Abstract
A single nucleotide polymorphism (SNP) rs6983267, located within the 8q24 region, is strongly associated with risk of colorectal and prostate cancer. It has been suggested that the mechanism of this association is related to differential interaction of TCF7L2 protein (previously known as TCF-4) with alleles of rs6983267, influencing the expression of a well-known oncogene, MYC, located 335 Kb telomeric. Here, we tested the correlation between mRNA expression of MYC and several alternatively spliced forms of TCF7L2 in 117 non-cancer colon samples. We observed a strong correlation (r = 0.60, p < 10-6) between expression of MYC and a unique splicing form of TCF7L2. The level of MYC expression in these samples was associated with expression of some TCF7L2 splicing forms but not with genotypes of rs6983267, or interaction of rs6983267 with TCF7L2 expression. These findings suggest that some splicing forms of TCF7L2 may be functionally important for regulation of MYC expression in colon tissue but this regulation is not directly dependent on rs6983267.
Findings
Recent genome-wide association studies (GWAS) have identified a single nucleotide polymorphism (SNP) rs6983267 within the 8q24 region associated with increased susceptibility to colorectal and prostate cancer [1-6]. Follow-up association studies have suggested that the same variant may also increase the risk for cancers of the kidney, thyroid and larynx [7,8]. The location of rs6983267 in the intergenic region 335 Kb upstream from the MYC gene, a well-known oncogene [9], generated a hypothesis that this SNP might be involved in a long-distance regulation of MYC expression. Located in a region with significant evolutionary conservation and enhancer potential [10-12], the SNP was predicted to affect a binding site for TCF7L2 [10,11], a key transcription factor in the WNT pathway. The risk allele G of rs6983267 was found to have a slightly stronger affinity to TCF7L2 in binding assays compared to the non-risk allele T, and stronger regulatory activity in luciferase reporter assays [10,11]. An analysis of long-range interactions showed that the region containing rs6983267 might be in physical proximity with MYC region [10]. These findings suggested that rs6983267 might be located within an enhancer element that interacts with TCF7L2 and regulates MYC expression [10,11]. MYC is a target gene of TCF7L2 [13-15] and its expression is regulated through two TCF7L2 binding sites within the MYC promoter [13]. No association has been found between rs6983267 and the mRNA expression of MYC in lymphoblastoid cell lines [11,16], normal and tumor colon samples [10,11,17-20], or with MYC immunostaining in colon tumors [5].
Previously, we performed a detailed study of TCF7L2 expression in several types of human tissue, including colon where we measured the expression of multiple assays targeting the majority of known splicing forms of TCF7L2 [21,22]. In the current study we sought to determine, whether the expression of TCF7L2 splicing forms we identified in non-cancer colon samples correlated with MYC expression and whether this expression was dependent on alleles of rs6983267 or interaction of rs6983267 with TCF7L2 expression.
We investigated non-cancer colon samples on the assumption that the effect of a germline genetic variation might be more easily detectable in conditions not affected by the effects of cancer or its treatment. The samples and the methods are described in Additional file 1. The mRNA expression of MYC was detected by sensitive quantitative reverse-transcriptase PCR (qRT-PCR) and 3 expression assays (Figure 1A.) The expression of MYC assays 1 and 2, corresponding to exons 2-3, and 1-2, respectively (RefSeq transcript NM_002467), was highly correlated (r = 0.95). MYC assay 3 targeted an alternative transcript initiated from a promoter P0 (GenBank accession number M13929) [23], however, expression of this transcript was very low (at a level of at least 100 times lower than of assays 1 and 2) and was not studied further. Expression of TCF7L2 was measured with 7 assays previously described (Figure 1B, Additional file 2) [21,22].
Figure 1.
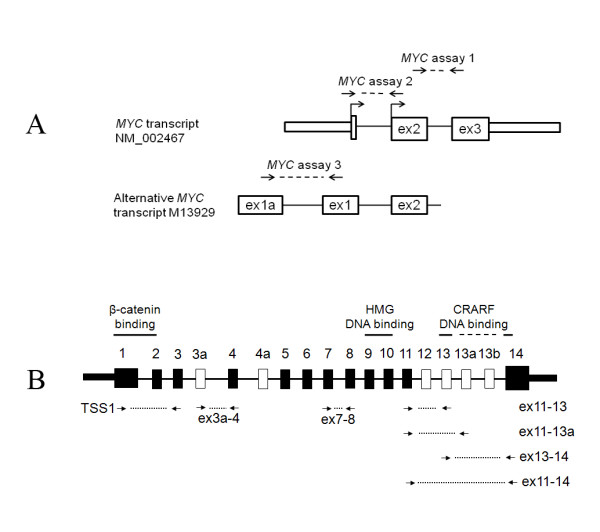
Location of MYC and TCF7L2 expression assays. A. MYC exons and 5' and 3'untranslated regions (UTRs) are marked by rectangles and two translation starts are marked by vertical lines and arrows. MYC assay1 is located over the junction of exons 2 and 3, and MYC assay 2 is located over the junction of exons 1 and 2. MYC assay 3 targets alternative transcript with an upstream exon. B. Constitutive exons of TCF7L2 are represented as black rectangles and alternative exons as white rectangles. Protein domains are indicated above corresponding exons: β-catenin binding domain is encoded by exons 1 and 2, high mobility group (HMG) DNA-binding domain is encoded by exons 9 and 10 and the CRARF DNA-binding domain is encoded by exons 13 and 14. Location of expression assays is indicated under corresponding exons and arrows show primer positions. The specificity of detection of particular splicing forms is achieved by probes located over exon junctions.
The strongest correlation between MYC and TCF7L2 expression was observed for assay "ex13-14" of TCF7L2 (r = 0.57- 0.60, p < 10-6), followed by assay "ex11-13" (r = 0.52-0.54, p < 10-6). The weakest correlation was detected for assay "ex11-13a" (r = 0.10 - 0.15, p = 0.12 - 0.28) (Table 1). These assays detect alternative splicing forms that include combinations of exons 11-13-14 and 11-13a-14 in the C-terminal end of the TCF7L2 transcripts (GenBank accession numbers FJ010174 and FJ010167). Both protein isoforms encoded by these splicing forms have long C-terminal reading frames (E-tails) with binding sites for the C-terminal binding protein (CtBP) involved in post-translational regulation of TCF7L2 expression [21,22]. Protein fragments encoded by the alternative exons 13 and 13a share 68% identity (17 amino acids of 25, Figure 2). The form with exons 11-13-14 encodes a 30-amino-acid highly conserved motif with a CRARF signature protein sequence, while in the form with exons 11-13a-14 this sequence is changed to CRALF (Figure 2). The CRARF protein sequence is also found in another member of TCF/LEF family of transcription factors, TCF-7 (former TCF-1) and in the ancestral drosophila TCF/pangolin protein [24]. The CRARF sequence serves as an additional DNA-binding domain and a strong transactivator of the WNT pathway [24,25]. The CRARF-form of TCF7L2 was shown to interact with two TCF7L2 binding sites within MYC promoter, TBE1 at -1156 bp and TBE2 at -589 bp upstream the first translation start site [13,24]. Therefore, we suggest that while both the CRARF and CRALF forms of TCF7L2 contain E-tails, only the CRARF form regulates MYC expression in the colon. A splicing form detected by assay "ex11-14" is the most common splicing form of TCF7L2 in all human tissues [21,22]. This splicing combination utilizes an alternative stop codon in the beginning of exon 14 resulting in a protein without the CtBP-binding domain. A somatic frameshift mutation in a polyA stretch within exon 14 found in colorectal cancer cell lines results in similar outcome - termination of protein by an alternative stop codon in the beginning of exon 14 [26,27]. We found a moderate correlation between assay "ex11-14" of TCF7L2 and MYC expression (Table 1). Next, we evaluated the levels of mRNA MYC expression in colon tissue in relation to genotypes of rs6983267. We observed significant effect of age, of several TCF7L2 splicing forms but no effect of rs6983267 alone or in interaction with TCF7L2 (Table 2).
Table 1.
Correlation between expression of MYC and TCF7L2 in colon samples
|
TCF7L2 assays |
MYC assay 1 | MYC assay 2 | ||
| r* | p-value# | r* | p-value# | |
| TCF7L2 TSS1 | 0.23 | 0.014 | 0.21 | 0.023 |
| TCF7L2 ex3a-4 | 0.17 | 0.070 | 0.15 | 0.12 |
| TCF7L2 ex7-8 | 0.38 | 2.4 × 10-5 | 0.34 | 2.1 × 10-4 |
| TCF7L2 ex11-13a | 0.15 | 0.12 | 0.10 | 0.28 |
| TCF7L2 ex11-14 | 0.40 | 1.2 × 10-5 | 0.34 | 2.5 × 10-4 |
| TCF7L2 ex11-13 | 0.54 | <10-6 | 0.52 | <10-6 |
| TCF7L2 ex13-14 | 0.60 | <10-6 | 0.57 | <10-6 |
*Spearman correlation coefficient and # two-sided p-values for MYC and TCF7L2 expression in 117 colon samples, p-values are not adjusted for multiple tests
Figure 2.
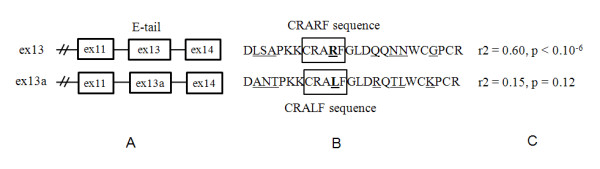
Detailed structure of C-terminal part of TCF7L2 gene. A. Combinations of exons 11, 13, 13a and 14 of TCF7L2 encoding proteins with long reading frames (E-tail). B. A form with alternative exon 13 encodes a protein sequence with CRARF motif; a form with alternative exons 13a encodes a protein sequence with CRALF motif, differences in amino acids in proteins encoded by exons 13 and 13a are underlined, CRARF and CRALF motifs are boxed. C. Expression of a splicing form with exon13 correlates with MYC expression (r = 0.60, p < 10-6), while expression of a splicing form with exon 13a does not correlate with MYC expression (r = 0.15, p = 0.12).
Table 2.
Association of rs6983267 with MYC expression in colon tissue
| factors | MYC assay 1, p-value * | MYC assay 2, p-value * |
| ars6983267 | 0.85 | 0.91 |
| bage | 0.040 | 0.043 |
| brs6983267 | 0.83 | 0.84 |
| age | 0.13 | 0.13 |
| Rs6983267 | 0.85 | 1.00 |
| TCF7L2 TSS1 | 0.054 | 0.076 |
| rs6983267*TCF7L2 TSS1 | 0.91 | 0.91 |
| age | 0.069 | 0.062 |
| Rs6983267 | 1.00 | 0.97 |
| TCF7L2 ex3a-4 | 0.36 | 0.52 |
| rs6983267*TCF7L2 ex3a-4 | 0.93 | 0.77 |
| Age | 0.31 | 0.39 |
| Rs6983267 | 0.75 | 0.28 |
| TCF7L2 ex7-8 | 0.0039 | 0.021 |
| rs6983267*TCF7L2 ex7-8 | 0.79 | 0.25 |
| age | 0.045 | 0.037 |
| Rs6983267 | 0.95 | 0.90 |
| TCF7L2 ex11-13a | 0.31 | 0.50 |
| rs6983267*TCF7L2 ex11-13a | 0.97 | 0.89 |
| age | 0.38 | 0.37 |
| Rs6983267 | 0.70 | 0.61 |
| TCF7L2 ex11-14 | 0.0008 | 0.013 |
| rs6983267*TCF7L2 ex11-14 | 0.57 | 0.50 |
| age | 0.13 | 0.19 |
| Rs6983267 | 0.22 | 0.41 |
| TCF7L2 ex11-13 | <0.0001 | <0.0001 |
| rs6983267*TCF7L2 ex11-13 | 0.29 | 0.48 |
| age | 0.25 | 0.27 |
| Rs6983267 | 0.28 | 0.30 |
| TCF7L2 ex13-14 | <0.0001 | <0.0001 |
| rs6983267*TCF7L2 ex13-14 | 0.35 | 0.35 |
*p-values for univariate regression analysis including 0, 1 and 2 alleles of rs6983267 and expression of MYC assays 1 and 2 in the presence of covariates -- age and level of expression of TCF7L2 assays or interaction between rs6983267 and TCF7L2 expression; p-values are not adjusted for multiple tests; a genotypes were available for 96 samples; b age and genotypes were available for 79 samples.
Our results show a strong role of TCF7L2 in regulation of MYC expression in colon, but not through rs6983267. Both TCF7L2 and MYC genes are expressed in colon and are important for maintaining proliferation of intestinal epithelium [28,29] (Additional file 3). Inactivation of the adenomatous polyposis coli (APC) tumor suppressor gene leads to formation of β-catenin/TCF7L2 complexes, constitutive activation of the WNT pathway and eventually colorectal cancer. Rare point mutations within TCF7L2 are also found in colorectal cancers [30]. The proliferative effect of TCF7L2 is achieved through its transcriptional regulation of several target genes such as MYC and CCND1 (Cyclin D1)[31]. Our results suggest that expression of MYC in colon tissue is most likely regulated by a splicing form of TCF7L2 encoding a protein with a potent transactivation CRARF-domain. However, we did not find any evidence for an effect of rs6983267 on TCF7L2 regulation of MYC expression. Of the family of TCF/LEF transcription factors, TCF7L2 has the highest expression in colon, but other members of this family may also be involved. Inactivation of TCF-7 (former TCF-1) leads to development of intestinal polyps [32]. Expression of LEF1 is found in tumors but not in normal colon tissue [33]. Each of these proteins can recognize the same TCF/LEF consensus binding site and, therefore, might bind alleles of rs6983267. High degree of similarity between TCF/LEF proteins may lead to cross-reactivity in chromatin immunoprecipitation (ChIP) assays. Thus, other TCF/LEF factors should also be examined for their effect on regulation of MYC expression.
In conclusion, SNP rs6983267 within the 8q24 region has been established as one of the strongest genetic risk factors for development of at least two types of cancer. Identification of functional mechanisms of this association is the highest priority of cancer genetics and would mean a significant step forward towards understanding of cancer pathogenesis and development of better diagnostic and therapeutic approaches. Our results provide new insights into the regulation of MYC expression by TCF7L2. However, further studies are needed to investigate alternative molecular mechanisms that can explain the association between rs6983267 and cancer risk.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
LPO designed and performed the study and wrote manuscript, JLH provided samples and wrote manuscript. Both authors read and approved the final manuscript.
Supplementary Material
Materials and methods. The data provided represent the materials, methods and statistical analysis used to study mRNA coexpression of MYC and TCF7L2.
Expression assays used in this study. primers, probes and TaqMan assay IDs.
Protein expression of MYC and TCF7L2 in normal human colon. A. Expression of MYC in normal human colon; B. Expression of TCF7L2 in normal human colon. Both proteins show glandular staining in colon epithelium. Images are courtesy of Protein Atlas http://www.proteinatlas.org
Acknowledgments
Acknowledgements
We thank Yi-Ping Fu for help with statistical analysis. The study was supported by the intramural research program of NCI/NIH (LPO) and by an R21 award to JLH (1R21DK078029-01). We would like to thank Cullan Welch, Nikki Usher, and Marj Carlson for their technical assistance with the tissue processing.
Contributor Information
Ludmila Prokunina-Olsson, Email: prokuninal@mail.nih.gov.
Jennifer L Hall, Email: jlhall@umn.edu.
References
- Tomlinson I, Webb E, Carvajal-Carmona L, Broderick P, Kemp Z, Spain S, Penegar S, Chandler I, Gorman M, Wood W, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet. 2007;39:984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- Haiman CA, Le Marchand L, Yamamato J, Stram DO, Sheng X, Kolonel LN, Wu AH, Reich D, Henderson BE. A common genetic risk factor for colorectal and prostate cancer. Nat Genet. 2007;39:954–956. doi: 10.1038/ng2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsson J, Sulem P, Manolescu A, Amundadottir LT, Gudbjartsson D, Helgason A, Rafnar T, Bergthorsson JT, Agnarsson BA, Baker A, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39:631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- Haiman CA, Patterson N, Freedman ML, Myers SR, Pike MC, Waliszewska A, Neubauer J, Tandon A, Schirmer C, McDonald GJ, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet. 2007;39:638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanke BW, Greenwood CM, Rangrej J, Kustra R, Tenesa A, Farrington SM, Prendergast J, Olschwang S, Chiang T, Crowdy E, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet. 2007;39:989–994. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, Orr N, Yu K, Chatterjee N, Welch R, Hutchinson A, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- Wokolorczyk D, Gliniewicz B, Sikorski A, Zlowocka E, Masojc B, Debniak T, Matyjasik J, Mierzejewski M, Medrek K, Oszutowska D, et al. A range of cancers is associated with the rs6983267 marker on chromosome 8. Cancer Res. 2008;68:9982–9986. doi: 10.1158/0008-5472.CAN-08-1838. [DOI] [PubMed] [Google Scholar]
- Park SL, Chang SC, Cai L, Cordon-Cardo C, Ding BG, Greenland S, Hussain SK, Jiang Q, Liu S, Lu ML, et al. Associations between variants of the 8q24 chromosome and nine smoking-related cancer sites. Cancer Epidemiol Biomarkers Prev. 2008;17:3193–3202. doi: 10.1158/1055-9965.EPI-08-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- Pomerantz MM, Ahmadiyeh N, Jia L, Herman P, Verzi MP, Doddapaneni H, Beckwith CA, Chan JA, Hills A, Davis M, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet. 2009;41:882–884. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuupanen S, Turunen M, Lehtonen R, Hallikas O, Vanharanta S, Kivioja T, Bjorklund M, Wei G, Yan J, Niittymaki I, et al. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genet. 2009;41:885–890. doi: 10.1038/ng.406. [DOI] [PubMed] [Google Scholar]
- Yeager M, Xiao N, Hayes RB, Bouffard P, Desany B, Burdett L, Orr N, Matthews C, Qi L, Crenshaw A, et al. Comprehensive resequence analysis of a 136 kb region of human chromosome 8q24 associated with prostate and colon cancers. Hum Genet. 2008;124:161–170. doi: 10.1007/s00439-008-0535-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- Muncan V, Sansom OJ, Tertoolen L, Phesse TJ, Begthel H, Sancho E, Cole AM, Gregorieff A, de Alboran IM, Clevers H, Clarke AR. Rapid loss of intestinal crypts upon conditional deletion of the Wnt/Tcf-4 target gene c-Myc. Mol Cell Biol. 2006;26:8418–8426. doi: 10.1128/MCB.00821-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- Houlston RS, Webb E, Broderick P, Pittman AM, Di Bernardo MC, Lubbe S, Chandler I, Vijayakrishnan J, Sullivan K, Penegar S, et al. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat Genet. 2008;40:1426–1435. doi: 10.1038/ng.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz MM, Ahmadiyeh N, Jia L, Herman P, Verzi MP, Doddapaneni H, Beckwith CA, Chan JA, Hills A, Davis M, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet. 2009 doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz MM, Beckwith CA, Regan MM, Wyman SK, Petrovics G, Chen Y, Hawksworth DJ, Schumacher FR, Mucci L, Penney KL, et al. Evaluation of the 8q24 prostate cancer risk locus and MYC expression. Cancer Res. 2009;69:5568–5574. doi: 10.1158/0008-5472.CAN-09-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuupanen S, Turunen M, Lehtonen R, Hallikas O, Vanharanta S, Kivioja T, Bjorklund M, Wei G, Yan J, Niittymaki I, et al. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genet. 2009 doi: 10.1038/ng.406. [DOI] [PubMed] [Google Scholar]
- Gruber SB, Moreno V, Rozek LS, Rennerts HS, Lejbkowicz F, Bonner JD, Greenson JK, Giordano TJ, Fearson ER, Rennert G. Genetic variation in 8q24 associated with risk of colorectal cancer. Cancer Biol Ther. 2007;6:1143–1147. doi: 10.1158/1535-7163.MCT-06-0555. [DOI] [PubMed] [Google Scholar]
- Prokunina-Olsson L, Kaplan LM, Schadt EE, Collins FS. Alternative splicing of TCF7L2 gene in omental and subcutaneous adipose tissue and risk of type 2 diabetes. PLoS One. 2009;4:e7231. doi: 10.1371/journal.pone.0007231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokunina-Olsson L, Welch C, Hansson O, Adhikari N, Scott LJ, Usher N, Tong M, Sprau A, Swift A, Bonnycastle LL, et al. Tissue-specific alternative splicing of TCF7L2. Hum Mol Genet. 2009;18:3795–3804. doi: 10.1093/hmg/ddp321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DL, Groudine M. Novel promoter upstream of the human c-myc gene and regulation of c-myc expression in B-cell lymphomas. Mol Cell Biol. 1986;6:3481–3489. doi: 10.1128/mcb.6.10.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atcha FA, Syed A, Wu B, Hoverter NP, Yokoyama NN, Ting JH, Munguia JE, Mangalam HJ, Marsh JL, Waterman ML. A unique DNA binding domain converts T-cell factors into strong Wnt effectors. Mol Cell Biol. 2007;27:8352–8363. doi: 10.1128/MCB.02132-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atcha FA, Munguia JE, Li TW, Hovanes K, Waterman ML. A new beta-catenin-dependent activation domain in T cell factor. J Biol Chem. 2003;278:16169–16175. doi: 10.1074/jbc.M213218200. [DOI] [PubMed] [Google Scholar]
- Cuilliere-Dartigues P, El-Bchiri J, Krimi A, Buhard O, Fontanges P, Flejou JF, Hamelin R, Duval A. TCF-4 isoforms absent in TCF-4 mutated MSI-H colorectal cancer cells colocalize with nuclear CtBP and repress TCF-4-mediated transcription. Oncogene. 2006;25:4441–4448. doi: 10.1038/sj.onc.1209471. [DOI] [PubMed] [Google Scholar]
- Duval A, Rolland S, Tubacher E, Bui H, Thomas G, Hamelin R. The human T-cell transcription factor-4 gene: structure, extensive characterization of alternative splicings, and mutational analysis in colorectal cancer cell lines. Cancer Res. 2000;60:3872–3879. [PubMed] [Google Scholar]
- Sansom OJ, Meniel VS, Muncan V, Phesse TJ, Wilkins JA, Reed KR, Vass JK, Athineos D, Clevers H, Clarke AR. Myc deletion rescues Apc deficiency in the small intestine. Nature. 2007;446:676–679. doi: 10.1038/nature05674. [DOI] [PubMed] [Google Scholar]
- Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters PJ, Clevers H. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet. 1998;19:379–383. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- Roose J, Huls G, van Beest M, Moerer P, Horn K van der, Goldschmeding R, Logtenberg T, Clevers H. Synergy between tumor suppressor APC and the beta-catenin-Tcf4 target Tcf1. Science. 1999;285:1923–1926. doi: 10.1126/science.285.5435.1923. [DOI] [PubMed] [Google Scholar]
- Hovanes K, Li TW, Munguia JE, Truong T, Milovanovic T, Lawrence Marsh J, Holcombe RF, Waterman ML. Beta-catenin-sensitive isoforms of lymphoid enhancer factor-1 are selectively expressed in colon cancer. Nat Genet. 2001;28:53–57. doi: 10.1038/88264. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Materials and methods. The data provided represent the materials, methods and statistical analysis used to study mRNA coexpression of MYC and TCF7L2.
Expression assays used in this study. primers, probes and TaqMan assay IDs.
Protein expression of MYC and TCF7L2 in normal human colon. A. Expression of MYC in normal human colon; B. Expression of TCF7L2 in normal human colon. Both proteins show glandular staining in colon epithelium. Images are courtesy of Protein Atlas http://www.proteinatlas.org


