot of the percentage of budded cells in W303-1A or JC9 strain is shown. (B) Microscopy analysis of mRFP-Gas1p. Cells were collected, fixed, and then washed with PBS. The fluorescence is present at the cell periphery, in the neck and bud scars as previously shown in Figure 2, but after 3.25 h of α-F only bud scars or septa are bright (arrows). Bud scars are on a focus plane that makes the cell fluorescence diffuse. Bar, 3 μm. (C) Immunoblot of total protein extracts (∼80 μg) from W303-1A and JC9 cells at different time from α-F addition. (D) Relative protein levels of Gas1p and mRFP-Gas1p determined by densitometric analysis of the bands of the immunoblot shown in C. Similar results were obtained in two independent experiments.
To further analyze this hypothesis, we determined the levels of the Gas1 and fusion proteins in total lysates at different time intervals after addition of α-F. As shown in the immunoblot of Figure 4C, Gas1p migrated as an ∼125-kDa polypeptide and mRFP-Gas1p as an ∼155-kDa band. After addition of α-F, the intensity of the 125-kDa band did not significantly change, whereas the 155-kDa band seemed to rapidly diminish being undetectable after 3.25 h of treatment. The level of Gas1 and mRFP-Gas1 polypeptides was normalized for the intensity of the actin band. As shown in Figure 4D, Gas1p remained approximately constant whereas mRFP-Gas1p levels decreased markedly during the treatment with α-F. Moreover, at time zero the steady-state level of mRFP-Gas1p was ∼50% the level of Gas1p, suggesting that mRFP-Gas1p is either less synthesized or degraded more rapidly than Gas1p in vegetative growth. Because GAS1 expression is turned off and its mRNA levels rapidly decrease during α-F treatment (Popolo et al., 1993a), these experiments point to an increased rate of degradation of mRFP-Gas1p compared with Gas1p. Therefore the reduction of mRFP-Gas1p in the cell contour observed in the microscopy analysis is more likely to be due to the removal of the protein from the plasma membrane caused by high turnover and to the absence of de novo synthesis.
We studied the localization of the fluorescent labeling that persisted during α-F treatment in more detail. We performed confocal microscopy analysis using Calcofluor to stain chitin (blue fluorescence) in cells expressing mRFP-Gas1p. During vegetative growth, mRFP-Gas1p was visible all over the cell with an increased concentration in the bud scars (Figure 5A). In cells treated for 3.25 h with α-F, the red fluorescence around the cell decreased but the signal colocalized with chitin in the bud scars remained intense (Figure 5A, bottom). These results indicate that mRFP-Gas1p undergoes a high turnover in the plasma membrane, whereas it is stabilized when localized to the bud scar. This particular behavior was specific to this fusion at the N-terminal end of Gas1p. We took advantage of it as a tool to further investigate the localization to the bud scar.
Figure 5.
mRFP-Gas1p colocalizes with chitin in the bud scars. (A) JC9 cells, carrying an integrated mRFP-GAS1 copy, were grown to log phase in YPD at 30°C and treated for 3.25 h with α-F. Cells were collected and analyzed without fixation with a confocal microscope. The images are the result of superimposed optical sections. Cells show CF staining of chitin (blue fluorescence) and mRFP-Gas1p (red fluorescence). In vegetative growth, mRFP-Gas1p was distributed at the cell periphery and also clearly colocalized with the bud scars as detected by the merging of the red and blue fluorescence, which produced a light pink color. In cells treated with α-F the red fluorescence in the periphery decreased but it was still very intense in the bud scars colocalizing with the chitin signal. Bar, 5 μm. (B) Flow chart of subcellular fractionation and cell wall treatments. (C) Immunoblot analysis with anti-Gas1p polyclonal antibodies of total protein extracts, soluble and membrane fractions from W303-1A and JC9 cells before (left) and after 3.25 of α-F treatment (right). Arrowhead: Gas1p; arrow: mRFP-Gas1p. A sharp band of about 100 kDa is a cross-reactive polypeptide. (D) Immunoblot of the cell wall fractions obtained and designated according to the scheme in C. Lanes 1 and 3, W303-1A cells; lanes 2 and 4, JC9 cells.
The Immobile Form of Gas1p Is Cross-Linked to the Cell Wall
We reasoned that if a fraction of mRFP-Gas1p was associated with the plasma membrane and another was covalently bound to the cell wall, in α-F–treated cells we should still detect mRFP-Gas1p bound to the cell wall fraction but not the fraction present in the plasma membrane. At time 0 and 3.25 h after α-F addition, different subcellular fractions and purified cell walls were obtained according to the scheme shown in Figure 5B. As shown in the immunoblot of Figure 5C, in untreated cells both Gas1p and mRFP-Gas1p were detected in the membrane fraction. After treatment with α-F, Gas1p was enriched in the membrane fraction whereas mRFP-Gas1 was not detectable in either the total or membrane fractions.
To determine whether Gas1 and mRFP-Gas1 proteins remain bound to the cell wall, we treated purified cell walls with a β(1,3)-glucanase preparation (laminarinase) or with pure exochitinase (fractions G and Exo, respectively, in Figure 5B). Before α-F addition, Gas1p and mRFPGas1p were readily released by both treatments (Figure 5D, lanes 1 and 2, G and Exo panels). This indicates that a pool of Gas1p is linked to β(1,3)-glucan and another to chitin. Similar results were obtained using Quantazyme, a recombinant β(1,3)-glucanase (data not shown). After treatment with α-F, both Gas1p and mRFP-Gas1p were still present in the β(1,3)-glucanase-released material, indicating that this pool is stably anchored to the cell wall (Figure 5D, lanes 3 and 4, G panel). Unexpectedly, neither protein was released by exochitinase (Figure 5D, lanes 3 and 4, Exo panel). We hypothesized that the pool bound to chitin could possibly be converted into an insoluble exochitinase-resistant form and new synthesis of the chitin-bound pool did not occur during α-F arrest. Because a CWP can be simultaneously linked to chitin and β(1,3)-glucan (Kollar et al., 1997), we treated the glucanase-resistant fraction with exochitinase and vice versa (fractions G-Exo and Exo-G in Figure 5B). The increased amount of Gas1p recovered in the G-Exo fraction (Figure 5D, lanes 3 and 4) supports the hypothesis that Gas1p linked to chitin could be further anchored to glucan in α-F–treated cells. However, because insoluble material still remained after the sequential digestions we cannot exclude that the chitin-bound Gas1p form, presumably present in the bud scars, could be resistant to the enzymatic extractions used.
These experiments indicate the existence of a new form of Gas1p linked to chitin. The complex is detected in proliferating cells but not in cell arrested in G1 by α-F treatment. Another fraction of Gas1p is linked to β(1,3)-glucan and extractable by β(1,3)-glucanase as reported previously (De Sampaio et al., 1999). This fraction is probably distributed all over the cell wall and corresponds to the faint signal detected by fluorescence microscopy after α-F arrest. Results supporting this hypothesis will be further described and discussed below (see Figure 9 and Discussion).
Figure 9.

Crh1 and Crh2 enzymes are required for the cross-linking of Gas1p at the chitin ring and bud scar. (A) Fluorescence microscopy of Gas1p-GFP in the strains ER374 (Crh+) and ER376 (crh1Δ crhΔ2). Gas1p-GFP is not present at the bud neck and bud scars. CF, calcofluor white staining of chitin. (B) Immunoblot of cell lysates with anti-Gas1p antibodies. Lane 1, BY4741; lane 2, crhlΔ crh2Δ (GRA007 strain); lane 3, crh1Δ crh2Δ gas1Δ. (C) Imunoblot of cell wall extracts with anti-Gaslp antibodies, G (laminarinase-extractable proteins) and Exo (exochitinase-extractable proteins). Lanes 1 and 3, BY4741; lanes 2 and 4, crh1Δ crh2Δ; lane 5, YPH499 and lane 6, chs3Δ mutant (ECY46-4-1B).
Chs3p-dependent Chitin Synthesis Mediates Gas1p-GFP Localization into the Chitin Ring
To study whether the anchoring of Gas1p to the chitin ring and bud scars requires chitin synthesis mediated by Chs3p—the enzyme responsible for the formation of the chitin ring—we examined the localization of Gas1p in a strain deleted for CHS3. As expected in chs3Δ cells, chitin ring and bud scars were absent, budding was abnormal, elongated bud necks were often observed and gave rise to protuberances after cell separation as previously described for chs3 null mutants (Shaw et al., 1991). In the chs3Δ mutant Gas1p-GFP was absent at both sides of the mother-daughter neck but was present all over the plasma membrane (Figure 6A, top). In addition, Gas1p-GFP was detected as a thin line inside the septum of binucleate cells where chitin synthesized by Chs2p forms the primary septum (Figure 6A, bottom). About one-third of the cells had an elongated neck region and the septum was curved as previously observed for chs3Δ mutants (Figure 6B; Shaw et al., 1991). In these cells, the line of the green fluorescence was also curved (Figure 6B). The same results were obtained when GAS1-GFP was on single or multicopy plasmids.
Figure 6.
Localization of Gas1p-GFP in chs3Δ mutant cells. (A) Top, a diploid cell of strain ER344 (chs3Δ) undergoing mitosis lacks Gas1p-GFP in the mother-daughter neck region. Bottom, a representative cell at citokinesis with Gas1p-GFP along the septum line. (B) Haploid cell of strain chs3Δ-GAS1-GFP shows a curved septum and an elongated neck. Gas1p-GFP is present at the septum. CF, calcofluor staining of chitin. Bar, 2.5 μm. (C and D) Confocal microscopy of wild-type and chs3Δ cells expressing Gas1p-GFP. The images are the result of superimposed optical sections. The merge of the green and blue fluorescence generates a light blue. (C) The wild-type diploid cell (strain ER342) shows a bud scar and a septum region. (D) A chs3Δ cell (strain ER344) shows a less brilliant septum region consistent with the lack of the chitin ring and Gas1p-GFP localization at the primary septum. The superimposition of CF fluorescence produced a double fluorescence at the cell contour which is due to the roundness of the cell. Bar, 10 μm.
To further refine the localization in the septum, Gas1p-GFP and chitin (blue fluorescence) were examined with the confocal microscope. In the wild-type cells, the septum was thick and very bright and the green and blue fluorescence overlapped (Figure 6C). The strong green signal resulted from the sum of the fluorescence of the primary septum and chitin ring. In the chs3 null mutant the septum seemed to be less bright with a uniform thickness and did not reach the cell contour in agreement with the lack of the chitin ring (Figure 6D). A merge of optical section series of the septum indicated that both chitin and Gas1p-GFP fluorescence form a disk in the septum (not shown in Figure 6). These results indicate that Gas1p is localized in the primary septum or in close proximity to it.
Localization of Gas1p-GFP in the chs2Δ and chs1Δ Mutants
To further explore the interaction of Gas1p with the chitin of the primary septum we determined the localization of Gas1p in a chs2Δ mutant. The lack of Chs2p is known to be compensated by the synthesis of a thick “remedial” septum that is amorphous and devoid of the trilaminar structure typical of a normal septum (Shaw et al., 1991). This septum is rich in chitin produced by Chs3p, which becomes essential for the survival of chs2Δ cells (Cabib and Schmidt, 2003). Remedial septa also contain cell wall material deposited in an orientation parallel to the mother-bud axis instead of perpendicular as occurs when the primary septum is present. As shown in Figure 7A, Gas1p-GFP exhibited a heterogeneous localization in a chs2Δ mutant strain. In cells which displayed an elongated septum, the fluorescence of Gas1p-GFP was faint, diffuse and distributed over the length of the septum. Gas1p-GFP was more abundant in thicker septa. Filaments with frayed ends were also detected as protruding from the bud scar (Figure 7A). This indicates that Gas1p-GFP accumulates in the remedial septa in aberrant arrangements probably as part of the cell wall material. Altogether, these observations led us to conclude that Gas1p-GFP localization in the septum is markedly disturbed in chs2Δ mutant cells. The effect could be due both to the absence of the deposition of chitin in the primary septum and to the synthesis of the remedial septum.
Figure 7.

Localization of Gas1p is perturbed in a chs2 null mutant. (A) CER5 (a chs2Δ mutant transformed with YEp24-GAS1-GFP) was analyzed during growth in YPD at 30°C. Short arrows indicate remedial septa with little Gas1p-GFP, arrowheads indicate thick remedial septa with bright Gas1p-GFP, the long arrow indicates an abnormal bud scar with filaments protruding and the thick arrows indicate necks with no Gas1p. (B and C) Cell lysates and proteins precipitated from the culture supernatants of the indicated strains, grown in YPD or in YPD buffered at pH 6.5, were analyzed by immunoblot with anti-Gas1p antibodies.
To further examine the interaction with chitin synthases, we analyzed the localization of Gas1p in a chs1Δ mutant but we could not detect any notable change in the major localization sites of Gas1p-GFP (Supplemental Figure S1).
Gas1p Is Released into the Medium in chs1Δ, chs2Δ, and chs3Δ Mutants
Our results suggest that Gas1p-GFP localization in the bud neck, bud scars, and septum is dependent on deposition of chitin. We reasoned that mutants deleted in genes encoding chitin synthases could release Gas1p into the growth medium. We monitored the presence of wild-type Gas1p in cell lysate and medium fraction from cell cultures of chs1Δ, chs2Δ, chs3Δ mutants and their isogenic strain. The results are shown in Figure 7B. Gas1p was present at similar levels in the cell lysates from the four strains. Interestingly, Gas1p was detected in the culture medium of chs1Δ, chs2Δ, and chs3Δ cells, whereas no appreciable amount of Gas1p was recovered from the parental strain. This indicates that the cross-linking of Gas1p to the cell wall depends on chitin synthesized by Chs2p (primary septum) or Chs3p (bud neck and scar) in accordance with the results shown in Figures 6 and 7A. Moreover the medium from the chs2Δ culture seemed to contain a higher amount of Gas1p compared with the other mutants.
Surprisingly, we also found Gas1p in the medium of a chs1Δ culture. It has been reported that chs1Δ daughter cells undergo lysis when the external pH becomes highly acidic, a condition that hyperactivates chitinase (Cabib et al., 1989). Therefore, we grew the cells both in YPD, where the pH drops during growth, and in YPD buffered at pH 6.5. At pH 6.5, Gas1p was still released into the medium although at a lower level with respect to cells grown in unbuffered YPD (Figure 7C). Thus, we can conclude that chitin synthases or their products are required for the binding of Gas1p to the chitin ring (bud scar), septum and also to repair chitin.
Conversion of Gas1p into a Transmembrane Protein Prevents Localization of Gas1p to the Chitin Ring, Bud Scars, and Primary Septum and Affects Cell Separation
To determine whether cross-linking at the chitin ring and septum depends on GPI, we replaced the portion encoding the C-terminal region of Gas1p with the transmembrane (TM) domain and cytosolic tail of Mid2p (see scheme in Figure 8A). The C-terminal portion of Gas1p included the Ser-rich region and the GPI-attachment signal. In this way, Gas1p was converted into a type I TM protein. Mid2p has a uniform distribution in the plasma membrane and acts as a sensor of cell wall stress, but it is not functional when devoid of the extracellular portion (Straede and Heinisch, 2007). The chimera mRFP-Gas1-Mid2p was detected as an ∼140-kDa polypeptide (data not shown). The growth rate (μ; h−1) of the W303-1A, gas1Δ strains and the gas1Δ strain expressing the mRFP-Gas1p or the mRFP-Gas1-Mid2 protein were 0.66, 0.24, 0.66, and 0.55, respectively, indicating that the chimera suppressed the slow-growth rate defect of the mutant although not as efficiently as mRFP-Gas1p. In cells expressing the chimera, we noticed an incomplete suppression of the morphological defects of gas1Δ cells. Approximately 15% of the cells remained attached in two, three or four cells groups, a phenotype not observed for cells expressing the mRFP-Gas1 full-length protein.
Figure 8.
Conversion to a TM-form excludes Gas1p from the septum region. (A) Scheme of the fusion of the mRFPGas1-Mid2. (B) Representative JC10 cell (left) showing mRFP-Gas1-Mid2p in the plasma membrane (arrowhead) and chitin (blue) in the chitin ring. A group of three unseparated cells (right) shows the gap between membranes (arrows). Bar, 5 μm. (C) Confocal microscopy analysis shows in more detail the dark thin line which divides the two cells. The intracellular red fluorescence is due to partial retention of the hybrid protein in the ER. Bar, 10 μm. (D) Flow cytometric analysis of the DNA content per cell of the indicated strains. In the histogram the percentage of cells in the various compartments is shown. (E) CF sensitivity assay of the indicated strains.
The hybrid protein was localized at the cell periphery demonstrating that it is transported to the cell surface (Figure 8B). In some cells, an accumulation of the hybrid Gas1p close to the bud neck was also visible but we have not been able to detect any localization in the bud scar indicating that the apparent accumulation could be due to the curvature of the plasma membrane (Figure 8B, left). Interestingly, in groups of two or more unseparated cells, the red fluorescence labeled the cell contour but a thin gap was present in the middle of the septum region (Figure 8B, right). To further confirm these observations we performed confocal microscopy analyses that clearly showed the presence of the protein in the plasma membrane and its absence at the chitin ring of undivided cells (Figure 8C). Because the localization of Gas1p at the chitin ring was missing we could also confirm the lack of Gas1p in the primary septum underneath. These results reveal that cross-linking of Gas1p to the entire septum region relies on the presence of GPI.
We further monitored the effects of mRFP-Gas1-Mid2p on cell separation. We measured the fraction of cells with a genomic DNA content of 1, between 1 and 2, 2, and more than 2 by flow cytometric analysis Cells that are not completely separated are counted as a single unit in this analysis. The profiles of the DNA content per cell are shown in Figure 8D. The percentage of cells with a DNA content >2C was ∼5.5 in cells expressing mRFP-Gas1p and increased to 32.4 in the gas1 mutant, whereas it was ∼13 in cells expressing mRFP-Gas1-Mid2p. Thus, the TM form of Gas1p only partially relieves the cell separation defect of gas1Δ cells. In conclusion, both Gas1p forms, the one cross-linked to the septum region and the plasma-membrane GPI-anchored form, concur in facilitating cell separation.
We further characterized the effect of the chimera on the ability to suppress the phenotypic defects of gas1Δ cells. As shown in Figure 8E, the protein suppressed the CF hypersensitivity of gas1Δ cells, indicating that the hybrid protein was functional.
Loss of GAS1 Affects the Width of the Neck Region
To explore the role of Gas1p localization in the chitin ring, we investigated whether the lack of Gas1p could affect the size of the neck region. The neck region is a crucial constriction between the mother and daughter cells. It has been shown that the width of the neck region normally remains constant throughout the cell cycle (Cabib, 2004). The diameters of the mother-daughter neck, expressed as mean (μm) ± SD, in the parental strain W303-1A, gas1Δ cells and strain JC10, expressing mRFP-Gas1-Mid2p, were ∼0.96 ± 0.16 (n = 15), 1.67 ± 0.29 (n = 20), and 1.45 ± 0.19 (n = 20), respectively. The enlargement of the bud neck suggests that the loss of Gas1p affects the rigidity of this region and that the cell wall-bound Gas1p contributes to this property.
Crh1 and Crh2 Transglycosylases Are Required for Immobilizing Gas1p into the Chitin Ring and Bud Scars
Crh1 and Crh2 enzymes have been proposed to assist the immobilization of specific GPI-CWPs to the bud scar through the binding of β(1,6)glucan to chitin (Cabib et al., 2008). Therefore, we analyzed the localization of Gas1p-GFP in the CRH1, CRH2 single and double null mutants. The isogenic strain showed the normal localization of Gas1p (Figure 9A, top). In the single crh1Δ and crh2Δ mutants the localization of Gas1p was similar to the wild type with the exception that the fluorescent signal in the bud scars was weaker in the crh2 single mutant (data not shown). Interestingly, in the double null mutant the fluorescence around the cell was intense whereas the green signal at the bud neck and bud scars was missing (Figure 9A, bottom). CF staining confirmed the presence of normal bud scars. This result indicates that Crh1 and Crh2 proteins are required for the immobilization of Gas1p-GFP to the chitin in the neck region and in the bud scars.
We also analyzed total lysates and cell wall fractions from exponentially growing cells. In a total lysate of the crh1Δ crh2Δ mutant, Gas1p was present at a level equivalent to the level of the parental strain (Figure 9B). The purified cell walls were treated with β(1,3)-glucanase (laminarinase) or pure exochitinase. The results are shown in Figure 9C. Gas1p was present in the glucanase-extractable material both in the parental strain and crh1Δ crh2Δ mutant indicating that the fraction of Gas1p readily extractable by β(1,3)-glucanase is not affected by the double deletion and, on the contrary, it seemed slightly increased (Figure 9C, lanes 1 and 2). Interestingly, Gas1p was released by exochitinase from the cells walls of the parental strain but not from the double mutant (Figure 9C, lanes 3 and 4). This experiment indicates that a putative GPI-Gas1p-β(1,6)-glucan-chitin complex is not produced in the absence of the Crh1and Crh2 proteins in agreement with the role of these enzymes in transferring chitin onto β(1,6)-glucan.
We hypothesized that the GPI-Gas1p-β(1,6)-glucan-chitin complex should not be extracted by exochitinase from chs3Δ cell walls. The digestion with exochitinase released Gas1p from the cell walls of the parental strain but not from the mutant (Figure 9C, lanes 5 and 6). Thus, both the lack of the cross-linking Crh enzymes or of the donor molecule abolishes the formation of the complex. As the Gas1p form released by exochitinase has an electrophoretic mobility very similar to that of the GPI-anchored Gas1p, the β(1,6)-glucan chain attached to Gas1p is likely to be very short.
As shown in Figure 9C (lanes 1 and 2), the electrophoretic mobility of the β(1,3)-glucanase-released form of Gas1p from BY4741 strain was heterogeneous (from 130 to >150 kDa). This could be consistent with the protein being covalently linked to β(1,3)-glucans with different accessibility to the enzymatic cleavage, to protein mannosylation or also with the presence of soluble chitin chains of different size. The digestion of the β(1,3)-glucanase-extractable material by exochitinase did not affect the electrophoretic mobility of Gas1p suggesting that soluble chitin chains are not linked to Gas1p unless they are very short (data not shown).
DISCUSSION
Localization of Fluorescent Versions of Gas1p in Yeast Cells
The results reported in this work indicate that Gas1p has four distinct localizations in vegetative growing cells: it is present at the cell periphery, in the chitin ring, at the primary septum and in the bud scars where it remains for several generations (Figure 11). At the plasma membrane, we could visualize Gas1p-GFP in microdomains and then confirm biochemically that the fusion protein is recovered in the lipid rafts fraction. This result is consistent with early biochemical studies that reported the presence of Gas1p in plasma membrane derived DRMs (Bagnat et al., 2000). The localization of Gas1p here described also provides a possible explanation of the static behavior of a truncated derivative of Gas1p fused to GFP used as a marker in studies of lipid rafts polarization (Bagnat and Simons, 2002; Valdez-Taubas and Pelham, 2003).
Figure 10.
Scheme summarizing the localization of Gas1p in vegetative cells. (A) In a new born cell Gas1p is in microdomains of the plasma membrane. (B) In a small-budded cell, Gas1p is in the plasma membrane but is also linked to the chitin ring presumably through a transglycoylase reaction. (C) In large-budded cells Gas1p is localized to or in close proximity to the primary septum. (D) After mother-daughter separation, Gas1p remains in the bud scar and distributed around the plasma membrane. Another pool of Gas1p probably dispersed all over the cell wall is not indicated. CR, chitin ring; PS, primary septum; BS, bud scar.
In a mass spectrometry analysis of the plasma-membrane DRMs, Gas1p, Gas3, Gas5p, and other enzymes involved in cell wall biogenesis, in particular Fks1p/Fks2p, Rho1p, and the GPI-mannoproteins Ecm33p and Crh2p, were identified (Aronova et al., 2007). This reinforces the notion that lipid rafts contribute to segregation and sorting membrane districts into which proteins specifically aggregate. A physical association of Gas1p with Fks1p/Fks2p and their regulatory subunit Rho1p could be of functional significance given that these enzymes could cooperate in the synthesis, extrusion and incorporation of β(1,3)-glucans into the expanding cell wall.
Our results confirm and extend previous reports regarding the existence of a cell wall form of Gas1p (De Sampaio et al., 1999; Yin et al., 2005). We showed that this is not simply a consequence of some leakiness in the mechanism of plasma membrane retention but a specific destination of the protein. A quantitative analysis also revealed that ∼6 × 103 copies of Gas1p per cell are cell wall bound (Yin et al., 2007). This corresponds to ∼20% of the total Gas1p typically present in a growing yeast cell (Popolo, unpublished data). Thus, the majority of the protein is localized in the plasma membrane but molecular mechanisms must exist to regulate the incorporation of a fraction of Gas1p molecules into the cell wall.
Mechanisms of Incorporation of Gas1 in Specific Sites of the Cell Wall
Evidence that cell wall anchoring of GPI proteins can occur in a selective manner was reported for three structural GPI-mannoproteins, Tip1, Cwp1 and Cwp2 (Smits et al., 2006). Cwp1p is localized at the birth scar whereas Tip1p and Cwp2p are incorporated in the mother cells and in small- to medium-sized buds, respectively. The localization of these proteins was shown to be completely determined by the timing of their transcription during the cell cycle (Smits et al., 2006). The synthesis of Gas1p is maximal at the G1-to-S transition, which is the moment of chitin ring formation at bud emergence (Popolo et al., 1993a; Ram et al., 1995). This pattern of expression could also be crucial for the incorporation of Gas1p into the cell wall but other mechanisms cannot be excluded. In fact Crh2p, which is synthesized throughout the cell cycle, is incorporated in a ring at the base of the bud by a mechanism that depends on actin cytoskeleton and septin ring organization (Rodriguez-Pena et al., 2002).
The analyses of purified cell walls from αF-arrested, crh1Δ crh2Δ and chs3Δ cells suggest that Gas1p is present in two pools in growing cells (Supplemental Figure S2). Pool A is represented by a GPI-Gas1p-β(1,6)-glucan complex that is linked to β(1,3)-glucan. This complex is readily extractable by treatment with β(1,3)-glucanase, usually yielding a diffuse band. Moreover this complex is present also in cells with deletions of CRH or CHS3 genes. Pool A is likely to be dispersed all over the cell wall. Pool B is represented by a GPI-Gas1p-β(1,6)-glucan-chitin complex as it is readily released by exochitinase treatment. To identify this new module, we used a pure exochitinase, as the commercial preparation contains proteases that degraded Gas1p. This complex is detected only in cells undergoing the budding cycle. The formation of such a complex requires Chs3-dependent chitin synthesis and Crh enzymes that probably capture the chitin chain during its polymerization and attach it to the β(1,3)-glucan side branch of β(1,6)-glucan linked to Gas1p (Supplemental Figure S2). Based on previous reports this side chain contains one to three glucose residues and is too short to be attacked by β(1,3)-glucanases (Kollar et al., 1997). Pool B is further linked to the main β(1,3)-glucan through the reducing end of the β(1,6)-glucan, yielding a form of Gas1p that is simultaneously linked to chitin and β(1,3)-glucan. Our data also suggest that in the bud scars a Pool B-derived structure could be present and be highly resistant to enzymatic extraction. This could be in accordance with the fact that bud scar is an area of the cell wall which remains after the action of hydrolytic enzymes (chitinase and glucanases) involved in cell separation and shows an exceptional stability over time.
Our work provides new evidence on in vivo substrates for Crh1 and Crh2 enzymes. The lack of linkage of Gas1p to the bud neck/bud scars in the crh1 crh2 null mutant together with the finding that Gas1p was not present in the material released from the cell wall by exochitinase support the notion that these enzymes transfer chitin to the β(1,6)-glucan linked to Gas1p. Thus, Gas1p is the first substrate of Crh1-Crh2 enzymes to be identified.
The lack of Gas1p localization to the chitin ring and its release in the medium in chs3 null mutants suggests that chitin is necessary for the attachment of the GPI-Gas1p-β(1,6)-glucan complex to the cell wall. As the Gas1p released into the medium in chs3 cells and also the Gas1p released by exochitinase in wild-type cells have an apparent molecular weight similar to that of GPI-containing Gas1p, we presume that the β(1,6)-glucan is very short. Chemical analysis of the linkage region between Gas1p and β(1,6)-glucan will be required to confirm the connection with the cell wall polysaccharides and characterize the structure of these interconnections.
The chitin made by Chs2p in the septum was shown to be free (Cabib and Duran, 2005). This is in apparent contrast with the Gas1p localization in the primary septum. Although we showed that the Gas1p colocalizes with the chitin of the primary septum, we cannot exclude the possibility that Gas1p is in close proximity to the chitin disk rather than being directly cross-linked to it. Gas1p could be part of the outermost mannoprotein layer of the secondary septa that are in strict contact with the chitin disk. The severe disturbance in the architecture of the septum in the chs2 null mutant could be responsible of the release of Gas1p into the medium. Immunoelectron microscopy will be an invaluable technique to investigate the localization of Gas1p in the primary and remedial septa in more detail.
As mentioned in the Introduction, Dfg5p and Dcw1p constitute an essential pair of putative mannosidases and candidates as catalysts of the transfer of the GPI-protein to the β(1,6)-glucan (Kitagaki et al., 2002). We found that Gas1p showed the usual locations in single dfg5Δ or dcw1Δ mutants, indicating that Dfg5p and Dcw1p are not required for the binding of Gas1p to the cell wall (Supplemental Figure S1). However these proteins could contribute to a common function and compensate for each other's absence.
Functional Significance of the Localization of Gas1p in Specific Sites of the Cell Wall
We attempted to elucidate the role of Gas1p at the chitin ring, a special structure in which growth is prevented and resistance must be high to drive the growth into the bud. Several hypotheses can be formulated for the role of Gas1p tethered at the neck region. Gas1p could assume an inactive conformation and prevent incorporation of new β(1,3)-glucan chains in the neck area. It should also be noted that Gas1p is highly glycosylated and contains seven intramolecular disulfide bonds which confer a high stability (Popolo et al., 2008). Thus, Gas1p could have a structural role and contribute to the resistance of the neck region and also act as a permeability barrier. Another attractive hypothesis is that Gas1p activity is required for a local remodeling of β(1,3)glucan or for promoting a loosening of the fibers to facilitate, at a later stage, the action of β(1,3)-glucanases, such as Eng1p (Baladron et al., 2002).
The availability of a TM-form of Gas1p provided a means to study the functional significance of Gas1p localization in the septum region. By expressing the mRFP-GAS1-MID2 fusion we could uncouple the functions of GPI-anchored and cell wall-bound Gas1p. We could determine that both forms contribute to bud neck size and facilitate cell separation in agreement with previous observations on the severe defects of gas1Δ cells in cell separation (Popolo et al., 1993b). As mentioned above, Gas1p is localized at—or in proximity to—the primary septum. Gas1p could be specifically targeted to the cell wall of this region to remodel β(1,3)glucan chains in preparation to cell separation.
Finally, it has been shown that gas1 null diploid mutants exhibit a random budding pattern (Ni and Snyder, 2001). It is still unclear whether Gas1p in the chitin ring could play an additional role in the marking of the previous budding site. It is also conceivable that the round shape of gas1 cells could affect the cytoskeleton and be indirectly responsible for the altered budding pattern.
Supplementary Material
ACKNOWLEDGMENTS
We thank Enrico Cabib, Vladimir Farkas and Javier Arroyo for helpful discussion and strains, Carlos Vazquez de Aldana and Maria de Medina Redondo for critical reading of the manuscript, Roberto Cavatorta for the preparation of the figures, and David Horner for English revision. We thank also Alessandro Nespoli for the assistance in flow cytometry. We are grateful to the National Institute of Advanced Industrial Science and Technology of Japan and Prof. Yoshifumi Jigami for the kind gift of pMF608. This work was partially supported by the European RTN project 512481 “CanTrain” and by the national P.R.I.N. grant 2005 (to L. P.). E. R. is a recipient of a fellowship from Fondo Sociale Europeo 415438. J. C. is a recipient of a Marie Curie contract in the frame of the Cantrain project.
Abbreviations used:
- α-F
α-factor
- CF
calcofluor white.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-11-1155) on September 30, 2009.
REFERENCES
- Aronova S., Wedaman K., Anderson S., Yates J., 3rd, Powers T. Probing the membrane environment of the TOR kinases reveals functional interactions between TORC1, actin, and membrane trafficking in Saccharomyces cerevisiae. Mol. Biol. Cell. 2007;18:2779–2794. doi: 10.1091/mbc.E07-03-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnat M., Keranen S., Shevchenko A., Simons K. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc. Natl. Acad. Sci. USA. 2000;97:3254–3259. doi: 10.1073/pnas.060034697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnat M., Simons K. Lipid rafts in protein sorting and cell polarity in budding yeast Saccharomyces cerevisiae. Biol. Chem. 2002;383:1475–1480. doi: 10.1515/BC.2002.169. [DOI] [PubMed] [Google Scholar]
- Baladron V., Ufano S., Duenas E., Martin-Cuadrado A. B., del Rey F., Vazquez de Aldana C. R. Eng1p, an endo-1,3-beta-glucanase localized at the daughter side of the septum, is involved in cell separation in Saccharomyces cerevisiae. Eukaryot. Cell. 2002;1:774–786. doi: 10.1128/EC.1.5.774-786.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabib E. The septation apparatus, a chitin-requiring machine in budding yeast. Arch. Biochem. Biophys. 2004;426:201–207. doi: 10.1016/j.abb.2004.02.030. [DOI] [PubMed] [Google Scholar]
- Cabib E., Blanco N., Grau C., Rodriguez-Pena J. M., Arroyo J. Crh1p and Crh2p are required for the cross-linking of chitin to beta(1-6)glucan in the Saccharomyces cerevisiae cell wall. Mol. Microbiol. 2007;63:921–935. doi: 10.1111/j.1365-2958.2006.05565.x. [DOI] [PubMed] [Google Scholar]
- Cabib E., Duran A. Synthase III-dependent chitin is bound to different acceptors depending on location on the cell wall of budding yeast. J. Biol. Chem. 2005;280:9170–9179. doi: 10.1074/jbc.M414005200. [DOI] [PubMed] [Google Scholar]
- Cabib E., Farkas V., Kosik O., Blanco N., Arroyo J., McPhie P. Assembly of the yeast cell wall. Crh1p and Crh2p act as transglycosylases in vivo and in vitro. J. Biol. Chem. 2008;283:29859–29872. doi: 10.1074/jbc.M804274200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabib E., Sburlati A., Bowers B., Silverman S. J. Chitin synthase 1, an auxiliary enzyme for chitin synthesis in Saccharomyces cerevisiae. J. Cell Biol. 1989;108:1665–1672. doi: 10.1083/jcb.108.5.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabib E., Schmidt M. Chitin synthase III activity, but not the chitin ring, is required for remedial septa formation in budding yeast. FEMS Microbiol. Lett. 2003;224:299–305. doi: 10.1016/S0378-1097(03)00477-4. [DOI] [PubMed] [Google Scholar]
- Carotti C., Ragni E., Palomares O., Fontane T., Tedeschi G., Rodriguez R., Latgé J. P., Vai M., Popolo L. Characterization of recombinant forms of the yeast Gas protein and identification of residues essential for glucanosyltransferase activity and folding. Eur. J. Biochem. 2004;271:3635–3645. doi: 10.1111/j.1432-1033.2004.04297.x. [DOI] [PubMed] [Google Scholar]
- de Groot P. W., de Boer A. D., Cunningham J., Dekker H. L., de Jong L., Hellingwerf K. J., de Koster C., Klis F. M. Proteomic analysis of Candida albicans cell walls reveals covalently bound carbohydrate-active enzymes and adhesins. Eukaryot. Cell. 2004;3:955–965. doi: 10.1128/EC.3.4.955-965.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Sampaio G., Bourdineaud J. P., Lauquin G. J. A constitutive role for GPI anchors in Saccharomyces cerevisiae: cell wall targeting. Mol. Microbiol. 1999;34:247–256. doi: 10.1046/j.1365-2958.1999.01585.x. [DOI] [PubMed] [Google Scholar]
- Fujita M., Yoko O. T., Jigami Y. Inositol deacylation by Bst1p is required for the quality control of glycosylphosphatidylinositol-anchored proteins. Mol. Biol. Cell. 2006;17:834–850. doi: 10.1091/mbc.E05-05-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti E., Popolo L., Vai M., Rota N., Alberghina L. O-linked oligosaccharides in yeast glycosyl phosphatidylinositol-anchored protein gp115 are clustered in a serine-rich region not essential for its function. J. Biol. Chem. 1994;269:19695–19700. [PubMed] [Google Scholar]
- Kitagaki H., Wu H., Shimoi H., Ito K. Two homologous genes, DCW1 (YKL046c) and DFG5, are essential for cell growth and encode glycosylphosphatidylinositol (GPI)-anchored membrane proteins required for cell wall biogenesis in Saccharomyces cerevisiae. Mol. Microbiol. 2002;46:1011–1022. doi: 10.1046/j.1365-2958.2002.03244.x. [DOI] [PubMed] [Google Scholar]
- Klis F. M., Boorsma A., De Groot P. W. Cell wall construction in Saccharomyces cerevisiae. Yeast. 2006;23:185–202. doi: 10.1002/yea.1349. [DOI] [PubMed] [Google Scholar]
- Kollar R., Petrakova E., Ashwell G., Robbins P. W., Cabib E. Architecture of the yeast cell wall. The linkage between chitin and beta(1->3)-glucan. J. Biol. Chem. 1995;270:1170–1178. doi: 10.1074/jbc.270.3.1170. [DOI] [PubMed] [Google Scholar]
- Kollar R., Reinhold B. B., Petrakova E., Yeh H. J., Ashwell G., Drgonova J., Kapteyn J. C., Klis F. M., Cabib E. Architecture of the yeast cell wall. Beta(1->6)-glucan interconnects mannoprotein, beta(1->)3-glucan, and chitin. J. Biol. Chem. 1997;272:17762–17775. doi: 10.1074/jbc.272.28.17762. [DOI] [PubMed] [Google Scholar]
- Lesage G., Bussey H. Cell wall assembly in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2006;70:317–343. doi: 10.1128/MMBR.00038-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage G., Sdicu A. M., Menard P., Shapiro J., Hussein S., Bussey H. Analysis of beta-1,3-glucan assembly in Saccharomyces cerevisiae using a synthetic interaction network and altered sensitivity to caspofungin. Genetics. 2004;167:35–49. doi: 10.1534/genetics.167.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinska K., Malinsky J., Opekarova M., Tanner W. Distribution of Can1p into stable domains reflects lateral protein segregation within the plasma membrane of living S. cerevisiae cells. J. Cell Sci. 2004;117:6031–6041. doi: 10.1242/jcs.01493. [DOI] [PubMed] [Google Scholar]
- Montijn R. C., Vink E., Muller W. H., Verkleij A. J., Van Den Ende H., Henrissat B., Klis F. M. Localization of synthesis of beta1,6-glucan in Saccharomyces cerevisiae. J. Bacteriol. 1999;181:7414–7420. doi: 10.1128/jb.181.24.7414-7420.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouyna I., Fontaine T., Vai M., Monod M., Fonzi W. A., Diaquin M., Popolo L., Hartland R. P., Latge J. P. Glycosylphosphatidylinositol-anchored glucanosyltransferases play an active role in the biosynthesis of the fungal cell wall. J. Biol. Chem. 2000;275:14882–14889. doi: 10.1074/jbc.275.20.14882. [DOI] [PubMed] [Google Scholar]
- Ni L., Snyder M. A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae. Mol. Biol. Cell. 2001;12:2147–2170. doi: 10.1091/mbc.12.7.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popolo L., Cavadini P., Vai M., Alberghina L. Transcript accumulation of the GGP1 gene, encoding a yeast GPI-anchored glycoprotein, is inhibited during arrest in the G1 phase and during sporulation. Curr. Genet. 1993a;24:382–387. doi: 10.1007/BF00351845. [DOI] [PubMed] [Google Scholar]
- Popolo L., Gilardelli D., Bonfante P., Vai M. Increase in chitin as an essential response to defects in assembly of cell wall polymers in the ggp1delta mutant of Saccharomyces cerevisiae. J. Bacteriol. 1997;179:463–469. doi: 10.1128/jb.179.2.463-469.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popolo L., Ragni E., Carotti C., Palomares O., Aardema R., Back J. W., Dekker H. L., de Koning L. J., de Jong L., de Koster C. G. Disulfide bond structure and domain organization of yeast β(1,3)-glucanosyltransferases involved in cell wall biogenesis. J. Biol. Chem. 2008;283:18553–18565. doi: 10.1074/jbc.M801562200. [DOI] [PubMed] [Google Scholar]
- Popolo L., Vai M. The Gas1 glycoprotein, a putative wall polymer cross-linker. Biochim. Biophys. Acta. 1999;1426:385–400. doi: 10.1016/s0304-4165(98)00138-x. [DOI] [PubMed] [Google Scholar]
- Popolo L., Vai M., Gatti E., Porello S., Bonfante P., Balestrini R., Alberghina L. Physiological analysis of mutants indicates involvement of the Saccharomyces cerevisiae GPI-anchored protein gp115 in morphogenesis and cell separation. J. Bacteriol. 1993b;175:1879–1885. doi: 10.1128/jb.175.7.1879-1885.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primig M., Williams R. M., Winzeler E. A., Tevzadze G. G., Conway A. R., Hwang S. Y., Davis R. W., Esposito R. E. The core meiotic transcriptome in budding yeasts. Nat. Genet. 2000;26:415–423. doi: 10.1038/82539. [DOI] [PubMed] [Google Scholar]
- Ragni E., Coluccio A., Rolli E., Rodriguez-Pena J. M., Colasante G., Arroyo J., Neiman A. M., Popolo L. GAS2 and GAS4, a pair of developmentally regulated genes required for spore wall assembly in Saccharomyces cerevisiae. Eukaryot. Cell. 2007a;6:302–316. doi: 10.1128/EC.00321-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragni E., Fontaine T., Gissi C., Latge J. P., Popolo L. The Gas family of proteins of Saccharomyces cerevisiae: characterization and evolutionary analysis. Yeast. 2007b;24:297–308. doi: 10.1002/yea.1473. [DOI] [PubMed] [Google Scholar]
- Ram A. F., Brekelmans S. S., Oehlen L. J., Klis F. M. Identification of two cell cycle regulated genes affecting the beta 1,3-glucan content of cell walls in Saccharomyces cerevisiae. FEBS Lett. 1995;358:165–170. doi: 10.1016/0014-5793(94)01418-z. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Pena J. M., Rodriguez C., Alvarez A., Nombela C., Arroyo J. Mechanisms for targeting of the Saccharomyces cerevisiae GPI-anchored cell wall protein Crh2p to polarised growth sites. J. Cell Sci. 2002;115:2549–2558. doi: 10.1242/jcs.115.12.2549. [DOI] [PubMed] [Google Scholar]
- Schmidt M., Varma A., Drgon T., Bowers B., Cabib E. Septins, under Cla4p regulation, and the chitin ring are required for neck integrity in budding yeast. Mol. Biol. Cell. 2003;14:2128–2141. doi: 10.1091/mbc.E02-08-0547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J. A., Mol P. C., Bowers B., Silverman S. J., Valdivieso M. H., Duran A., Cabib E. The function of chitin synthases 2 and 3 in the Saccharomyces cerevisiae cell cycle. J. Cell Biol. 1991;114:111–123. doi: 10.1083/jcb.114.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K., Vaz W. L. Model systems, lipid rafts, and cell membranes. Annu. Rev. Biophys. Biomol. Struct. 2004;33:269–295. doi: 10.1146/annurev.biophys.32.110601.141803. [DOI] [PubMed] [Google Scholar]
- Smits G. J., Schenkman L. R., Brul S., Pringle J. R., Klis F. M. Role of cell cycle-regulated expression in the localized incorporation of cell wall proteins in yeast. Mol. Biol. Cell. 2006;17:3267–3280. doi: 10.1091/mbc.E05-08-0738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straede A., Heinisch J. J. Functional analyses of the extra- and intracellular domains of the yeast cell wall integrity sensors Mid2 and Wsc1. FEBS Lett. 2007;581:4495–4500. doi: 10.1016/j.febslet.2007.08.027. [DOI] [PubMed] [Google Scholar]
- Vai M., Gatti E., Lacana E., Popolo L., Alberghina L. Isolation and deduced amino acid sequence of the gene encoding gp115, a yeast glycophospholipid-anchored protein containing a serine-rich region. J. Biol. Chem. 1991;266:12242–12248. [PubMed] [Google Scholar]
- Valdez-Taubas J., Pelham H. R. Slow diffusion of proteins in the yeast plasma membrane allows polarity to be maintained by endocytic cycling. Curr. Biol. 2003;13:1636–1640. doi: 10.1016/j.cub.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Valdivieso M. H., Ferrario L., Vai M., Duran A., Popolo L. Chitin synthesis in a gas1 mutant of Saccharomyces cerevisiae. J. Bacteriol. 2000;182:4752–4757. doi: 10.1128/jb.182.17.4752-4757.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warenda A. J., Konopka J. B. Septin function in Candida albicans morphogenesis. Mol. Biol. Cell. 2002;13:2732–2746. doi: 10.1091/mbc.E02-01-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q. Y., de Groot P. W., de Jong L., Klis F. M., De Koster C. G. Mass spectrometric quantitation of covalently bound cell wall proteins in Saccharomyces cerevisiae. FEMS Yeast Res. 2007;7:887–896. doi: 10.1111/j.1567-1364.2007.00272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q. Y., de Groot P. W., Dekker H. L., de Jong L., Klis F. M., de Koster C. G. Comprehensive proteomic analysis of Saccharomyces cerevisiae cell walls: identification of proteins covalently attached via glycosylphosphatidylinositol remnants or mild alkali-sensitive linkages. J. Biol. Chem. 2005;280:20894–20901. doi: 10.1074/jbc.M500334200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.