Abstract
Background
The Nme family, previously known as Nm23 or NDPK, is involved in various molecular processes including tumor metastasis and some members of the family, but not all, exhibit a Nucleoside Diphosphate Kinase (NDPK) activity. Ten genes are known in humans, in which some members have been extensively studied. In non-mammalian species, the Nme protein family has received, in contrast, far less attention. The picture of the vertebrate Nme family remains thus incomplete and orthology relationships with mammalian counterparts were only partially characterized. The present study therefore aimed at characterizing the Nme gene repertoire in vertebrates with special interest for teleosts, and providing a comprehensive overview of the Nme gene family evolutionary history in vertebrates.
Results
In the present study, we present the evolutionary history of the Nme family in vertebrates and characterize the gene family repertoire for the first time in several non-mammalian species. Our observations show that vertebrate Nme genes can be separated in two evolutionary distinct groups. Nme1, Nme2, Nme3, and Nme4 belong to Group I while vertebrate Nme5, Nme6, Nme7, Nme8, and Nme9 belong to Group II. The position of Nme10 is in contrast more debatable due to its very specific evolutionary history. The present study clearly indicates that Nme5, Nme6, Nme7, and Nme8 originate from duplication events that occurred before the chordate radiation. In contrast, Nme genes of the Group I have a very different evolutionary history as our results suggest that they all arise from a common gene present in the chordate ancestor. In addition, expression patterns of all zebrafish nme transcripts were studied in a broad range of tissues by quantitative PCR and discussed in the light of the function of their mammalian counterparts.
Conclusion
This work offers an evolutionary framework that will pave the way for future studies on vertebrate Nme proteins and provides a unified vertebrate Nme nomenclature that is consistent with the nomenclature in use in mammals. Based on protein structure and expression data, we also provide new insight into molecular functions of Nme proteins among vertebrates and raise intriguing questions on the roles of Nme proteins in gonads.
Background
The first descriptions of Nucleoside Diphosphate Kinase (NDPK) activity, that corresponds to the phosphoryl transfer from a nucleoside triphosphate to a nucleoside diphosphate, were made in pigeon breast muscle [1] and yeast [2]. Sequences encoding for proteins with putative [3] or experimentally validated [4-6] NDPK activity were subsequently identified. These proteins, originally named NDPK based on their NDPK activity, belong to the Nme protein family according to current official gene nomenclature [7-10]. These proteins "expressed in non-metastatic cell", and thus named Nme, were also previously known as Nm23 proteins. In humans, the NME family is composed of ten genes and some of the proteins, but not all, exhibit NDPK activity.
Nme genes were first identified in mouse [11] and in the fruit fly Drosophila melanogaster [12] in which they drew attention for their surprising implication in tumor metastasis process [11] and in normal fly development [12] respectively. Soon, several orthologs of these genes were identified in other organisms ranging from the bacteria Escherichia coli [13] to humans [14]. They were subsequently studied for their role as tumor metastasis suppressor or enhancer depending on the cancer type. To date, ten genes displaying partial or complete NDPK domains have been identified in humans (reviewed in [15]). Proteins of this family were classified into two groups based on sequence characteristics and NDPK activity [15]. Group I Nme proteins (Nme1 to 4) display a particularly well conserved domain and active site, whereas Group II Nme proteins (Nme5 to 10) display highly divergent domains and all of them, except Nme6, lack NDPK activity [15]. In fish and amphibians, proteins of the Nme family have been implicated in key developmental processes in the oocyte or embryo [16-18]. However, the Nme proteins repertoire remains uncharacterized in almost all non-mammalian vertebrates. In teleost fish, only two Nme sequences were reported [18,19]. In non-mammalian species, the picture of the Nme family remains fuzzy and the orthology relationships of reported Nme proteins with their mammalian counterparts were only partially characterized [18,20]. Therefore, the evolutionary process which gave rise to such a complex gene family remains poorly understood and requires a complete characterization that will pave the way for future investigations of the roles of Nme proteins in vertebrates.
In the present study, we describe the evolutionary history of the Nme gene family in chordates and provide, for the first time, a comprehensive characterization of the Nme gene repertoire in vertebrates.
Results and Discussion
Evolutionary history of Nme gene family in vertebrates
Nucleoside disphosphate (NDP) kinase activity is ubiquitously found in organisms from bacteria to humans. In humans, ten NME genes exist that have been separated in two groups based on their amino-acid sequence [15]. These two groups originate from a gene duplication of a single NDPK ancestor gene that probably occurred before or around the metazoan radiation [21]. As indicated above, the evolutionary history of vertebrate Nme proteins has received very little attention as most existing studies focused on mammalian proteins or on specific members of the family [15,18,20,21]. Some information is however available in cellular slime molds [22], drosophila and C. elegans [21]. In contrast, available data in chordates and non-mammalian vertebrate species are extremely limited apart from the report of several Nme sequences [18-20].
A two group classification
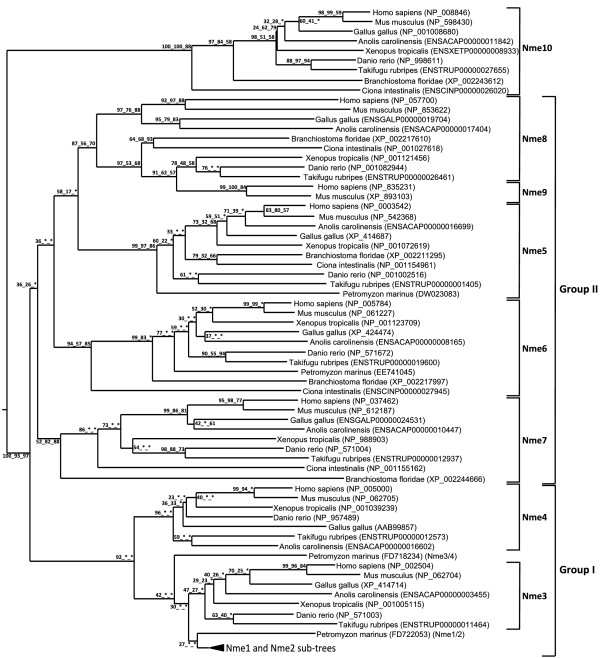
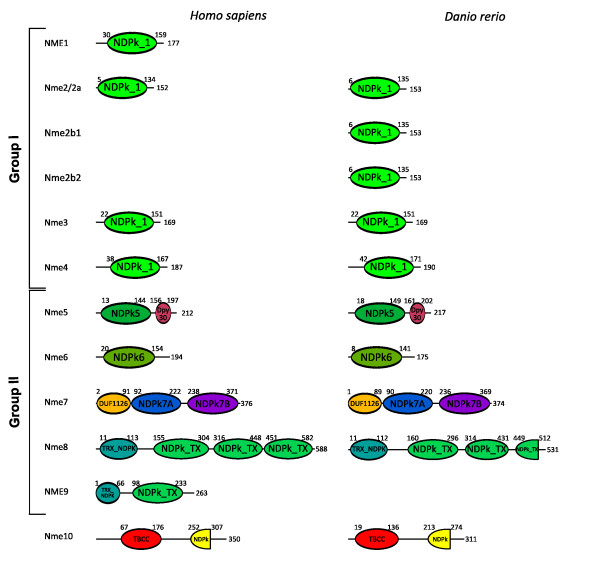
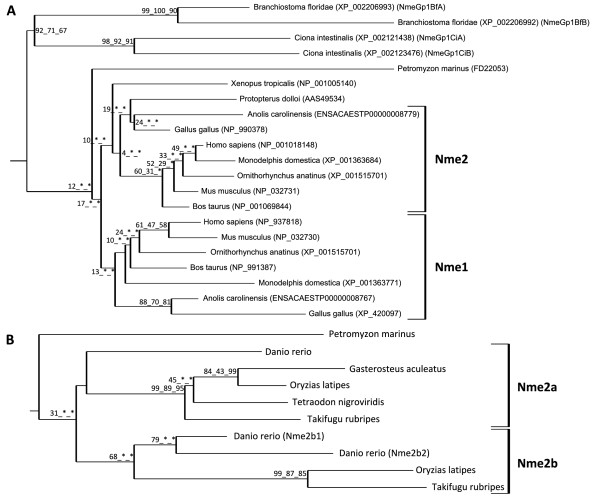
The phylogenetic analysis of Nme proteins (Fig. 1) shows two strongly supported distinct clusters. Nme1, Nme2, Nme3, and Nme4 belong to the Group I cluster while Nme5, Nme6, Nme7, Nme8 and Nme9 belong to the Group II cluster. Within each group, all Nme subtypes are also distinctly separated from each other, with the exception of Nme9 sequences that are only found in eutherians and appear to be closely related to Nme8 sequences (Fig. 1). The analysis of the domain structure of Nme proteins using the NCBI Conserved Domain Database [23] clearly demonstrates the existence of two distinct groups among Nme1 to 9 proteins (Fig. 2) that clearly possess distinct domains. Proteins of the Group I (Nme1 to 4, Table 1) display a single type NDPk_1 domain while proteins of the Group II (Nme5 to 9, Table 2) display a single or several NDPk domains of different types, associated or not with extra-domains. For all Nme, the sequence structure, including domain(s) nature(s), length or position in the sequence, as well as the exon-intron structure (Fig. 3A &4), is highly conserved between human and zebrafish (Danio rerio) proteins. Together, our results on exon-intron structure, protein domains, and phylogenetic analysis, clearly indicate that the separation of vertebrate Nme1 to Nme9 proteins in two groups that has been proposed in mammals [15] is also valid for all vertebrates.
Figure 1.
Phylogenetic reconstruction of the Nme protein family in chordates. The phylogenetic tree was constructed from a single multiple alignment. Bootstrap values for neighbor joining, maximum parsimony, and maximum likelihood methods, respectively, are indicated for each node. * indicates that the node does not exist in the corresponding tree. The consensus tree was calculated with the FIGENIX [87] automated phylogenomic annotation pipeline. Nme1-Nme2 subtree was removed from the main tree and studied separately (see Figure 5) for tetrapods and teleosts because of high sequence similarity and different evolutionary history. For each sequence, NCBI or Ensembl accession number and species are shown.
Figure 2.
Proteins domains of human and zebrafish Nme proteins. The Genbank Conserved Domain Database was used for protein domain characterization [23]. Amino acid numbers corresponding to the beginning and the end of each domain are indicated and the total amino acid length number of the protein is shown at the end of each protein.
Table 1.
Group I Nme proteins: names and symbols by species, accession numbers and corresponding chromosomal location
| Species | Name | Other names | GenBank Acc # | Ensembl Acc # | Localisation | Position | |
| C. intestinalis | NmeGp1CiA | NDK B | XP_002123476 | ENSCINP00000011619 | Chr 8q | 5,908,347-5,908,808 | |
| C. intestinalis | NmeGp1CiB | XP_002121438 | ENSCINP00000002194 | Chr 2q | 7,888,026-7,888,562 | ||
| NmeGp1 | B. floridae | NmeGp1BfA | XP_002206993 | Chr Un | 116,102,546-116,104,838 | ||
| B. floridae | NmeGp1BfB | XP_002206992 | Chr Un | 116,092,575-116,094,429 | |||
| P. marinus | Nme2 | FD722053* | |||||
| P. marinus | Nme3/4 | FD718234* | Contig24671 | 6,345-8,952 | |||
| H. sapiens | NME1 | NDK A; NM23-H1; GAAD | NP_937818 | ENSP00000337060 | Chr 17 | 46,585,919-46,594,449 | |
| M. musculus | Nme1 | NDK A; Nm23-M1 | NP_032730 | ENSMUSP00000021220 | Chr 11 | 93,820,547-93,829,574 | |
| B. taurus | Nme1 | NDK A; NDKA2; NBR-A | NP_991387 | ENSBTAP00000006104 | Chr 19 | 36,634,791-36,645,841 | |
| Nme1 | M. domestica | Nme1 | LOC100012868 | XP_001363771 | ENSMODP00000015750 | Chr 2 | 184,090,725-184,104,175 |
| O. anatinus | Nme1 | ENSOANP00000018628 | SuperContig 18222 | 1,054-2,769 | |||
| G. gallus | Nme1 | Nm23A | XP_420097 | ENSGALP00000011811 | Chr18 | 9,930,932-9,933,309 | |
| A. carolinensis | Nme1 | ENSACAESTP00000008767 | Scaffold_268 | 1,710,978-1,717,896 | |||
| H. sapiens | NME2 | NDK B; NM23-H2; PUF | NP_001018149 | ENSP00000376888 | Chr 17 | 46,598,821-46,604,103 | |
| M. musculus | Nme2 | NDK B; Nm23-M2 | NP_032731 | ENSMUSP00000021217 | Chr 11 | 93,811,130-93,817,195 | |
| B. taurus | Nme2 | NDK B; PUF | NP_001069844 | Chr 19 | 36,625,295-36,629,092 | ||
| Nme2 | M. domestica | Nme2 | NDK B | XP_001363684 | ENSMODP00000015743† | Chr 2 | 184,076,307-184,081,978 |
| O. anatinus | Nme2 | ENSOANP00000018629 | SuperContig 18222 | 6,818-8,864 | |||
| G. gallus | Nme2 | CNDPK; NDK_CHICK | NP_990378 | ENSGALP00000034078 | Chr18 | 5,062,096-5,064,054 | |
| A. carolinensis | Nme2 | ENSACAESTP00000008779 | Scaffold_268 | 1,724,830-1,729,637 | |||
| X. tropicalis | Nme2 | NME1 | NP_001005140 | ENSXETP00000024764 | Scaffold_673 | 77,640-81,451 | |
| H. sapiens | NMELV | NME1-NME2 | NP_001018146 | ENSP00000376894 | Chr 17 | 49,230,997-49,249,103 | |
| P. troglodytes | NmeLV | NME1-NME2 | XP_511889 | ENSPTRP00000044657 | Chr 17 | 50,152,238-50,171,443 | |
| Nme-LV | E. caballus | NmeLV | NME1-NME2 | XP_001499951 | ENSECAP00000019175† | Chr 11 | 26,692,478-26,707,469 |
| B. taurus | NmeLV | NDKB-BOVIN | ENSBTAP00000041066 | Chr 19 | 36,625,292-36,645,867 | ||
| O. anatinus | NmeLV | NME1-NME2 | XP_001515701 | SuperContig18222 | 1057-8751 | ||
| A. carolinensis | NmeLV | ENSACAP00000002161 | Scaffold_268 | 1,706,438-1,729,637 | |||
| D. rerio | Nme2a | NM23B; nme2l | NP_956264 | ENSDARP00000064338 | Chr 20 | 5,310,868-5,319,057 | |
| O. latipes | Nme2a | ENSORLP00000018429 | Chr 19 | 21,821,808-21,827,808 | |||
| Nme2a | G. aculeatus | Nme2a | ENSGACP00000020117 | Scaffold_48 | 706,609-708,477 | ||
| T. nigroviridis | Nme2a | CAF90396† | ENSTNIP00000005730 | Chr Un_random | 34,843,841-34,848,064 | ||
| T. rubripes | Nme2a | ENSTRUP00000014520 | Scaffold_29 | 678,695-680,960 | |||
| D. rerio | Nme2b1 | nme2; nme1; ndpkz1; NM23B | NP_571001 | ENSDARP00000074169† | Scaffold Zv7_NA1913 | 1,772-2,772 | |
| Nme2b | D. rerio | Nme2b2 | Ndpkz2 | NP_571002 | Scaffold Zv7_NA1913 | 8,750-9,346 | |
| O. latipes | Nme2b | NDKA; GAAD; NME1-NME2 | ENSORLP00000023099 | Scaffold1014 | 323-3,121 | ||
| T. rubripes | Nme2b | NDKA; GAAD | ENSTRUP00000001870 | Scaffold_307 | 158,498-160,002 | ||
| H. sapiens | NME3 | NDPKC; DR-nm23; NM23-H3 | NP_002504 | ENSP00000219302 | Chr 16 | 1,760,323-1,761,711 | |
| M. musculus | Nme3 | NDPKC; DR-nm23; Nm23-M3 | NP_062704 | ENSMUSP00000024978 | Chr 17 | 25,033,459-25,034,448 | |
| G. gallus | Nme3 | NDPKC; DR-nm23 | XP_414714 | ENSGALP00000003531 | Chr 14 | 13,989,877-13,991,647 | |
| A. carolinensis | Nme3 | NDPKC; NDK 3; DR-nm23 | ENSACAP00000003455 | Scaffold_1065 | 123,674-128,641 | ||
| Nme3 | X. tropicalis | Nme3 | MGC89980 | NP_001005115 | ENSXETP00000022770 | Scaffold_27 | 933,363-937,018 |
| D. rerio | Nme3 | ndpkz3; NDPK-Z3 | NP_571003 | ENSDARP00000075112 | Chr 3 | 13,184,712-13,194,308 | |
| O. latipes | Nme3 | NDK 3; NDPKC; DR-nm23 | ENSORLP00000014699 | Chr 8 | 15,106,871-15,109,573 | ||
| G. aculeatus | Nme3 | NDK 3; NDPKC; DR-nm23 | ENSGACP00000017241 | GroupXI | 12,674,486-12,677,154 | ||
| T. rubripes | Nme3 | NDK 3; NDPKC; DR-nm23 | ENSTRUP00000011464 | Scaffold_294 | 107,469-108,977 | ||
| T. nigroviridis | Nme3 | NDK 3; NDPKC; DR-nm23 | CAG02649 | ENSTNIP00000014528 | Chr Un_random | 6,812,965-6,814,848 | |
| H. sapiens | NME4 | NDPKD; NM23-H4 | NP_005000 | ENSP00000219479 | Chr 16 | 387,193-390,754 | |
| M. musculus | Nme4 | NDPKD; Nm23-M4 | NP_062705 | ENSMUSP00000025007 | Chr 17 | 26,228,682-26,232,433 | |
| G. gallus | Nme4 | NDPKD | AAB99857 | Chr 14 | 13,763,108-13,764,673 | ||
| A. carolinensis | Nme4 | ENSACAP00000016602 | scaffold_1361 | 31,704-32,514 | |||
| Nme4 | X. tropicalis | Nme4 | NP_001039239 | ENSXETP00000022726 | Scaffold_27 | 1,305,654-1,312,714 | |
| D. rerio | Nme4 | zgc:56482 | NP_957489 | ENSDARP00000060403 | Chr 3 | 14,178,447-14,192,697 | |
| O. latipes | Nme4 | NDPKD | ENSORLP00000014089 | Chr 8 | 14,902,635-14,905,718 | ||
| G. aculeatus | Nme4 | NDPKD | ENSGACP00000016999 | GroupXI | 12,533,938-12,535,079 | ||
| T. rubripes | Nme4 | NDPKD | ENSTRUP00000012573 | Scaffold_112 | 81,060-82,895 | ||
| T. nigroviridis | Nme4 | NDPKD | CAG12673 | ENSTNIP00000022136 | Chr Un_random | 7,826,243-7,828,703 | |
Protein names were retrieved from Genbank, Ensembl, iHOP and ZFIN. Location was obtained using Ensembl genome browser, or by UCSC Genome Bioinformatics BLAT when not available on Ensembl. *, sequence is an EST. †, incomplete sequence.
Table 2.
Group II Nme proteins: names and symbols by species, accession numbers and corresponding chromosomal location
| Species | Name | Other names | GenBank Acc # | Ensembl Acc # | Localisation | Position | |
| C. intestinalis | Nme5 | ci-ndk/dpy26 | NP_001154961 | ENSCINP00000008954† | Chr 7q | 1,031,987-1,036,756 | |
| B. floridae | Nme5 | XP_002211295 | Chr Un | 731,166,868-731,169,160 | |||
| P. marinus | Nme5 | DW023083* | Contig18268 | 2,311-13,400 | |||
| H. sapiens | NME5 | NDK-H5; NM23-H5; IPIA-β | NP_003542 | ENSP00000265191 | Chr 5 | 137,478,761-137,503,031 | |
| M. musculus | Nme5 | NDK-M5; Nm23-M5 | NP_542368 | ENSMUSP00000078269 | Chr 18 | 34,722,295-34,738,760 | |
| Nme5 | G. gallus | Nme5 | NDP kinase homolog 5; IPIA-β | XP_414687 | ENSGALP00000022919 | Chr 13 | 14,517,448-14,548,478 |
| A. carolinensis | Nme5 | NDP kinase homolog 5; IPIA-β | ENSACAP00000016699 | Scaffold_29 | 3,794,500-3,804,710 | ||
| X. tropicalis | Nme5 | NDPK homolog 5; IPIA-β | NP_001072619 | ENSXETP00000008322 | Scaffold_65 | 2,613-8,494 | |
| D. rerio | Nme5 | zgc:92812 | NP_001002516 | ENSDARP00000060997 | Chr 14 | 55,814,542-55,821,523 | |
| O. latipes | Nme5 | NDP kinase homolog 5; IPIA-β | ENSORLP00000006672 | Chr 10 | 13,043,372-13,046,033 | ||
| G. aculeatus | Nme5 | NDP kinase homolog 5; IPIA-β | ENSGACP00000023932 | GroupIV | 11,477,537-11,479,481 | ||
| T. rubripes | Nme5 | NDP kinase homolog 5; IPIA-β | ENSTRUP00000001405 | Scaffold_126 | 261,197-263,042 | ||
| T. nigroviridis | Nme5 | NDP kinase homolog 5; IPIA-β | CAG01205 | ENSTNIP00000013421 | Chr 1 | 943,305-945,715 | |
| C. intestinalis | Nme6 | NDK 6; IPIA-α | XP_002129729 | ENSCINP00000027945† | Scaffold_1779 | 5,509-6,021 | |
| B. floridae | Nme6 | XP_002217997 | Chr Un | 307,935,668-307,942,172 | |||
| P. marinus | Nme6 | EE741045* | |||||
| H. sapiens | NME6 | IPIA-α; NDK-H6; NM23-H6 | NP_005784 | ENSP00000307125 | Chr 3 | 48,310,595-48,317,852 | |
| M. musculus | Nme6 | NDK-6; Nm23-M6 | NP_061227 | ENSMUSP00000035053 | Chr 9 | 109,735,308-109,745,475 | |
| Nme6 | G. gallus | Nme6 | NDK 6; IPIA-α | XP_424474 | ENSGALP00000015875 | Chr 1 | 96,907-98,947 |
| A. carolinensis | Nme6 | NDK 6; IPIA-α | ENSACAP00000008165 | Scaffold_2735 | 3,138-10,113 | ||
| X. tropicalis | Nme6 | NDK 6; IPIA-α | NP_001123709 | ENSXETP00000034257 | Scaffold_857 | 287,305-293,502 | |
| D. rerio | Nme6 | Ndpkz6 | NP_571672 | ENSDARP00000094574 | Chr 20 | 19,668,343-19,681,489 | |
| O. latipes | Nme6 | NDK 6; IPIA-α | ENSORLP00000020366 | Chr 24 | 13,630,759-13,633,495 | ||
| G. aculeatus | Nme6 | NDK 6; IPIA-α | ENSGACP00000014120 | GroupXVIII | 10,683,098-10,684,634 | ||
| T. rubripes | Nme6 | NDK 6; IPIA-α | ENSTRUP00000019600 | Scaffold_72 | 321,734-323,012 | ||
| T. nigroviridis | Nme6 | NDK 6; IPIA-α | CAG09120† | ENSTNIP00000019439 | Chr 14 | 5,693,013-5,693,908 | |
| C. intestinalis | Nme7 | NDK/DM44 | NP_001155162 | ENSCINP00000025129† | Chr 1p | 3,423,461-3,424,231 | |
| B. floridae | Nme7 | XP_002244666 | Chr Un | 788,282,087-788,292,883 | |||
| H. sapiens | NME7 | NDK-7; NM23-H7 | NP_037462 | ENSP00000356785 | Chr 1 | 167,368,399-167,603,810 | |
| M. musculus | Nme7 | NDK-7; Nm23-M7 | NP_612187 | ENSMUSP00000027862 | Chr 1 | 166,237,803-166,334,805 | |
| G. gallus | Nme7 | NDK-7 | ENSGALP00000024531 | Chr 1 | 87,015,645-87,088,484 | ||
| Nme7 | A. carolinensis | Nme7 | NDK-7 | ENSACAP00000008165 | Scaffold_2735 | 3,138-10,113 | |
| X. tropicalis | Nme7 | MGC75677 | NP_988903 | ENSXETP00000005150 | Scaffold_169 | 1,646,165-1,680,298 | |
| D. rerio | Nme7 | Ndpkz4; Ndpkz7 | NP_571004 | ENSDARP00000073091 | Chr 6 | 20,659,126-20,718,810 | |
| O. latipes | Nme7 | DK039970* | ENSORLP00000019661† | Chr 4 | 29,680,976-29,697,346 | ||
| G. aculeatus | Nme7 | NDK 7 | ENSGACP00000018067 | GroupVIII | 17,708,704-17,717,807 | ||
| T. rubripes | Nme7 | NDK 7 | ENSTRUP00000012937 | Scaffold_13 | 352,791-361,358 | ||
| T. nigroviridis | Nme7 | NDK 7 | ENSTNIP00000004782 | Chr 1 | 10,762,024-10,774,089 | ||
| C. intestinalis | Nme8 | TXNDC3; CiIC3 | NP_001027618 | ENSCINP00000013583 | Chr 9q | 3,694,058-3,703,935 | |
| B. floridae | Nme8 | XP_002217610 | Chr Un | 302,074,149-302,085,811 | |||
| H. sapiens | NME8 | TXNDC3; CILD6; NM23-H8; SPTRX2 | NP_057700 | ENSP00000199447 | Chr 7 | 37,854,724-37,906,525 | |
| M. musculus | Nme8 | Txndc3; Sptrx2 | NP_853622 | ENSMUSP00000089358 | Chr 13 | 19,736,950-19,789,629 | |
| G. gallus | Nme8 | TXNDC3 | XP_426021 | ENSGALP00000019704 | Chr 2 | 46,221,676-46,240,899 | |
| Nme8 | A. carolinensis | Nme8 | TXNDC3; Sptrx-2 | ENSACAP00000017404 | Scaffold_28 | 5,222,555-5,265,400 | |
| X. tropicalis | Nme8 | TXNDC6; Txl-2 | NP_001121456 | ENSXETP00000002355 | Scaffold_664 | 444,670-467,036 | |
| D. rerio | Nme8 | zgc:162216 | NP_001082944 | Chr13 | 17,124,355-17,152,394 | ||
| O. latipes | Nme8 | TXNDC6; Txl-2 | ENSORLP00000020619 | Chr 21 | 22,030,361-22,038,307 | ||
| G. aculeatus | Nme8 | TXNDC6; Txl-2 | ENSGACP00000008024 | GroupXVI | 11,682,650-11,688,299 | ||
| T. rubripes | Nme8 | TXNDC6; Txl-2 | ENSTRUP00000026461 | Scaffold_46 | 902,376-907,179 | ||
| T. nigroviridis | Nme8 | TXNDC6; Txl-2 | CAG09297 | ENSTNIP00000019567 | Chr 2 | 14,973,997-14,979,390 | |
| H. sapiens | Nme9 | TXNDC6, TXL-2 | NP_835231 | ENSP00000321929 | Chr 3 | 137,980,279-138,048,205 | |
| Nme9 | M. musculus | Nme9 | TXL-2 | XP_893103 | Chr 9 | 99,360,108-99,371,350 | |
| B. taurus | Nme9 | TXNDC6 | NP_001069083 | ENSBTAP00000017071 | Chr 1 | 132,828,887-132,856,623 | |
| C. intestinalis | Nme10 | XRP2 | XP_002121234† | ENSCINP00000026020 | Chr 14q | 3,012,876-3,017,751 | |
| B. floridae | Nme10 | XP_002243612 | Chr Un | 778,883,396-778,885,823 | |||
| H. sapiens | NME10 | RP2; TBCCD2 | NP_008846 | ENSP00000218340 | Chr X | 46,696,347-46,741,793 | |
| M. musculus | Nme10 | Rp2h | NP_598430 | ENSMUSP00000111049 | Chr X | 19,941,607-19,982,781 | |
| G. gallus | Nme10 | RP2; XRP2_CHICK | NP_001008680 | ENSGALP00000026942 | Chr 1 | 134,314,101-134,332,289 | |
| Nme10 | A. carolinensis | Nme10 | XRP2 | ENSACAP00000011842 | Scaffold_571 | 329,765-343,988 | |
| X. tropicalis | Nme10 | XRP2 | ENSXETP00000008933 | Scaffold_253 | 956,431-969,596 | ||
| D. rerio | Nme10 | RP2 | NP_998611 | ENSDARP00000065116 | Chr 6 | 40,312,188-40,323,321 | |
| O. latipes | Nme10 | XRP2 | ENSORLP00000020082 | Chr 4 | 30,358,786-30,362,942 | ||
| G. aculeatus | Nme10 | RP2 | ENSGACP00000018275 | GroupVIII | 18,001,999-18,006,009 | ||
| T. rubripes | Nme10 | XRP2 | ENSTRUP00000027655 | Scaffold_13 | 677,342-680,474 | ||
| T. nigroviridis | Nme10 | XRP2 | CAG01390 | ENSTNIP00000013571 | Chr 1 | 11,019,475-11,022,175 | |
Protein names were retrieved from Genbank, Ensembl, iHOP and ZFIN. Location was obtained using Ensembl genome browser, or by UCSC Genome Bioinformatics BLAT when not available on Ensembl. *, sequence is an EST. †, incomplete sequence.
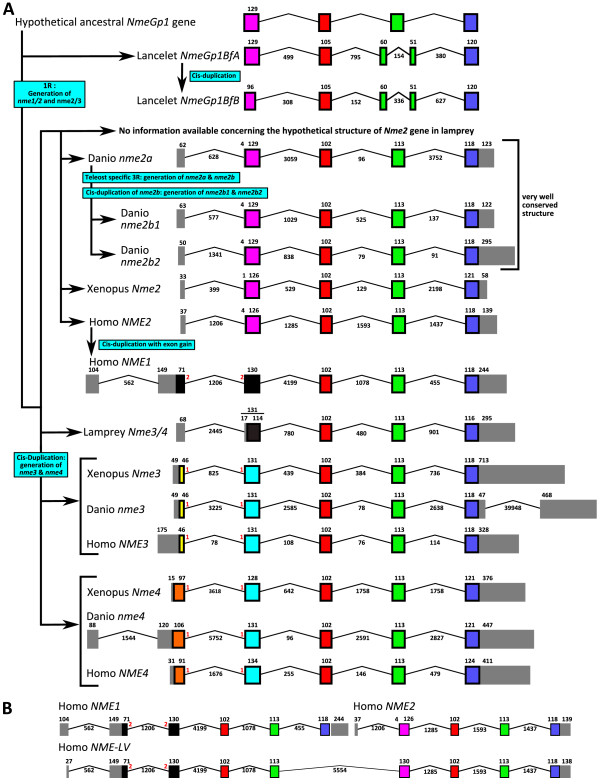
Figure 3.
Intron-Exon structure of Group I Nme genes in chordates. (A) Intron-Exon structure and evolutionary depiction of Group I Nme gene structure among chordate lineage. (B) Intron-Exon structure of human NMELV transcript compared to Intron-Exon structure of human NME1 and NME2. Intron-exon structure was obtained using Ensembl database, or, when no information was available, by Blat of protein and cDNA sequences on genomes assemblies. Boxes correspond to exons. Non-coding exons are shown in grey. The size of introns and exons in nucleotides is shown. Introns are not drawn to scale.
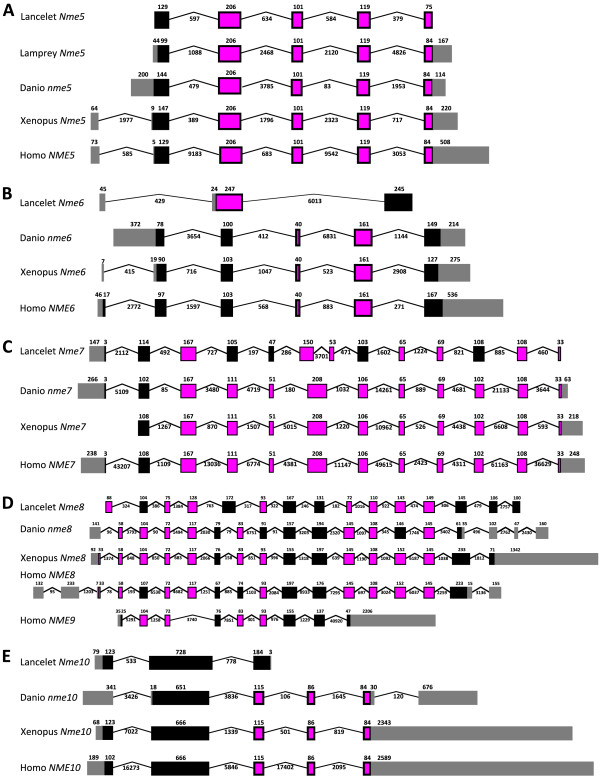
Figure 4.
Intron-Exon structure of Group II Nme genes in chordates. Intron-Exon structure of Nme5 (A), Nme6 (B), Nme7 (C), Nme8/NME9 (D), and Nme10 (E) genes structure among chordates. Intron-exon structure was obtained using Ensembl database, or, when no information was available, by Blat of protein and cDNA sequences on genomes assemblies. Boxes correspond to exons. Non-coding exons are shown in grey. The size of introns and exons in nucleotides is shown. Introns are not drawn to scale.
Nme10, the outgroup of the family
Nme10 protein, previously named X-linked Retinitis Pigmentosa 2 (XRP-2), is the most recently identified member of the Nme family and vertebrate Nme10 proteins form a specific group as shown by the phylogenetic analysis (Fig. 1). It is also noteworthy that sequence identities between prochordates and vertebrates range from 34.5% to 58.2%, indicating a high divergence between prochordate and vertebrate proteins in comparison to the high sequence identity observed among vertebrates species (i.e. 60.9% to 93%) [See Additional file 1]. The protein domain analysis reveals that all vertebrate Nme10 only possess a partial NDPk domain (Fig. 2), which is not present in either Ciona (Ciona intestinalis) or lancelet (Branchiostoma floridae) Nme10 proteins (data not shown). The comparison of the exon-intron structure of the Nme10 gene between lancelet and vertebrates (Fig. 4E) clearly shows that the addition of the partial NDPk domain in vertebrates is associated with a different number of exons in the 3' end of the gene. Together, these observations suggest that a partial NDPk domain was inserted in the Nme10 gene before the gnathostome radiation. As the current status of the lamprey genome preliminary assembly did not allow us to identify any Nme10-related gene in lamprey we are currently unable to provide a better evaluation of the timing of the insertion of this NDPk fragment into the Nme10 gene in the vertebrate lineage. In summary, our observations clearly show that Nme10, in contrast to all other vertebrate Nme proteins, is characterized by a recent incorporation of an NDPk domain. However, because of the gene nomenclature used in mammals [24], we suggest to name this gene Nme10 in vertebrates. In contrast, the classification of this gene in the Group II is more debatable in the light of its totally different evolutionary history.
Nme5, Nme6, Nme7 and Nme8 originate from duplication events that occurred prior to the chordate radiation
We have been able to identify Nme5, Nme6, Nme7, and Nme8 proteins in ciona and lancelet as well as in all investigated vertebrate species, with the exception of the lamprey in which Nme7 and Nme8 could not be found in the current genome preliminary assembly. While we cannot rule out that Nme7 and Nme8 have been lost in lamprey, it is also possible that the preliminary status of the genome assembly and the relatively low sequencing coverage (5.9X) can explain why we have been unable to identify these genes. It should however be stressed that both domain (Fig. 2) and exon-intron structure (Fig. 4A-D) of Nme5, Nme6, Nme7, and Nme8 are particularly well conserved among chordates, with the exception of lancelet Nme6 gene that displays a very specific exon-intron structure. In addition, Nme5, Nme6, Nme7 and Nme8 proteins exhibit high degree of identity among chordates [See Additional files 1, 2, and 3]. In addition, the orthology relationships among species are also clearly supported by the phylogenetic analysis for each protein subtype (Fig 1). Together with existing data on the origin of Group II Nme proteins [21], our observations indicate that Nme5, Nme6, Nme7, and Nme8 genes originate from duplication events that occurred before the chordate radiation.
Nme9, a novel eutherian Nme8-related protein
The Nme9 protein was recently characterized and classified as a member of Group II [24,25]. Thus far, Nme9 has only been found in human, mouse and cow databases but not in any non-mammalian vertebrate species (Table 2). The human NME9 protein contains a Thioredoxin domain (TRX_NDPk) and an NDPk_TX domain that are also found in the N-terminus region of the human NME8 protein (Fig. 2). Similarly, NME8 and NME9 display a similar exon-intron structure in the 5'-region of the gene (Fig. 4D). It is also noteworthy that Nme8 and Nme9 genes are located on different chromosomes in both humans and mice. Based on these observations, we hypothesize that Nme9 originates from an incompletely translocated duplication of the Nme8 gene. The position of human and mouse Nme9 sequences in the phylogenetic analysis support the strong relationship between Nme9 and Nme8 (Fig. 1). The position of Nme9 sequences within the Nme8/Nme9 subtree is in contrast inconsistent with the above hypothesis. The possibility that prochordate, teleost, and amphibian Nme8 proteins would be more closely related to mammalian Nme9 proteins than to mammalian Nme8 proteins can however be ruled out by the highly conserved exon-intron structure (Fig. 4D) and domain organization (Fig. 2) of the Nme8 gene among chordates. Altogether, these results clearly indicate that Nme9 belongs to the Group II of the Nme proteins. Given that Nme9 gene could only be found in eutherians our data suggest that Nme9 arose from a duplication event that occurred after the separation of eutherian and metatherian groups.
Vertebrate Nme proteins of the Group I
In mammals, the Group I Nme is composed of Nme1, Nme2, Nme3 and Nme4 and orthologs could be identified in both anole lizard and chicken. The situation is in contrast much more complex for amphibians, teleosts, lamprey and prochordates as discussed below.
Gnathostome Nme3 and Nme4 originate from an Nme3/4 vertebrate ancestor
In Xenopus tropicalis, as well as in all studied teleost, orthologs of amniotes Nme3 and Nme4 proteins could be identified (Fig. 1). The phylogenetic analysis of Group I Nme proteins reveals a strongly supported divergence of Nme4 from other Nme of the Group I (Fig. 1). At the amino-acid level, Nme4 proteins exhibit sequence identities ranging from 40.2 to 85.1% among vertebrates [See Additional file 2]. Nme4 protein domain structure is also very well conserved between human and zebrafish as the domain size is equal in both species (130 aa) even though some minor differences exist in pre- and post-domain length (Fig. 2). Similarly, Nme4 exon-intron structure is also very well conserved in Xenopus, zebrafish and human, and differences only concern exon size in the pre-domain coding region (Fig. 3A). The phylogenetic analysis also suggests that Nme3 proteins are divergent from Nme1/Nme2 (Fig 1). Nme3 proteins display sequence identities ranging from 58.4 to 84.1% among vertebrates [See Additional file 2]. The Nme3 protein domain structure (Fig. 2) is identical in humans and zebrafish. Similarly an identical exon-intron structure (Fig. 3A) was observed in Xenopus tropicalis, human and zebrafish nme3 genes. Together, these observations strongly suggest that despite the low support values of the Nme3 branch on the phylogenetic tree (Fig. 1), orthologs of mammalian Nme3 proteins can be found in teleosts and amphibians. This conclusion is further supported by the phylogenetic analysis carried out using all available teleost Nme sequences regardless of the genome sequencing status of the species [See Additional file 4] in which high bootstrap values support the Nme3 branch.
In contrast to teleosts, amphibians and mammals, only one Nme3/Nme4-related sequence could be found in lamprey. Interestingly, the phylogenetic analysis suggests that this sequence is related to both Nme3 and Nme4 groups (Fig. 1). The exon-intron structure of this Nme3/Nme4-related lamprey gene reveals similarities with both Nme3 and Nme4 genes (Fig. 3A). Interestingly, when adding non-coding and coding parts, the size of the second exon of the lamprey Nme3/Nme4-related gene is exactly the same as the size of the second exon of Xenopus Nme3, zebrafish Nme3, human Nme3, and zebrafish Nme4. It should also be noted that for both Nme3 and Nme4, the first intron is inserted after the first base of a codon. Finally, it is noteworthy that Nme3 and Nme4 genes are always located on the same chromosome (Table 1) at very close locations in mammals, chicken, Xenopus and teleosts. Altogether, these observations suggest that, in the vertebrate ancestor, for whom the lamprey is the most closely related descendant, only one Nme3/Nme4-related gene existed. We hypothesize that this ancestor Nme3/Nme4 gene gained a start codon in the first exon after the separation of cyclostomes and gnathostomes lineages. Nme3 and Nme4 subsequently arose from a cis-duplication of this gene that occurred before or around teleost radiation. The Nme3/Nme4-related gene found in lamprey was thus named Nme3/4 to reflect its phylogenetic relationship with Nme3 and Nme4 genes.
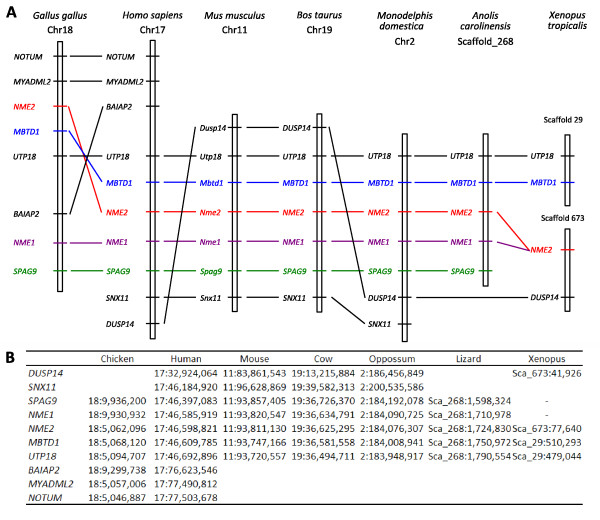
An amniote specific cis-duplication of Nme1/2 ancestor gene
In contrast to Nme3 and Nme4, orthologs of both human NME1 and NME2 can only be found in amniotes and form two clusters corresponding to Nme2 and Nme1 proteins respectively (Fig. 5A). In Xenopus tropicalis and lungfish (Protopterus dolloi), only one Nme1/Nme2-related protein was identified as shown by the phylogenetic analysis. No Nme1-like cDNA was found among the 1.2 million Xenopus tropicalis ESTs available in public databases (August 2009). Within amniotes, Nme1 and Nme2 are always located on the same chromosome (Table 1). Furthermore, in mammals and lizard, Nme1 and Nme2 are always located next to each other (Fig. 6). In addition, the synteny analysis of Nme1 and Nme2 in tetrapods demonstrated that conserved genes in the vicinity of human NME1 and NME2 genes could be identified among all studied amniote species (Fig. 6). In chicken, we hypothesize that a chromosomal inversion of the chromosomic part located between Nme1 and Myadl2 resulted in the separation of the two genes. In amniotes, Nme2 and Nme1 are always linked to Mbtd1 and Spag9. In Xenopus tropicalis, the synteny conservation in the vicinity of Nme2 is less clear (Fig. 6). Nevertheless, note that Dusp14 is in the vicinity of Nme2 among all tetrapods with the exception of chicken and anole lizard. Altogether, these observations suggest that, in all studied amniote species, Nme1 and Nme2 are co-orthologs of Xenopus tropicalis Nme1/Nme2-related gene, and that a cis-duplication event of the ancestor gene occurred before or around amniote radiation. This observation is in total agreement with the conclusions made by Ishikawa and coworkers [21] indicating that rat and human NME1 and NME2 resulted from a cis-duplication of a common ancestor gene. This is also consistent with the previously made hypothesis of a duplication of the ancestor gene that occurred after the separation of tetrapods and fish lineages and after amphibians and amniotes divergence [18,20]. However, we cannot rule out that the cis-duplication of Nme1/Nme2-ancestor gene occurred before amphibian radiation. In that case, the duplication would have been followed by the loss of Nme1 in amphibians. However, no trace of an Nme1 gene could be found on Xenopus tropicalis genomic sequence between Nme2 and Dusp4 genes (Fig. 6). This observation would thus be in favor of the hypothesis of duplication of the Nme1/Nme2 ancestor gene after amphibian radiation.
Figure 5.
Phylogenetic analysis of Nme1 and Nme2 proteins. Tetrapods Nme1 and Nme2 (A) and teleost Nme2 (B) phylogenetic trees were constructed from separate multiple alignments. Bootstrap values for neighbor-joining, maximum parsimony, and maximum likelihood methods, respectively, are indicated for each node. * indicates that the node does not exist in the corresponding tree. The consensus tree was calculated with the FIGENIX automated phylogenomic annotation pipeline [87]. For each sequence, accession number and species name are shown.
Figure 6.
Conserved synteny around Nme1 and Nme2 loci in tetrapods. The syntenic relationships of genes in the vicinity of human NME1 and NME2 linked genes were established using CASSIOPE. For clarity reasons, only selected genes are shown. (A) Graphical view of syntenic relationships in chicken, human, mouse, cow, opossum, and anole lizard Nme1 and Nme2 loci vicinity. Only one Nme2 was found in Xenopus tropicalis (B) Location of each marker on the corresponding numbered chromosome or scaffold.
Mammalian Nme2 is most closely related to the Nme1/Nme2 ancestor gene
Comparison of the primary structure of Nme1 and Nme2 reveals that both proteins are highly conserved among amniotes with mean amino-acid (aa) sequence identities of 83,1% and 88.5% respectively [See Additional file 5]. It is also noteworthy that Nme2 is more conserved than Nme1 among vertebrates. The phylogenetic analysis suggests that both lungfish (Protopterus dolloi) and Xenopus Nme1/Nme2-related proteins would be more closely related to amniote Nme2 than to Nme1 (Fig. 5A). In addition, the exon-intron structure of Xenopus Nme1/Nme2-related gene is highly similar to human NME2 exon-intron structure (Fig. 3A). This highly conserved exon-intron structure is also found in zebrafish (Fig. 3A). In contrast, human NME1 exon-intron structure is different from human NME2 and Xenopus sequences as it exhibits an additional exon at the 5' end of the gene. Together, these observations indicate that NME2 is most similar to the ancestor gene while NME1 exhibits a different exon-intron structure. For this reason, the Xenopus tropicalis Nme1/Nme2-related gene was named Nme2. This name was thus also used for Nme1/Nme2-related genes found in teleosts and lamprey.
The NmeLV form
Using the different sequence databases available in amniotes, a long variant transcript, corresponding to a read-trough transcript of Nme1 and Nme2 genes can be found in human, chimpanzee, horse, cow, platypus, and anole lizard (Table 1). In contrast, this read-through transcript could not be found in chicken in which a chromosomal inversion resulted in the separation of Nme1 and Nme2 genes on the chromosome. Interestingly, the human transcript is composed of the first four exons of NME1 and all NME2 exons (Fig. 3B). To date, the corresponding protein, Nme Long Variant (NmeLV) has only been studied in humans [26] and no information is available in other species.
Nme2a and Nme2b in teleosts probably emerged from 3R genome duplication and Nme2a is most similar to the vertebrate ancestor
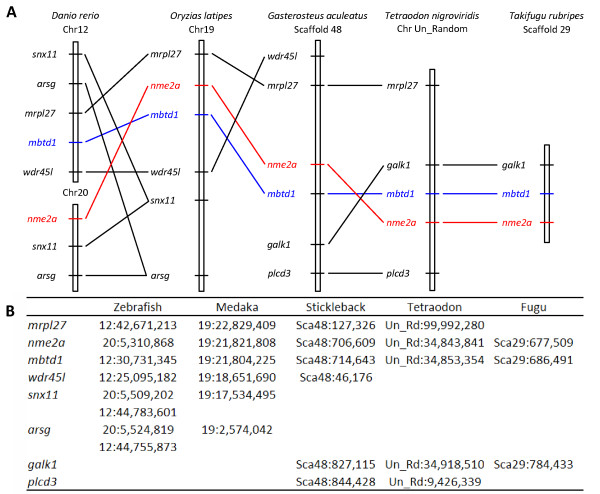
In studied teleost species, the number of Nme1/2-related genes varies from 1 to 3 depending on the species (Fig. 5B). As indicated above, these genes have been named nme2 because they are most similar to the Nme2 gene (Fig. 2 &3A). The phylogenetic analysis revealed that nme2a is present in the five teleost species with complete genome sequence, whereas nme2b genes could not be found in stickleback and tetraodon (Fig. 5B). In contrast, a single Nme2b protein was found in medaka (Oryzias latipes), and fugu (Takifugu rubripes) while, the phylogentic tree clearly indicates a further duplication of the nme2b gene in zebrafish resulting in two distinct proteins termed Nme2b1 and Nme2b2. The phylogenic analysis also suggests that Nme2a and Nme2b are co-orthologs of the lamprey Nme2. This further confirms that the lamprey Nme2 gene could be a direct descendant of the Nme2 ancestor gene (Fig. 5B). In addition, zebrafish Nme2a, Nme2b1, and Nme2b2 have exactly the same protein domain structure, with the same total length and the same NDPk_1 domain located at the same position (Fig. 2). Similarly, zebrafish nme2a, nme2b1, and nme2b2 have exactly the same coding exon structure (Fig. 3A). As previously indicated, the exon-intron structure is well conserved among vertebrate Nme2 genes and clearly distinct from the Nme1 gene. Conserved genes in the vicinity of nme2a gene in teleosts were identified among studied species by a synteny conservation study (Fig. 7). For medaka, stickleback (Gasterosteus aculeatus), tetraodon (Tetraodon nigroviridis), and fugu, the synteny is well conserved and the mbtd1 gene was found in the vicinity of the nme2a gene in agreement to what is observed in tetrapods (Fig. 7). Interestingly, Nakatani et al [27], demonstrated that medaka chromosome 19, on which is located nme2a, is orthologous to a part of human chromosome 17, on which NME1 and NME2 are located. In addition, the primary structure appears to be more conserved for Nme2a in comparison to Nme2b as they display 73.9 and 67.7% mean aa identities respectively [See Additional file 5]. Altogether, these observations suggest that among teleost nme2 genes, nme2a is most similar to the ancestor gene. In teleost, the nme2b gene was not found in tetraodon and stickleback, thus indicating a possible loss of this gene in both species. Furthermore, for all studied teleosts displaying nme2a and nme2b, the two paralogous genes are always located on different chromosomes or scaffolds (Table 1). Interestingly, the fugu nme2b gene is associated to a paralog of mbtd1 (data not shown), suggesting that the duplication event from which nme2a and nme2b arose in teleost is linked to the teleost-specific third round of whole genome duplication (3R). The phylogenetic analysis performed using all available Nme2 sequences in teleosts [See Additional file 6] would be in favor of this hypothesis as numerous other teleost species from different genders such as seabream (Sparus aurata), pike (Esox lucius), seabass (Dicentrarchus labrax), black cod (Anoploma fimbria), and grouper (Epinepheles coioides) exhibit nme2a and nme2b genes. Finally, it is noteworthy that, in contrast to nme2, gene duplicates resulting from 3R whole genome duplication were not retained for other teleost nme genes.
Figure 7.
Conserved synteny around the nme2a locus in teleost fish. Syntenic relationships of genes in the vicinity of nme2a gene was established using Ensembl orthology informations [83]. (A) Graphical view of syntenic relationships in zebrafish, medaka, stickleback, tetraodon and fugu nme2a locus vicinity. (B) Location of each marker on the corresponding numbered chromosome or scaffold.
nme2b1 and nme2b2 emerged from a cis-duplication of nme2b
In contrast to nme2a, very little information is available on the position of nme2b genes in teleosts as they are all located on scaffolds. In zebrafish, it should nevertheless be noted that nme2b1 and nme2b2 genes are located in tandem on the same scaffold (Table 2). This suggests a cis-duplication event of zebrafish nme2b ancestor gene from which nme2b1 and nme2b2 genes arose.
The Nme gene repertoire in the vertebrate ancestor
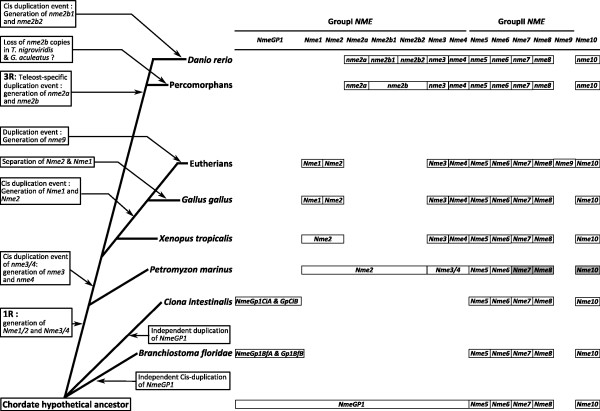
In order to better characterize the putative Nme gene repertoire of the vertebrate ancestor, we have analyzed Nme-related sequences available in the two prochordate Ciona intestinalis and Branchiostoma floridae. As discussed above, orthologs for Nme5, Nme6, Nme7, Nme8 and Nme10 could be identified, thus indicating that these genes emerged before chordate radiation (Fig. 1). Concerning Group I Nme, two sequences could be found in both species. In the lancelet, the genome second assembly available from the Joint Genome Institute [28], clearly shows that only two Group I Nme genes are present in the lancelet genome. The phylogenetic analysis (Fig. 5A), clearly indicates that the two lancelet sequences are closely related to each other but clearly divergent from Ciona intestinalis, lamprey and tetrapod Nme1/Nme2 sequences. Similarly, the two Ciona intestinalis sequences are closely related to each other but highly divergent from other Nme1/Nme2 sequences. In this species, both genes are located on different chromosomes whereas in the lancelet they are located in tandem on the same chromosome. Altogether, these observations suggest that the Group I Nme gene pair arose from a cis-duplication of an ancestor gene in lancelet, whereas emergence of the two Group I Nme genes in ciona is more likely to be explained by a duplication followed by a translocation event. We thus hypothesize that in each species, the two genes result from an independent duplication event of an ancestor gene common to all chordates. This would be consistent with the number of Group I Nme genes in lamprey, as generation of Nme2 and Nme3/4 can be explained by the first round of whole genome duplication (1R) which occurred early in the vertebrate lineage [27]. The ancestor gene, from which emerged all Group I Nme, was thus named NmeGroupI (NmeGp1) (Fig. 8).
Figure 8.
Schematic depiction of Nme genes repertoire in chordates. For each species, the repertoire of Nme genes is shown with the exception of lamprey in which Nme7, Nme8, and Nme10 could not be found in the genome preliminary assembly. These three genes were thus shaded in grey.
Expression and putative functions of Nme proteins
Nme1/2-related proteins
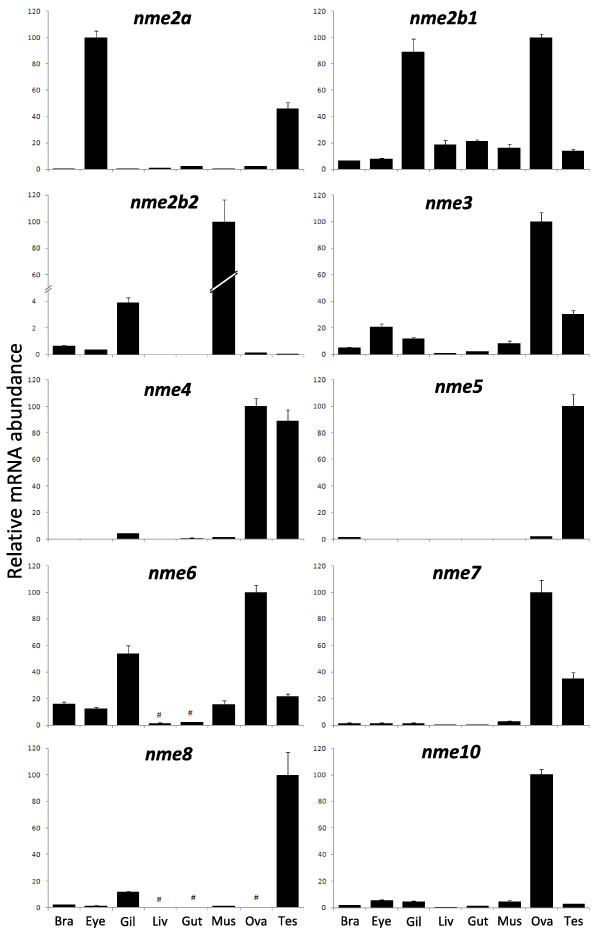
Given its role in metastatic dissemination, the Nme1 protein, has been extensively studied in humans and rodents [15,24]. A significant amount of data is also available for Nme2 [29]. Homologs of human genes were identified in several vertebrate species, such as rodents [6,30], cow [31], Xenopus laevis [20], zebrafish [19], salmon [18]; and non-vertebrate species such as scallop [32], drosophila [33], Dictyostelium discoideum [5], Myxococcus Xanthus [3], Schizosaccharomyces pombe [34] and various plants [35]. The orthology relationship of these Nme1/2-related proteins with human counterparts was not, however, always thoroughly characterized. Nme1/2-related proteins, as all Group I Nme, display a single NDPk_1 domain (Fig. 2), and various enzymatic assays demonstrated its kinase activity in different species [4,20,30-32,36]. According to our observations (Fig. 9A), the zebrafish Nme2 proteins display all the key residues for enzyme structure and activity [37,38] thus suggesting that Nme2 protein could exhibit a NDPK activity. Nme2 is widely expressed in adult tissues as shown in rat [39] and mouse [40,41]. During mouse embryogenesis, Nme2 protein accumulation is coincident with the functional differentiation of multiple organs [42]. No data are available about tissue expression of Nme2 in adult Xenopus. During Xenopus laevis early development, Nme2 transcripts cannot be detected before mid-blastula transition (MBT) but are expressed in differentiating tissues at later stages, thus suggesting an implication in cell differentiation and proliferation [17]. Our tissue distribution study has shown that the three nme2 zebrafish genes have very different tissue expression patterns (Fig. 10). In a previous study, an nme2 homolog was cloned in zebrafish [19]. This transcript, initially named nme23-b, corresponds to nme2b1 and was found to be expressed in hepatopancreas, head, ovary, and intestine by northern blot analysis. These observations are in total agreement with the broad tissues distribution of nme2b1 with a predominant expression in ovary and gills (Fig. 10) reported in the present study. In contrast to what is observed for nme2b1, zebrafish nme2a and nme2b2 have very specific tissue distributions (Fig. 10). It should however be stressed that, despite the extremely high expression in muscle, nme2b2 is also significantly expressed in all assayed tissues. Similarly nme2a expression is also weakly detected in all tissues in addition to the strong expression observed in eyes and testis. In Atlantic salmon, an nme2-related mRNA, belonging to the nme2a sub-family [See Additional file 6], is highly expressed in brain, and during early development it could not be detected before the end of gastrulation [18]. Altogether, the tissue distribution of the three zebrafish nme2 genes suggests that nme2a and nme2b genes have undergone specialization after duplication of a common ancestor nme2 gene [43]. Interestingly, Cañestro et al [44] recently demonstrated that in the case of the loss of one paralog after a duplication event, the surviving paralog can display combined expression pattern of both paralogs kept in another species. In the light of this conclusion, it would be interesting to study nme2 expression in species that lack the nme2b copy. Human NME2 was first identified as the PuF transcription factor that recognizes a nuclease hypersensitive site (NHE) motif in the c-myc promoter and stimulates transcription [29,45,46]. NME2 transcriptional activation of c-myc gene by binding to its promoter was confirmed in mouse [47] and Xenopus laevis [20]. Furthermore, Awd, the drosophila NME2 homolog, is required for proper differentiation and tissues morphology [12]. Thus, NME2 expression pattern during embryogenesis is consistent with implication in cell proliferation and differentiation. In addition, human NME2 may associate with estrogen receptor-β and is able to modulate estrogen-induced gene transcription [48]. Implication of NME2 in regulation of gene expression has also been demonstrated for other genes implicated in several biological processes including nuclease activity (for review see [49]). Altogether, available data suggest that vertebrates Nme2 proteins are involved in a wide variety of cellular processes that require further investigations.
Figure 9.
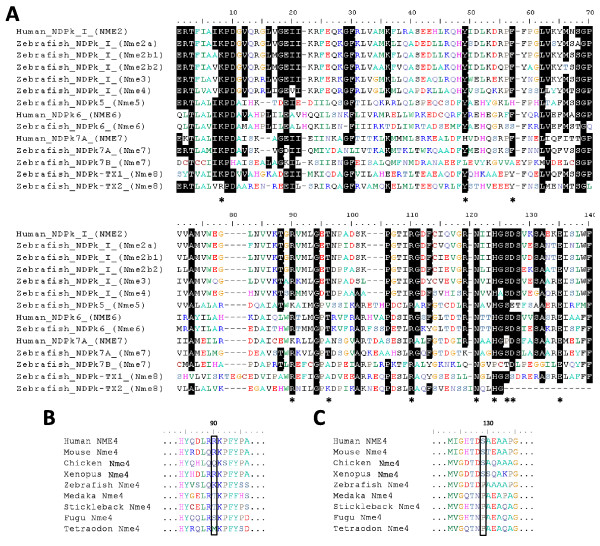
Sequence alignment of zebrafish and human Nme protein domains and highlight on Nme4 specificity. (A) Nme protein domains were identified using NCBI Conserved Domain Database [23]. Domains were aligned using MUSCLE [86], and graphic view was generated using BioEdit V. 7.0.9 software. Residues identified as important for kinase function catalytic mechanism are indicated by an * according to X-ray structure information on human NME2 [37] and Lascu and Gonin review [38] on the catalytic mechanism of NDP Kinase. (B) Mitochondrial membrane linkage triad with central Arginine90. (C) Proline to Serine mutation restricted to tetrapods. Sequences accession numbers are listed in Tables 1 and 2.
Figure 10.
Tissue distribution of zebrafish nme mRNAs. Tissue expressions of zebrafish nme2a, nme2b1, nme2b2, nme3, nme4, nme5, nme6, nme7, nme8, and nme10. Real-time PCR analysis was conducted using total RNA originating from the following tissues sampled in three different sexually-mature females: brain (Bra), eyes (Eye), gills (Gil), hepatopancreas (Liv), intestine (Gut), muscle (Mus), ovary (Ova), and three different mature males: testis (Tes). For each tissue, three separate reverse transcription (RT) reactions were carried out using separate RNA samples originating from three different fish. RT reactions were pooled and use to run real-time PCR in quadruplicates. Mean and SD are shown (n = 4). #, Expression levels not significantly different from background signal at p < 0.05. For all genes, relative abundance is expressed in percentage of highest tissue expression after signal normalization by 18S gene expression.
Nme3
The Nme3 protein has been characterized in humans [50-52] and mice [53]. Nme3, as all the proteins of the Group I, displays a single NDPk_1 domain (Fig. 2). In humans, enzymatic activity could not be measured using the full length recombinant protein [36], but a truncated recombinant protein displayed kinase activity similar to that of the NME1 and NME 2 proteins [52]. We show here that zebrafish Nme3 possesses all the residues necessary for enzyme structure and activity [37,38] (Fig. 9A). Together, these observations would suggest an NDPk activity of the zebrafish Nme3. Zebrafish tissue distribution analysis showed that nme3 is expressed in all studied tissues with the strongest expression in the ovary, and a lower, but significant, expression in testis, eye and gills (Fig. 10). To our knowledge, the strong ovarian expression of nme3 has never been reported in vertebrates in a non-malignant context. In contrast, existing data indicate that human NME3 is ubiquitously expressed in non-metastatic tissues with a particularly strong expression in specific structures of the brain [15]. During mouse organogenesis, Nme3 is preferentially expressed in the nervous and sensory system [54], whereas in adult mouse, transcripts are found ubiquitously distributed with higher expression in brain and liver [53]. During Xenopus laevis embryogenesis, it was shown that Nme3 was predominantly expressed in the head region [55]. To date, very little is known about NME3 function in a non-malignant context. It was shown that over-expression of NME3 gene in 32Dc13 peripheral blood cells inhibited differentiation into granulocytes and caused apoptosis [50], without requiring NDPk enzymatic activity [56]. In addition, it was shown that NME3 induces morphological changes associated with neural differentiation in neuroblastoma cells [57] and that it could act on cell motility by enhancing the amount of integrin β [58]. In the Xenopus laevis it was shown that Nme3 was highly expressed in the ciliary marginal zone of the retina and involvement of Nme3 in cell fate determination during retinogenesis was therefore suggested [55]. It was also shown that NME3 was an estrogen-responsive gene in the context of mammary tumors [59]. To date, no information is available on the physiological or cellular functions of Nme3 in teleosts. However, an implication in cell differentiation, proliferation and apoptosis can be hypothesized.
Nme4
Nme4 protein has been characterized in humans [60], mouse [53], pigeon [61] and Xenopus laevis [55]. Nme4, as all Group I Nme, is composed of a single NDPk_1 domain (Fig. 2). Zebrafish Nme4 possesses all the residues necessary for enzyme structure and kinase activity [37,38] (Fig. 9A). In humans, the enzymatic activity of NME4 was experimentally confirmed [36,62]. As reported here (Fig. 9C), all studied Nme4 tetrapod proteins naturally display a serine residue at position 129, equivalent to the lethal Killer of prune (K-pn) mutation of the drosophila [12]. It was previously shown that the presence of Serine129 residue has local structural effects that weaken subunit interactions and decreases hexamer stability [62]. Strikingly, teleost Nme4 sequences do not display the Serine129, but display the Proline129 shared by all other Group I Nme members (Fig. 9C). The presence of this mutation in tetrapod proteins that cannot be found in any studied teleost species suggests that this mutation appeared just after the sarcopterigian radiation. It was recently shown that human NME4 binds the inner mitochondrial membrane and couples nucleotide transfer with respiration [63]. The binding property to mitochondrial membranes is due to electrostatic interactions between the central Arginine90 of a triad of basic residue and anionic phospholipids [63]. A basic residue equivalent to Arg90 can also be found in mouse, Xenopus tropicalis and zebrafish Nme4 (Fig. 9B). Tetraodon Nme4 possesses a hydrophobic methionine and might be able to electrostatically interact with anionic phospholipids too. In contrast, chicken and other studied teleost Nme4 sequences display a hydrophilic residue in position 90. This could suggest that these Nme4 are unable to interact with anionic phospholipids. It has been shown that pigeon Nme4, also displaying a hydrophilic 90-residue, is located in the mitochondrial matrix [61]. Many functions such as nucleotide supply, functional interactions with Krebs cycle succinyl thiokinase, catabolism of short chain fatty acids [64,65] and, more recently, GTP synthesis in relationship with iron homeostasis [66] have been proposed. In the present study, we report that zebrafish nme4 is highly and predominantly expressed in gonads, weakly expressed in gills, and barely detectable in other studied tissue (Fig. 10). In contrast, human NME4 was shown to be widely distributed and expressed in a tissue-dependant manner with a moderate expression in liver, muscle and ovary and a low expression in testis and brain [60]. In mouse, Nme4 was only detectable in heart, liver and kidney [53]. In Xenopus laevis, Nme4 is predominantly expressed in the head region and an indirect regulation of retinal gliogenesis by Nme4 was demonstrated [55]. The gonad-predominant expression of nme4 reported here, if confirmed in other teleost species, could suggest a different function of fish Nme4 in gonads in comparison to mammalian Nme4. However, a Relative Rate Test [67] did not reveal a significantly different evolutionary rate between tetrapods and fish (p = 0.70). This suggests that observed differences in expression patterns reported above are not linked to different evolutionary rates.
Nme5
Nme5 sequences have been characterized in humans [68] and mouse [69]. The zebrafish Nme5, as human NME5, is composed of an NDPk5 domain followed by a Dpy-30 domain (Fig. 2). In agreement with previous observations made in human and mouse [69], the zebrafish NDPk5 domain also lacks three of the eleven residues deemed crucial for enzyme structure and activity [37,38] (Fig. 9A). The lack of kinase activity was confirmed using human recombinant proteins [36,68]. However, a pronounced 3'→ 5' exonuclease activity was measured for human NME5 [36]. In zebrafish, nme5 was predominantly expressed in testis and detected at low levels in brain and ovary (Fig. 10). Our results are in total agreement with data obtained in humans [68] and mouse [69] in which a predominant testis expression was observed. Low expression levels were also detected in human brain and kidney [68] while a low expression of the mouse transcript was detected in ovary, heart, kidney, and brain [69]. In human testis, NME5 gene expression is located in spermatogonia and early spermatocytes [68], whereas expression appears at pachytene stages in mouse [69]. A marked delay in protein expression can be observed as Nme5 protein is only found in the flagella of spermatids and spermatozoa, adjacent to the central pair and outer doublets of axonemal microtubules [70]. Functionally, murine Nme5 protein might be involved in late spermiogenesis by increasing the ability of late-stage spermatids to eliminate reactive oxygen species [69,71]. Together, our observations suggest that, within Group II, the Nme5 protein of vertebrates probably lacks NDPK activity and might have evolved towards testicular functions, possibly in germ cells.
Nme6
To date, NME6 has only been sequenced and characterized in humans [72,73]. Zebrafish Nme6 displays a single NDPk6 domain, also found in the human protein [72,73] (Fig. 2). In contrast to human NME6, the zebrafish Nme6 lacks one of the eleven residues deemed crucial for enzyme structure and activity, i.e. Phenylalanine58, but display a Phe in position 59 [37,38](Fig. 9A)., Using E. coli recombinant proteins, it was shown that human NDPk6 domain exhibited a kinase activity [73]. This observation was, however, not confirmed in another study [36]. Zebrafish nme6 is expressed in all studied tissues apart from hepatopancreas and intestine, and the highest expression levels were observed in ovary and gills (Fig. 10). Our results are consistent with previous RT-PCR results showing that NME6 was expressed in every human tissue, with strongest expression in ovary/placenta, muscle and intestine [72,73]. Very little is known about NME6 function or expression in a non-malignant context. However, it has been hypothesized that NME6 protein was partially colocalized with mitochondria and that over expression in SAOS2 cells resulted in growth suppression and generation of multinucleated cells. Thus, NME6 may play a role in regulation of cell growth and cell cycle progression [73]. All together, our results suggest that zebrafish Nme6 could possess kinase activity and might have conserved a crucial role in cell cycle, growth or development.
Nme7
To date, very little is known about human NME7 [15]. The zebrafish Nme7, as human NME7, contains a DUF1126 domain, belonging to the DM10 family, and an NDPk_7A and an NDPk_7B domain (Fig. 2). Very little is known about the function of DUF1126 domain and its DM10 family. However, it was suggested that this domain family may act as flagellar NDPk regulatory modules or as units specifically involved in axonemal targeting or assembly [74]. In contrast to the human NDPk_7A domain, the zebrafish domain displays all the residues deemed crucial for enzyme structure and activity [37,38] (Fig. 9A). In addition, human and zebrafish NDPk_7B domain respectively lack 3 and 5 residues deemed crucial for enzyme structure and activity [37,38](Fig. 9A). Yoon et al [36] confirmed the lack of kinase activity in human NME7 but reported a marked exonuclease activity. Zebrafish nme7 is predominantly expressed in gonads and only a weak expression can be found in other studied tissue (Fig. 10). Our results are consistent with human NME7 expression which is predominantly expressed in testis and expressed at significant levels in ovary and brain [15].
Nme8 and Nme9
To date, Nme8 protein has only been described in humans and mice and was called SPTRX2 for its resemblance with another protein, SPTRX1, also displaying a thioredoxin domain [25,75]. An orthologous gene was also characterized in Ciona intestinalis [76]. Proteins of this family are made of one thioredoxin domain (TRX_NDPK) followed by three tandemly repeated NDP kinase domains (NDPk_TX) (Fig. 2). Nme8 protein domain structure is very well conserved between human and zebrafish, with the exception of the third zebrafish NDPk-TX domain which is truncated. NME9 protein was also only described in humans [77] and displays a thioredoxin domain associated to a NDPk_TX domain (Fig. 2). Despite their thioredoxin domain, no thioredoxin activity, corresponding to a general protein-disulfide reductase, could be detected neither in Nme8 [24] nor in Nme9 [77]. Our results also show that the zebrafish NDPk_TX domains lacks crucial amino acids for kinase activity [37,38] (Fig. 9A) and are consistent with several enzymatic studies [25,36,77]. Similarly to NME5 and 7, human NME8 exhibits exonuclease activity [36]. Zebrafish nme8 is highly and predominantly expressed in testis and significantly detected in gills in comparison to all other tissues (Fig. 10). This observation is in complete agreement with existing data in mammals [15,25,75]. As previously reported, NME8 protein have domain arrangement similarities with sea urchin IC1, a member of the dynein intermediate chain [25,76,78]. The functional implication of NME8 in sperm axonemal organization was suggested [75,76] and key role of NME8 in flagellar anomalies and primary ciliary dyskinesia was disclosed [79]. Human NME9 was also described as highly expressed in testis but also in lung and other ciliated cell containing tissue and able to associate with microtubules [77]. Together, these observations suggest that zebrafish Nme8 might also be implicated in testicular function, possibly in axonemal organization.
Nme10
Nme10, also called XRP2, is the most recently described member of the Nme family and was only characterized in human and mouse [80]. Vertebrate Nme10 proteins display a TBCC (Tubulin-specific chaperone protein co-factor C) domain and a partial NDPk domain (Fig. 2). The TBCC domain acts as a GTPase activating protein (GAP) for β-tubulin [24]. The zebrafish partial NDPk domain lacks many crucial amino acids for kinase activity, in particular the catalytic histidine [37,38]. The lack of NDPk activity in human NME10 was confirmed by enzymatic assay [80]. Similarly to NME5, 7 and 8, NME10 exhibits exonuclease activity [80]. Zebrafish nme10 is predominantly expressed in the ovary and only a weak expression can be found in other studied tissue (Fig. 10). In humans and mice, Nme10 was found to be expressed in a wide variety of tissues [81]. Strong ovarian expression was however never reported as no study used ovarian tissue to study Nme10 expression. In humans, mutation in the NME10 gene induce Retinitis Pigmentosa, the major form of heritable blindness [80]. Interestingly, the partial NDPk domain of NME10 protein may have important function as most disease-related mutations of the NME10 gene concern this part of the protein [81]. Furthermore, the human NME10 protein, shown to be mainly located into the cytoplasm, undergoes re-localization into the nucleus when cells are treated with DNA damaging agent inducing oxidative stress, thus suggesting a participation in DNA repairing reactions [80]. The roles of Nme10 in fish and all other non-mammalian species are currently unknown and deserve specific studies. The ovarian-predominant expression, if confirmed in other species, is rather intriguing as it could suggest a major role of Nme10 in oogenesis.
Conclusion
In the present study, we provide a comprehensive overview of the evolutionary history of the Nme family in vertebrates (Fig. 8). We also provide a characterization of the Nme gene repertoire in several vertebrate species including non-mammalian species and propose a gene nomenclature that is consistent with existing mammalian nomenclature. Our observations show that vertebrate Nme genes can be separated in two evolutionary distinct groups. Nme1, Nme2, Nme3, and Nme4 belong to the Group I while vertebrate Nme5, Nme6, Nme7, Nme8, and Nme9 belong to the Group II. The position of Nme10 in the Group II is in contrast more debatable due to its very specific evolutionary history and the recent incorporation of an NDPk domain, before or around the gnathostome radiation. The present study clearly indicates that Nme5, Nme6, Nme7, and Nme8 originate from duplication events that occurred before the chordate radiation. Finally, we show that Nme9 is a mammalian-specific protein closely related to Nme8 that arose from the cis-duplication of the Nme8/Nme9 ancestor gene after the separation of eutherians and metatherians. In contrast to the Group II, Nme genes of the Group I have a totally different evolutionary history. Our observations suggest that a single Group I gene ancestor was present in the chordate ancestor genome. The first round of whole genome duplication (1R) then resulted in two distinct genes named Nme2 and Nme3/4 that can be found in the lamprey genome. In contrast, no duplicates seem to have been retained after the second round of whole genome duplication (2R). We provide evidence that the Nme3/4 gene was cis-duplicated, thus resulting in Nme3 and Nme4 genes that can be found in all investigated gnathostome genomes. Our analyses also suggest that the Nme1 gene found in mammals, chicken and lizard results from the duplication of the Nme2 gene that occurred after amphibian radiation. In teleosts, the third round of whole genome duplication (3R) resulted in the apparition of two paralogous genes, nme2a and nme2b. While nme2a could be found in all teleost genomes, nme2b underwent different fates depending on the species. Finally, based on protein structure and tissue expression of zebrafish nme genes, we provide new insights in tissue specificity and molecular functions of Nme proteins in vertebrates and raise intriguing questions on the role of Nme protein in the vertebrate gonads.
Methods
Sequence analysis
All Nme sequences were identified using the following genome assemblies: zebrafish (Danio rerio, Assembly ZV7), medaka (Oryzias latipes, Assembly MEDAKA1), stickleback (Gasterosteus aculeatus, Assembly BROAD S1), tetraodon (Tetraodon nigroviridis, Assembly V.7), fugu (Takifugu rubripes, Assembly V.4), Xenopus (Xenopus tropicalis, Assembly V.4.1), anole lizard (Anolis carolinensis, AnoCar1.0 Assembly), chicken (Gallus gallus, Assembly V.2.1), mouse (Mus musculus, Assembly NCBI m37), human (Homo sapiens, Assembly NCBI 36), lamprey (Petromyzon marinus, Preliminary assembly 5.9X), Ciona intestinalis (Assembly V.2.0) and lancelet (Branchiostoma floridae, Assembly V.2.0). A large number of sequences were obtained from NCBI NR database using human or zebrafish protein sequence as a query [82]. When more than one sequence was obtained, the RefSeq and/or the longest one were preferentially selected. When sequences were not available in NR database, BLASTP on Ensembl database [83], BLAT on UCSC Genome Bioinformatics [84,85] and TBLASTN on EST_OTHERS database on Genbank [82] were used. For cow (Bos Taurus, Assembly Btau_4.0), opossum (Monodelphis domestica, Assembly MonDom5) and platypus (Ornithorhynchus anatinus, Assembly Ornithorhynchus_anatinus-5.0) only sequences corresponding Nme1 and Nme2 proteins were searched for. In mammalian species, a read-through transcript over Nme1 and Nme2 genes, named NmeLV, was recently identified [26]. Protein sequences corresponding to this transcript were not kept in the phylogenetic reconstruction as they displayed in their sequence the complete Nme2 protein sequence, thus leading to uninformative additional information. However, sequences from human, chimpanzee (Pan troglodytes), horse (Equus caballus), cow, platypus and anole lizard were found as reported in Table 1. Chromosomal localization of Nme genes was performed using Ensembl genome browser, or with UCSC Genome Bioinformatics BLAT when not available on Ensembl. Sequences for each Nme family were aligned by using MUSCLE [86] with default multiple alignment parameters and identity matrix were obtained with BioEdit 7.0.9 software. Intron-exon structure was obtained through Ensembl database, or, when no information was available, by species genome assembly Blat of protein and RNA sequences to get coding and non-coding intron-exon structure. The protein domain structure of Nme proteins was compared between human and zebrafish using Genbank Conserved Domain Database [23]. Domains defined by GenBank Conserved Domain Database were extracted from total protein sequence and aligned using MUSCLE.
Phylogenetic analyses of Nme proteins
Phylogenetic reconstructions were performed using the automated genomic annotation platform FIGENIX [87]. All protein sequences of the Nme family were added to a single multiple alignment to assess their phylogenetic relationships. Sequence alignment was performed automatically by FIGENIX pipeline using MUSCLE. Alignment of sequences of different length and repeated domains present some difficulties due to domains similarities. Therefore, concerning sequences displaying repeated domains, alignment was performed using the part of the sequence showing the highest homology with sequences displaying a single domain. The sequence alignment used for phylogenetic analysis of the whole family is given in Additional file 7. The pipeline used is based on three different methods of phylogenetic tree reconstruction, i.e. Neighbour Joining, Maximum Parsimony, and Maximum likelihood and a midpoint-rooted consensus tree was built. Bootstrapping was carried out with 1000 replications. Bootstrap values are reported for each method when a node exists as identical in the three trees. However, sometimes a node only exist in one or two methods, and therefore * indicates that this node does not exist in the corresponding tree. The Nme1-Nme2 subtree was removed from the main tree and studied separately between tetrapods and teleosts because of different evolutionary history and high similarities leading to non-usable phylogenetic reconstruction.
Relative Rate Test
For Nme4, a higher evolutionary rate between tetrapods and teleost was hypothesized according to major differences in expression patterns. A Relative Rate Test was therefore performed using the Plasmodium falciparum Nme protein [GenBank: XP_001350376] as an outgroup and using the RRTree software [67]. Input alignment file was generated using MUSCLE. RRTree is a user-friendly program for comparing substitution rates between lineages of protein or DNA sequences, relative to an outgroup. Genetic diversity is taken into account through the use of sequences from several species.
Synteny analysis
The synteny relationships of Nme1 and Nme2 members over tetrapods genomes were analyzed using CASSIOPE (Clever Agent System for Synteny Inheritance and Other Phenomena in Evolution) [88]. Briefly, CASSIOPE integrates two important steps in a single automated process: (1) the phylogeny: orthologous/paralogous genes are determined by the aggregation of three phylogenetic methods using the Figenix plateform [87]. Additionally, phylogenetic information allows reconstruction of the evolutionary history and thereby a more accurate ancestral genome reconstruction (2) a statistical test: CASSIOPE therefore utilizes a specific statistical test to assess the significance of the predicted, conserved gene clusters on chromosomes. CASSIOPE does not perform synteny analysis on Scaffolds. As most teleost nme2 genes are located on Scaffolds, synteny analyses of nme2a and nme2b members in fish was thus conducted manually using Ensembl database putative orthology relationships [89].
Zebrafish tissues sampling
Investigations were conducted according to the international guiding principles for the use and care of laboratory animals and in compliance with French and European regulations on animal welfare (DDSV approval #35-31). Three mature female zebrafish were obtained from the fish rearing facilities at INRA-SCRIBE (Rennes, France), over anesthetized and tissues immediately sampled, snap-frozen in liquid nitrogen and conserved at -80°C until RNA extraction. Testis samples were also obtained from three different males.
Real-Time PCR analyses
For each tissue sample, total RNA was isolated using Tri-Reagent® (Molecular Research Center, Cincinnati, OH) according to the manufacturer's instructions. Reverse transcription (RT) was performed as previously described [90] using 2 μg of RNA for each sample with M-MLV enzyme and Random Primers (Promega, Madison, WI). For each studied tissue, cDNA originating from three individual fish were pooled and subsequently used for real-time PCR. Control reactions were run without reverse transcriptase and used as negative control in the real-time PCR study. Quantitative RT-PCR experiments were performed using an Applied Biosystems StepOnePlus. RT products, including control reactions, were diluted to 1/25, and 4 μl was used for each real-time PCR. All q-RT-PCR reactions were performed in quadruplicates. Real-time PCR was performed using a real-time PCR kit provided with a Fast-SYBR® Green fluorophore (Applied Biosystems) with either 200 or 300 nM of each primer. In order to avoid genomic DNA contamination bias, primers were designed on exon junctions. Primer sequences are listed in Additional file 8. The relative abundance of target cDNA within a sample set was calculated from serially diluted cDNA pool (standard curve) using Applied Biosystem StepOne™ V.2.0 software. After amplification, a fusion curve was obtained to validate the amplification of a single PCR product. The fusion curves obtained showed that each primer pair used was specific of a single nme transcript. Normalization of gene expression by 18S and ef1a resulted in similar results. Before further analysis, real-time PCR data were normalized using 18S transcript abundance in samples diluted to 1/2000 and with 100 nM of each primer. The control reactions were used to calculate background expression level for each gene to identify tissues exhibiting expression levels significantly higher than background.
Authors' contributions
TD performed the experiments, produced the figures and drafted the manuscript. PP participated to the phylogenetic reconstruction and in the writing of the manuscript. JB participated in experiments and data analysis. CF and JB conceived and coordinated the study and participated in the writing of the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Identity matrices for Nme8 and Nme10 among chordates. For Nme8 and Nme10, each protein was compared to all cognate chordates proteins. Multiple alignments were performed with MUSCLE and identity matrices generated by BioEdit 7.0.9 software.
Identity matrices for Nme3 to Nme5 among chordates. For Nme3 and Nme4, each protein was compared to all cognate vertebrate proteins, and to all cognate chordate proteins for Nme5. Multiple alignments were performed with MUSCLE and identity matrices generated by BioEdit 7.0.9 software.
Identity matrices for Nme6 and Nme7 among chordates. For Nme6 and Nme7, each protein was compared to all cognate chordates proteins. Multiple alignments were performed with MUSCLE and identity matrices generated by BioEdit 7.0.9 software.
Phylogenetic reconstruction of the Nme protein family in teleosts. Phylogenetic tree was constructed from a single multiple alignment. Bootstrap values for neighbour joining, maximum parsimony, and maximum likelihood methods, respectively, are indicated for each node. * indicates that the node does not exist in the corresponding tree. The consensus tree was calculated using the FIGENIX [87] automated phylogenomic annotation pipeline. Nme1-Nme2 subtree was removed from the main tree and studied separately. For each sequence, NCBI or Ensembl accession number and species name are shown.
Identity matrices for Nme1 and Nme2 among vertebrates. Fish Nme2, tetrapods Nme1 and tetrapods Nme2 were studied separately. Multiple alignments were performed with MUSCLE and identity matrices generated by BioEdit 7.0.9 software.
Phylogenetic reconstruction of Nme2 proteins in teleosts. Teleost Nme2 phylogenetic trees were constructed from separate multiple alignments. Bootstrap values for neighbor joining, maximum parsimony, and maximum likelihood methods, respectively, are indicated for each node. * indicates that the node does not exist in the corresponding tree. The consensus tree was calculated with the FIGENIX automated phylogenomic annotation pipeline [87]. For each sequence, accession number and species name are shown.
Alignment of chordate Nme proteins. Sequence alignment generated and used by FIGENIX for chordate Nme protein phylogenetic reconstruction.
Primer used for the real-time PCR study. For each target gene, abbreviated names, GenBank accession number of the corresponding zebrafish sequence and primer sequences are shown.
Acknowledgments
Acknowledgements
TD received an INRA - IFREMER PhD fellowship. Authors thank Alexis Fostier for helpful discussions, Frederic Borel for fish rearing, Juan Martin Traverso for zebrafish tissue collection and Olivier Chabrol for his help in using FIGENIX and CASSIOPE softwares.
Contributor Information
Thomas Desvignes, Email: thomas.desvignes@rennes.inra.fr.
Pierre Pontarotti, Email: Pierre.Pontarotti@univ-provence.fr.
Christian Fauvel, Email: Christian.Fauvel@ifremer.fr.
Julien Bobe, Email: Julien.Bobe@rennes.inra.fr.
References
- Krebs HA, Hems R. Some reactions of adenosine and inosine phosphates in animal tissues. Biochim Biophys Acta. 1953;12:172–180. doi: 10.1016/0006-3002(53)90136-X. [DOI] [PubMed] [Google Scholar]
- Berg P, Jok Lik WK. Transphosphorylation between Nucleoside Polyphosphates. Nature. 1953;172:1008–1009. doi: 10.1038/1721008a0. [DOI] [PubMed] [Google Scholar]
- Munoz-Dorado J, Inouye M, Inouye S. Nucleoside diphosphate kinase from Myxococcus xanthus. I. Cloning and sequencing of the gene. J Biol Chem. 1990;265:2702–2706. [PubMed] [Google Scholar]
- Munoz-Dorado J, Inouye S, Inouye M. Nucleoside diphosphate kinase from Myxococcus xanthus. II. Biochemical characterization. J Biol Chem. 1990;265:2707–2712. [PubMed] [Google Scholar]
- Lacombe ML, Wallet V, Troll H, Veron M. Functional cloning of a nucleoside diphosphate kinase from Dictyostelium discoideum. J Biol Chem. 1990;265:10012–10018. [PubMed] [Google Scholar]
- Kimura N, Shimada N, Nomura K, Watanabe K. Isolation and characterization of a cDNA clone encoding rat nucleoside diphosphate kinase. J Biol Chem. 1990;265:15744–15749. [PubMed] [Google Scholar]
- Hoffmann R, Valencia A. A gene network for navigating the literature. Nat Genet. 2004;36:664. doi: 10.1038/ng0704-664. [DOI] [PubMed] [Google Scholar]
- Bult CJ, Eppig JT, Kadin JA, Richardson JE, Blake JA, the Mouse Genome Database Group The Mouse Genome Database (MGD): mouse biology and model systems. Nucl Acids Res. 2008;36:D724–D728. doi: 10.1093/nar/gkm961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUGO Gene Nomenclature Committee at the European Bioinformatics Institute http://www.genenames.org
- Twigger SN, Shimoyama M, Bromberg S, Kwitek AE, Jacob HJ. The Rat Genome Database, update 2007--easing the path from disease to data and back again. Nucleic Acids Res. 2007;35:D658–D662. doi: 10.1093/nar/gkl988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeg PS, Bevilacqua G, Kopper L, Thorgeirsson UP, Talmadge JE, Liotta LA, et al. Evidence for a Novel Gene Associated With Low Tumor Metastatic Potential. J Natl Cancer Inst. 1988;80:200–204. doi: 10.1093/jnci/80.3.200. [DOI] [PubMed] [Google Scholar]
- Biggs J, Tripoulas N, Hersperger E, Dearolf C, Shearn A. Analysis of the lethal interaction between the prune and Killer of prune mutations of Drosophila. Genes & Development. 1988;2:1333–1343. doi: 10.1101/gad.2.10.1333. [DOI] [PubMed] [Google Scholar]
- Hama H, Almaula N, Lerner CG, Inouye S, Inouye M. Nucleoside diphosphate kinase from Escherichia coli; its overproduction and sequence comparison with eukaryotic enzymes. Gene. 1991;105:31–36. doi: 10.1016/0378-1119(91)90510-I. [DOI] [PubMed] [Google Scholar]
- Rosengard AM, Krutzsch HC, Shearn A, Biggs JR, Barker E, Margulies IMK, et al. Reduced Nm23/Awd protein in tumour metastasis and aberrant Drosophila development. Nature. 1989;342:177–180. doi: 10.1038/342177a0. [DOI] [PubMed] [Google Scholar]
- Lacombe ML, Milon L, Munier A, Mehus JG, Lambeth DO. The human Nm23/nucleoside diphosphate kinases. Journal of Bioenergetics and Biomembranes. 2000;32:247–258. doi: 10.1023/A:1005584929050. [DOI] [PubMed] [Google Scholar]
- Kim SY, Ferrell JE, Chae SK, Lee KJ. Inhibition of progesterone-induced Xenopus oocyte maturation by Nm23. Cell Growth & Differentiation. 2000;11:485–490. [PubMed] [Google Scholar]
- Ouatas T, Selo M, Sadji Z, Hourdry J, Denis H, Mazabraud A. Differential expression of nucleoside diphosphate kinases (NDPK/NM23) during Xenopus early development. International Journal of Developmental Biology. 1998;42:43–52. [PubMed] [Google Scholar]
- Murphy M, Harte T, McInerney J, Smith TJ. Molecular cloning of an Atlantic salmon nucleoside diphosphate kinase cDNA and its pattern of expression during embryogenesis. Gene. 2000;257:139–148. doi: 10.1016/S0378-1119(00)00374-7. [DOI] [PubMed] [Google Scholar]
- Lee JS, Lee SH. Cloning and characterization of cDNA encoding zebrafish Danio rerio NM23-B gene. Gene. 2000;245:75–79. doi: 10.1016/S0378-1119(00)00037-8. [DOI] [PubMed] [Google Scholar]
- Ouatas T, Abdallah B, Gasmi L, Bourdais J, Postel E, Mazabraud A. Three different genes encode NM23 nucleoside diphosphate kinases in Xenopus laevis. Gene. 1997;194:215–225. doi: 10.1016/S0378-1119(97)00160-1. [DOI] [PubMed] [Google Scholar]
- Ishikawa N, Shimada N, Takagi Y, Ishijima Y, Fukuda M, Kimura N. Molecular evolution of nucleoside diphosphate kinase genes: Conserved core structures and multiple-layered regulatory regions. Journal of Bioenergetics and Biomembranes. 2003;35:7–18. doi: 10.1023/A:1023433504713. [DOI] [PubMed] [Google Scholar]
- Troll H, Winckler T, Lascu I, Muller N, Saurin W, Veron M, et al. Separate nuclear genes encode cytosolic and mitochondrial nucleoside diphosphate kinase in Dictyostelium discoideum. J Biol Chem. 1993;268:25469–25475. [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, et al. CDD: specific functional annotation with the Conserved Domain Database. Nucl Acids Res. 2009;37:D205–D210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissan M, Dabernat S, Peuchant E, Schlattner U, Lascu I, Lacombe ML. The mammalian Nm23/NDPK family: from metastasis control to cilia movement. Mol Cell Biochem. 2009;329:51–62. doi: 10.1007/s11010-009-0120-7. [DOI] [PubMed] [Google Scholar]
- Sadek CM, Damdimopoulos AE, Pelto-Huikko M, Gustafsson JA, Spyrou G, Miranda-Vizuete A. Sptrx-2, a fusion protein composed of one thioredoxin and three tandemly repeated NDP-kinase domains is expressed in human testis germ cells. Genes Cells. 2001;6:1077–1090. doi: 10.1046/j.1365-2443.2001.00484.x. [DOI] [PubMed] [Google Scholar]
- Valentijn LJ, Koster J, Versteeg R. Read-through transcript from NM23-H1 into the neighboring NM23-H2 gene encodes a novel protein, NM23-LV. Genomics. 2006;87:483–489. doi: 10.1016/j.ygeno.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Nakatani Y, Takeda H, Kohara Y, Morishita S. Reconstruction of the vertebrate ancestral genome reveals dynamic genome reorganization in early vertebrates. Genome Research. 2007;17:1254–1265. doi: 10.1101/gr.6316407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOE Joint Genome Institute http://www.jgi.doe.gov/
- Postel EH, Berberich SJ, Flint SJ, Ferrone CA. Human c-myc transcription factor PuF identified as nm23-H2 nucleoside diphosphate kinase, a candidate suppressor of tumor metastasis. Science. 1993;261:478–480. doi: 10.1126/science.8392752. [DOI] [PubMed] [Google Scholar]
- Urano T, Takamiya K, Furukawa K, Shiku H. Molecular cloning and functional expression of the second mouse nm23/NDP kinase gene, nm23-M2. Febs Letters. 1992;309:358–362. doi: 10.1016/0014-5793(92)80807-S. [DOI] [PubMed] [Google Scholar]
- Abdulaev NG, Karaschuk GN, Ladner JE, Kakuev DL, Yakhyaev AV, Tordova M, et al. Nucleoside Diphosphate Kinase from Bovine Retina: Purification, Subcellular Localization, Molecular Cloning, and Three-Dimensional StructureΓ Çá. Biochemistry. 1998;37:13958–13967. doi: 10.1021/bi980853s. [DOI] [PubMed] [Google Scholar]
- Shi XZ, Zhao XF, Wang JX. Molecular cloning and analysis of function of nucleoside diphosphate kinase (NDPK) from the scallop Chlamys farreri. Biochemistry (Mosc) 2008;73:686–692. doi: 10.1134/S0006297908060096. [DOI] [PubMed] [Google Scholar]
- Biggs J, Hersperger E, Steeg PS, Liotta LA, Shearn A. A Drosophila gene that is homologous to a mammalian gene associated with tumor metastasis codes for a nucleoside diphosphate kinase. Cell. 1990;63:933–940. doi: 10.1016/0092-8674(90)90496-2. [DOI] [PubMed] [Google Scholar]
- Izumiya H, Yamamoto M. Cloning and Functional Analysis of the ndk1 Gene Encoding Nucleoside-diphosphate Kinase in Schizosaccharomyces pombe. J Biol Chem. 1995;270:27859–27864. doi: 10.1074/jbc.270.46.27859. [DOI] [PubMed] [Google Scholar]
- Escobar Galvis ML, Hakansson G, Alexciev K, Knorpp C. Cloning and characterisation of a pea mitochondrial NDPK. Biochimie. 1999;81:1089–1096. doi: 10.1016/S0300-9084(99)00353-3. [DOI] [PubMed] [Google Scholar]
- Yoon JH, Singh P, Lee DH, Qiu J, Cai S, O'Connor TR, et al. Characterization of the 3' - 5' Exonuclease Activity Found in Human Nucleoside Diphosphate Kinase 1 (NDK1) and Several of Its Homologues. Biochemistry. 2005;44:15774–15786. doi: 10.1021/bi0515974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb PA, Perisic O, Mendola CE, Backer JM, Williams RL. The Crystal Structure of Human Nucleoside Diphosphate Kinase, NM23-H2. Journal of Molecular Biology. 1995;251:574–587. doi: 10.1006/jmbi.1995.0457. [DOI] [PubMed] [Google Scholar]
- Lascu I, Gonin P. The catalytic mechanism of nucleoside diphosphate kinases. J Bioenerg Biomembr. 2000;32:237–246. doi: 10.1023/A:1005532912212. [DOI] [PubMed] [Google Scholar]
- Shimada N, Ishikawa N, Munakata Y, Toda T, Watanabe K, Kimura N. A second form (beta isoform) of nucleoside diphosphate kinase from rat. Isolation and characterization of complementary and genomic DNA and expression. J Biol Chem. 1993;268:2583–2589. [PubMed] [Google Scholar]
- Dabernat S, Larou M, Masse K, Hokfelt T, Mayer G, Daniel JY, et al. Cloning of a second nm23-M1 cDNA: expression in the central nervous system of adult mouse and comparison with nm23-M2 mRNA distribution. Brain Res Mol Brain Res. 1999;63:351–365. doi: 10.1016/S0169-328X(98)00300-3. [DOI] [PubMed] [Google Scholar]
- Dabernat S, Larou M, Masse K, Dobremez E, Landry M, Mathieu C, et al. Organization and expression of mouse nm23-M1 gene. Comparison with nm23-M2 expression. Gene. 1999;236:221–230. doi: 10.1016/S0378-1119(99)00288-7. [DOI] [PubMed] [Google Scholar]
- Lakso M, Steeg PS, Westphal H. Embryonic Expression of Nm23 During Mouse Organogenesis. Cell Growth & Differentiation. 1992;3:873–879. [PubMed] [Google Scholar]
- Zhang J. Evolution by gene duplication: an update. Trends in Ecology & Evolution. 2003;18:292–298. doi: 10.1016/S0169-5347(03)00033-8. [DOI] [Google Scholar]
- Canestro C, Catchen JM, Rodriguez-Mari A, Yokoi H, Postlethwait JH. Consequences of lineage-specific gene loss on functional evolution of surviving paralogs: ALDH1A and retinoic acid signaling in vertebrate genomes. PLoS Genet. 2009;5:e1000496. doi: 10.1371/journal.pgen.1000496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berberich SJ, Postel EH. PuF/NM23-H2/NDPK-B transactivates a human c-myc promoter-CAT gene via a functional nuclease hypersensitive element. Oncogene. 1995;10:2343–2347. [PubMed] [Google Scholar]
- Stahl JA, Leone A, Rosengard AM, Porter L, King CR, Steeg PS. Identification of a Second Human nm23 Gene, nm23-H2. Cancer Res. 1991;51:445–449. [PubMed] [Google Scholar]
- Arnaud-Dabernat S, Masse K, Smani M, Peuchant E, Landry M, Bourbon PM, et al. Nm23-M2/NDP kinase B induces endogenous c-myc and nm23-M1/NDP kinase A overexpression in BAF3 cells. Both NDP kinases protect the cells from oxidative stress-induced death. Exp Cell Res. 2004;301:293–304. doi: 10.1016/j.yexcr.2004.07.026. [DOI] [PubMed] [Google Scholar]
- Rayner K, Chen YX, Hibbert B, White D, Miller H, Postel EH, et al. Discovery of NM23-H2 as an estrogen receptor beta-associated protein: role in estrogen-induced gene transcription and cell migration. J Steroid Biochem Mol Biol. 2008;108:72–81. doi: 10.1016/j.jsbmb.2007.07.006. [DOI] [PubMed] [Google Scholar]
- Postel EH, Berberich SJ, Rooney JW, Kaetzel DM. Human NM23/nucleoside diphosphate kinase regulates gene expression through DNA binding to nuclease-hypersensitive transcriptional elements. J Bioenerg Biomembr. 2000;32:277–284. doi: 10.1023/A:1005541114029. [DOI] [PubMed] [Google Scholar]
- Venturelli D, Martinez R, Melotti P, Casella I, Peschle C, Cucco C, et al. Overexpression of DR-nm23, a protein encoded by a member of the nm23 gene family, inhibits granulocyte differentiation and induces apoptosis in 32Dc13 myeloid cells. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:7435–7439. doi: 10.1073/pnas.92.16.7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez R, Venturelli D, Perrotti D, Veronese ML, Kastury K, Druck T, et al. Gene Structure, Promoter Activity, and Chromosomal Location of the DR-nm23 Gene, a Related Member of the nm23 Gene Family. Cancer Res. 1997;57:1180–1187. [PubMed] [Google Scholar]
- Erent M, Gonin P, Cherfils J, Tissier P, Raschella G, Giartosio A, et al. Structural and catalytic properties and homology modelling of the human nucleoside diphosphate kinase C, product of the DRnm23 gene. Eur J Biochem. 2001;268:1972–1981. doi: 10.1046/j.1432-1327.2001.2076.doc.x. [DOI] [PubMed] [Google Scholar]
- Masse K, Dabernat S, Bourbon PM, Larou M, Amrein L, Barraud P, et al. Characterization of the nm23-M2, nm23-M3 and nm23-M4 mouse genes: comparison with their human orthologs. Gene. 2002;296:87–97. doi: 10.1016/S0378-1119(02)00836-3. [DOI] [PubMed] [Google Scholar]
- Amrein L, Barraud P, Daniel JY, Perel Y, Landry M. Expression patterns of nm23 genes during mouse organogenesis. Cell Tissue Res. 2005;322:365–378. doi: 10.1007/s00441-005-0036-9. [DOI] [PubMed] [Google Scholar]
- Mochizuki T, Bilitou A, Waters C, Hussain K, Zollo M, Ohnuma Si. Xenopus NM23-X4 regulates retinal gliogenesis through interaction with p27Xic1. Neural Development. 2009;4:1. doi: 10.1186/1749-8104-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturelli D, Cesi V, Ransac S, Engelhard A, Perrotti D, Calabretta B. The Nucleoside Diphosphate Kinase Activity of DRnm23 Is Not Required for Inhibition of Differentiation and Induction of Apoptosis in 32Dcl3 Myeloid Precursor Cells. Experimental Cell Research. 2000;257:265–271. doi: 10.1006/excr.2000.4899. [DOI] [PubMed] [Google Scholar]
- Negroni A, Venturelli D, Tanno B, Amendola R, Ransac S, Cesi V, et al. Neuroblastoma specific effects of DR-nm23 and its mutant forms on differentiation and apoptosis. Cell Death Differ. 2000;7:843–850. doi: 10.1038/sj.cdd.4400720. [DOI] [PubMed] [Google Scholar]
- Amendola R, Martinez R, Negroni A, Venturelli D, Tanno B, Calabretta B, et al. DR-nm23 gene expression in neuroblastoma cells: relationship to integrin expression, adhesion characteristics, and differentiation. J Natl Cancer Inst. 1997;89:1300–1310. doi: 10.1093/jnci/89.17.1300. [DOI] [PubMed] [Google Scholar]
- Kamalakaran S, Radhakrishnan SK, Beck WT. Identification of Estrogen-responsive Genes Using a Genome-wide Analysis of Promoter Elements for Transcription Factor Binding Sites. J Biol Chem. 2005;280:21491–21497. doi: 10.1074/jbc.M409176200. [DOI] [PubMed] [Google Scholar]
- Milon L, RousseauMerck MF, Munier A, Erent M, Lascu I, Capeau J, et al. nm23-H4, a new member of the family of human nm23 nucleoside diphosphate kinase genes localised on chromosome 16p13. Human Genetics. 1997;99:550–557. doi: 10.1007/s004390050405. [DOI] [PubMed] [Google Scholar]
- Lambeth DO, Mehus JG, Ivey MA, Milavetz BI. Characterization and Cloning of a Nucleoside-diphosphate Kinase Targeted to Matrix of Mitochondria in Pigeon. J Biol Chem. 1997;272:24604–24611. doi: 10.1074/jbc.272.39.24604. [DOI] [PubMed] [Google Scholar]
- Milon L, Meyer P, Chiadmi M, Munier A, Johansson M, Karlsson A, et al. The Human nm23-H4 Gene Product Is a Mitochondrial Nucleoside Diphosphate Kinase. J Biol Chem. 2000;275:14264–14272. doi: 10.1074/jbc.275.19.14264. [DOI] [PubMed] [Google Scholar]
- Tokarska-Schlattner M, Boissan M, Munier A, Borot C, Mailleau C, Speer O, et al. The Nucleoside Diphosphate Kinase D (NM23-H4) Binds the Inner Mitochondrial Membrane with High Affinity to Cardiolipin and Couples Nucleotide Transfer with Respiration. J Biol Chem. 2008;283:26198–26207. doi: 10.1074/jbc.M803132200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs HA, Wiggins D. Phosphorylation of adenosine monophosphate in the mitochondrial matrix. Biochem J. 1978;174:297–301. doi: 10.1042/bj1740297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambeth DO. What is the function of GTP produced in the Krebs citric acid cycle? IUBMB Life. 2002;54:143–144. doi: 10.1080/15216540214539. [DOI] [PubMed] [Google Scholar]
- Gordon D, Lyver E, Lesuisse E, Dancis A, Pain D. GTP in the mitochondrial matrix plays a crucial role in organellar iron homoeostasis1. Biochem J. 2006;400:163–168. doi: 10.1042/BJ20060904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson-Rechavi M, Huchon D. RRTree: Relative-Rate Tests between groups of sequences on a phylogenetic tree. Bioinformatics. 2000;16:296–297. doi: 10.1093/bioinformatics/16.3.296. [DOI] [PubMed] [Google Scholar]
- Munier A, Feral C, Milon L, Pinon VPB, Gyapay G, Capeau J, et al. A new human nm23 homologue (nm23-H5) specifically expressed in testis germinal cells. Febs Letters. 1998;434:289–294. doi: 10.1016/S0014-5793(98)00996-X. [DOI] [PubMed] [Google Scholar]
- Hwang KC, Ok DW, Hong JC, Kim MO, Kim JH. Cloning, sequencing, and characterization of the murine nm23-M5 gene during mouse spermatogenesis and spermiogenesis. Biochemical and Biophysical Research Communications. 2003;306:198–207. doi: 10.1016/S0006-291X(03)00916-1. [DOI] [PubMed] [Google Scholar]
- Munier A, Serres C, Kann ML, Boissan M, Lesaffre C, Capeau J, et al. Nm23/NDP kinases in human male germ cells: role in spermiogenesis and sperm motility? Experimental Cell Research. 2003;289:295–306. doi: 10.1016/S0014-4827(03)00268-4. [DOI] [PubMed] [Google Scholar]
- Choi YJ, Cho SK, Hwang KC, Park C, kim JH, Park SB, et al. Nm23-M5 mediates round and elongated spermatid survival by regulating GPX-5 levels. Febs Letters. 2009;583:1292–1298. doi: 10.1016/j.febslet.2009.03.023. [DOI] [PubMed] [Google Scholar]
- Mehus JG, Deloukas P, Lambeth DO. NME6: a new member of the nm23/nucleoside diphosphate kinase gene family located on human chromosome 3p21.3. Human Genetics. 1999;104:454–459. doi: 10.1007/s004390050987. [DOI] [PubMed] [Google Scholar]
- Tsuiki H, Nitta M, Furuya A, Hanai N, Fujiwara T, Inagaki M, et al. A novel human nucleoside diphosphate (NDP) kinase, Nm23-H6, localizes in mitochondria and affects cytokinesis. J Cell Biochem. 1999;76:254–269. doi: 10.1002/(SICI)1097-4644(20000201)76:2<254::AID-JCB9>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- King SM. Axonemal protofilament ribbons, DM10 domains, and the link to juvenile myoclonic epilepsy. Cell Motil Cytoskeleton. 2006;63:245–253. doi: 10.1002/cm.20129. [DOI] [PubMed] [Google Scholar]
- Miranda-Vizuete A, Tsang K, Yu Y, Jimenez A, Pelto-Huikko M, Flickinger CJ, et al. Cloning and Developmental Analysis of Murid Spermatid-specific Thioredoxin-2 (SPTRX-2), a Novel Sperm Fibrous Sheath Protein and Autoantigen. J Biol Chem. 2003;278:44874–44885. doi: 10.1074/jbc.M305475200. [DOI] [PubMed] [Google Scholar]
- Padma P, Hozumi A, Ogawa K, Inaba K. Molecular cloning and characterization of a thioredoxin/nucleoside diphosphate kinase related dynein intermediate chain from the ascidian, Ciona intestinalis. Gene. 2001;275:177–183. doi: 10.1016/S0378-1119(01)00661-8. [DOI] [PubMed] [Google Scholar]
- Sadek CM, Jimenez A, Damdimopoulos AE, Kieselbach T, Nord M, Gustafsson JA, et al. Characterization of Human Thioredoxin-like 2. A novel microtubule-binding thioredoxin expressed predominantly in the cilia of lung airway epithelium and spermatid manchette and axoneme. J Biol Chem. 2003;278:13133–13142. doi: 10.1074/jbc.M300369200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa K, Takai H, Ogiwara A, Yokota E, Shimizu T, Inaba K, et al. Is outer arm dynein intermediate chain 1 multifunctional? Mol Biol Cell. 1996;7:1895–1907. doi: 10.1091/mbc.7.12.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duriez B+, Duquesnoy P, Escudier E, Bridoux AM, Escalier D, Rayet I, et al. A common variant in combination with a nonsense mutation in a member of the thioredoxin family causes primary ciliary dyskinesia. Proceedings of the National Academy of Sciences. 2007;104:3336–3341. doi: 10.1073/pnas.0611405104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Qiu J, Cai S, Chen Y, Cheetham ME, Shen B, et al. The retinitis pigmentosa-mutated RP2 protein exhibits exonuclease activity and translocates to the nucleus in response to DNA damage. Experimental Cell Research. 2006;312:1323–1334. doi: 10.1016/j.yexcr.2005.12.026. [DOI] [PubMed] [Google Scholar]
- Schwahn U, Lenzner S, Dong J, Feil S, Hinzmann B, van Duijnhoven G, et al. Positional cloning of the gene for X-linked retinitis pigmentosa 2. Nat Genet. 1998;19:327–332. doi: 10.1038/1214. [DOI] [PubMed] [Google Scholar]
- NCBI Basic Local Alignment Search Tool (BLAST) http://blast.ncbi.nlm.nih.gov/Blast.cgi [DOI] [PubMed]
- Hubbard TJP, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, et al. Ensembl 2009. Nucl Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The Human Genome Browser at UCSC. Genome Research. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. BLAT-The BLAST-Like Alignment Tool. Genome Research. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucl Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouret P, Vitiello V, Balandraud N, Gilles A, Pontarotti P, Danchin E. FIGENIX: Intelligent automation of genomic annotation: expertise integration in a new software platform. BMC Bioinformatics. 2005;6:198. doi: 10.1186/1471-2105-6-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rascol V, Levasseur A, Chabrol O, Grusea S, Gouret P, Danchin E, et al. CASSIOPE: An expert system for conserved regions searches. BMC Bioinformatics. 2009;10:284. doi: 10.1186/1471-2105-10-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilella AJ, Severin J, Ureta-Vidal A, Heng L, Durbin R, Birney E. EnsemblCompara GeneTrees: Complete, duplication-aware phylogenetic trees in vertebrates. Genome Research. 2009;19:327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobe J, Nguyen T, Jalabert B. Targeted Gene Expression Profiling in the Rainbow Trout (Oncorhynchus mykiss) Ovary During Maturational Competence Acquisition and Oocyte Maturation. Biol Reprod. 2004;71:73–82. doi: 10.1095/biolreprod.103.025205. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Identity matrices for Nme8 and Nme10 among chordates. For Nme8 and Nme10, each protein was compared to all cognate chordates proteins. Multiple alignments were performed with MUSCLE and identity matrices generated by BioEdit 7.0.9 software.
Identity matrices for Nme3 to Nme5 among chordates. For Nme3 and Nme4, each protein was compared to all cognate vertebrate proteins, and to all cognate chordate proteins for Nme5. Multiple alignments were performed with MUSCLE and identity matrices generated by BioEdit 7.0.9 software.
Identity matrices for Nme6 and Nme7 among chordates. For Nme6 and Nme7, each protein was compared to all cognate chordates proteins. Multiple alignments were performed with MUSCLE and identity matrices generated by BioEdit 7.0.9 software.
Phylogenetic reconstruction of the Nme protein family in teleosts. Phylogenetic tree was constructed from a single multiple alignment. Bootstrap values for neighbour joining, maximum parsimony, and maximum likelihood methods, respectively, are indicated for each node. * indicates that the node does not exist in the corresponding tree. The consensus tree was calculated using the FIGENIX [87] automated phylogenomic annotation pipeline. Nme1-Nme2 subtree was removed from the main tree and studied separately. For each sequence, NCBI or Ensembl accession number and species name are shown.
Identity matrices for Nme1 and Nme2 among vertebrates. Fish Nme2, tetrapods Nme1 and tetrapods Nme2 were studied separately. Multiple alignments were performed with MUSCLE and identity matrices generated by BioEdit 7.0.9 software.
Phylogenetic reconstruction of Nme2 proteins in teleosts. Teleost Nme2 phylogenetic trees were constructed from separate multiple alignments. Bootstrap values for neighbor joining, maximum parsimony, and maximum likelihood methods, respectively, are indicated for each node. * indicates that the node does not exist in the corresponding tree. The consensus tree was calculated with the FIGENIX automated phylogenomic annotation pipeline [87]. For each sequence, accession number and species name are shown.
Alignment of chordate Nme proteins. Sequence alignment generated and used by FIGENIX for chordate Nme protein phylogenetic reconstruction.
Primer used for the real-time PCR study. For each target gene, abbreviated names, GenBank accession number of the corresponding zebrafish sequence and primer sequences are shown.