Abstract
Genetic alterations that activate the mitogen-activated protein kinase (MAP kinase) pathway occur commonly in cancer. For example, the majority of melanomas harbor mutations in the BRAF oncogene, which are predicted to confer enhanced sensitivity to pharmacologic MAP kinase inhibition (e.g., RAF or MEK inhibitors). We investigated the clinical relevance of MEK dependency in melanoma by massively parallel sequencing of resistant clones generated from a MEK1 random mutagenesis screen in vitro, as well as tumors obtained from relapsed patients following treatment with AZD6244, an allosteric MEK inhibitor. Most mutations conferring resistance to MEK inhibition in vitro populated the allosteric drug binding pocket or α-helix C and showed robust (≈100-fold) resistance to allosteric MEK inhibition. Other mutations affected MEK1 codons located within or abutting the N-terminal negative regulatory helix (helix A), which also undergo gain-of-function germline mutations in cardio-facio-cutaneous (CFC) syndrome. One such mutation, MEK1(P124L), was identified in a resistant metastatic focus that emerged in a melanoma patient treated with AZD6244. Both MEK1(P124L) and MEK1(Q56P), which disrupts helix A, conferred cross-resistance to PLX4720, a selective B-RAF inhibitor. However, exposing BRAF-mutant melanoma cells to AZD6244 and PLX4720 in combination prevented emergence of resistant clones. These results affirm the importance of MEK dependency in BRAF-mutant melanoma and suggest novel mechanisms of resistance to MEK and B-RAF inhibitors that may have important clinical implications.
Keywords: BRAF, drug resistance, MAP kinase, melanoma
Approximately one-third of all cancers harbor genetic alterations that aberrantly upregulate mitogen-activated protein kinase (MAPK)-dependent signal transduction (1). In the MAPK pathway, RAS oncoproteins activate RAF, MEK, and ERK kinases to direct key cell proliferative and survival signals. When rendered constitutively active by genetic mutation, the MAP kinase pathway is believed to confer “oncogene dependency” (2), an excessive reliance on its dysregulated activity for tumor viability. Therefore, protein kinases within this signaling cascade offer promising targets for novel anticancer therapeutics.
In melanoma, uncontrolled MAP kinase pathway activity is nearly ubiquitous and occurs most commonly through gain-of-function mutations involving codon 600 of the B-RAF kinase (3) (BRAFV600E; 50–70% of cases). Considerable preclinical evidence has associated the BRAFV600E mutation with heightened sensitivity to pharmacologic inhibition of RAF or MEK kinases (4, 5). Although early clinical trials of RAF and MEK inhibitors failed to show a substantial benefit (6, 7), recent phase I studies of selective RAF inhibitors have shown promising results in patients with BRAF-mutant tumors (8, 9). Thus, optimizing therapeutic efficacy while avoiding or bypassing the emergence of resistance to MAP kinase pathway inhibition will likely gain increasing importance in melanoma and other MAP kinase-driven cancers.
Here, we describe the use of random mutagenesis and massively parallel sequencing to identify mutations within MEK kinase that promote resistance to pharmacologic MEK inhibition. This deep sequencing approach was also leveraged to interrogate melanomas derived from patients treated with the investigational MEK inhibitor AZD6244. Structural and functional characterization of the resulting MEK1 resistance mutations highlighted two major types of resistance to allosteric MEK inhibition, one of which confers cross-resistance to selective B-RAF inhibition. These results may therefore inform a clinical understanding of resistance to MEK and RAF inhibition, as well as approaches to prevent its emergence during targeted treatment.
Results
Comprehensive Identification of MEK1 Resistance Mutations in Vitro.
To begin to characterize resistance to MAP kinase pathway inhibition in the context of BRAFV600E melanoma, we used a random mutagenesis screen (10) together with massively parallel sequencing (11) to discover the spectrum of variants associated with resistance to allosteric MEK inhibition in vitro. We expressed a saturating cDNA library of MEK1 mutations in A375 melanoma cells, which harbor the BRAFV600E mutation and are highly sensitive to MEK inhibition. After culturing these cells for 4 weeks in the presence of a diarylamine MEK inhibitor (12, 13) (either 1.5 μM AZD6244 or 2 μM CI-1040), resistant clones emerged, ≈1,000 of which were pooled and characterized en masse by massively parallel sequencing (SI Text for a full description). An additional 100 clones were sequenced by the Sanger method. A complete set of mutant MEK1 alleles and the methods of identification are listed in Table S1. This combined analysis of ≈1,100 resistant clones vastly exceeded the scope of prior mutagenesis studies (14) and offered a comprehensive framework for unbiased characterization of MEK1-mediated drug resistance.
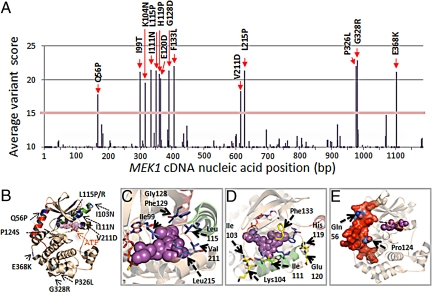
The landscape of MEK1 mutations that emerged in the presence of AZD6244 is shown in Fig. 1A. Similar results were obtained from mutatgenesis experiments using CI-1040 (Fig. S1A). We investigated the distribution of candidate resistance alleles across each MEK1 nucleotide position and mapped the resulting amino acid substitutions within the three-dimensional structure of the full-length MEK1 kinase domain (15) (PDB code: 3EQC) (Fig. 1B; see also Fig. S1B for a larger view).
Fig. 1.
Candidate MEK1 resistance mutations. (A) The average variant score of candidate mutations across the MEK1 coding sequence from the AZD6244 mutagenesis screen (based on one lane of Illumina sequencing) is shown. The corresponding amino acid substitutions from high scoring mutations (>15%) are indicated in bold. (B) The locations of putative MEK1 inhibitor resistance alleles are indicated within the crystal structure of MEK1 (blue). Both ATP and an arylamine MEK inhibitor (PD318088; purple) are shown to bind MEK1; this inhibitor chemotype binds a hydrophobic pocket adjacent to the ATP binding site. Helix C (green) and helix A (red) are indicated. Mutations within the MEK1 allosteric binding pocket (C), along or adjacent to helix C, are shown in green (D) or within/interfacing with helix A, are shown in red as a protein surface (E).
Putative MEK1 resistance alleles segregated into two classes: “primary” mutations clustering within or directly perturbing the allosteric binding pocket and “secondary” mutations that resided outside of the drug binding region. Primary MEK1 resistance alleles could be further subdivided into mutations situated directly within the arylamine binding pocket (e.g., I99T, L115P/R, G128D, F129L, V211D, and L215P; Fig. 1C) and a distinct set located in the second protein shell, along and adjacent to the C helix (e.g., I103N, K104N, I111N, H119P, E120D, F133L; Fig. 1D). Notably, the MEK1 hydrophobic pocket includes residues from both α-helix C and the activation loop. Binding of arylamine inhibitors within this pocket prevents the structural reorganization of α-helix C and other motifs, which generates a catalytically active MEK1 conformation. Thus, primary mutations may cause resistance either by direct interference or through altered C helix conformation.
The secondary class of resistance mutations populated two regions of MEK1. One region includes the N-terminal negative regulatory domain known as helix A (15) (e.g., Q56P) and a proline proximal to the C helix (P124) that abuts helix A (Fig. 1E). P124 mutations were identified either by Sanger sequencing of individual resistant clones (P124S) or by deep sequencing (P124Q), albeit at an average variant score that fell beneath the initial threshold for detection (Table S1). The proximity of Q56 and P124 to the N-terminal negative regulatory domain (Fig. 1 B and E) suggests a resistance mechanism that involves upregulation of intrinsic MEK1 kinase activity. Toward this end, the Q56P and P124S mutations closely mimic T55P and P124L, respectively—two germline variants observed in patients with cardio-facio-cutaneous (CFC) syndrome (15, 16) that confer aberrant MEK activation. The D67N mutation, which was identified in the CI-1040 mutagenesis screen (Fig. S1A), also occurs as a germline CFC variant (17) and has occasionally been detected as a somatic cancer mutation (18). The other secondary region involves the C-terminal kinase domain (e.g., G328R, P326L, and E368K); the functional significance of these mutations is currently unknown. On the basis of these findings, we reasoned that on-target resistance to allosteric MEK inhibition may arise through reduction of drug binding affinity or enhanced intrinsic MEK1 activation.
Functional Validation of MEK1 Resistance Alleles.
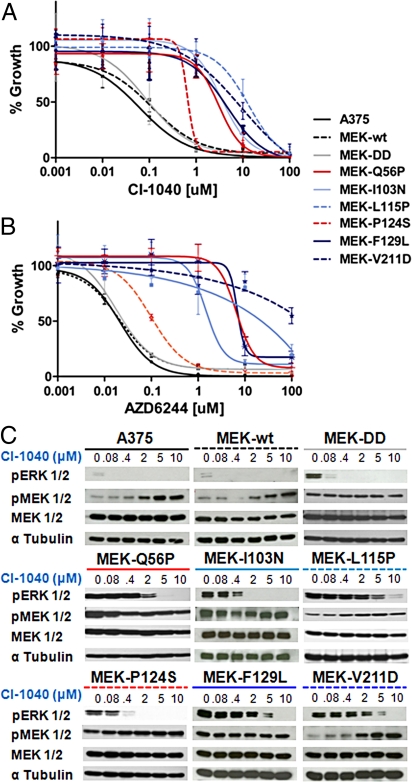
To confirm the functional effects of putative MEK1 resistance alleles identified by random mutagenesis, we introduced several representative mutations into the sequence of wild-type MEK1 and expressed the mutant cDNAs in parental A375 melanoma cells. All variant constructs were expressed at comparable levels (Fig. S1C). As expected, A375 cells expressing wild-type MEK1 (MEK-WT) or a constitutively active variant (MEK-DD) showed CI-1040 GI50 values comparable to that of wild-type A375 cells (≈100 nM; Fig. 2A). In contrast, all primary resistance alleles examined (I103N, L115P, F129L, and V211D) increased CI-1040 and AZD6244 GI50 values by 50- to 1,000-fold (Fig. 2 A and B). The secondary resistance allele Q56P also conferred ≈100-fold resistance to MEK inhibition (Fig. 2 A and B), whereas the effect of the P124S allele was significant but less pronounced (4- to 10-fold; Fig. 2 A and B and Table S2). The Q56P mutation also increased MEK kinase activity in vitro (Fig. S2). Several other resistance alleles had little effect on kinase activity in vitro (Fig. S2); however, subtle kinase effects in vivo cannot be excluded.
Fig. 2.
Functional characterization of MEK1 resistance mutations identified in vitro. Growth inhibition curves of parental A375 (solid black), A375 cells expressing primary or secondary MEK1 resistance alleles, a constitutive active MEK variant (MEK-DD; grey), or wild-type MEK1 (hatched black) are shown for CI-1040 (A) or AZD6244 (B). (C) The levels of pERK1/2, pMEK1/2, MEK1/2, and α-tubulin are shown for A375 cells expressing MEK1 mutations following 16-hour incubation with CI-1040 at 10 μM, 5 μM, 2 μM, 0.4 μM, 0.08 μM, and 0 μM.
Biochemical studies of MEK inhibition corroborated the pharmacologic results above. Treatment with CI-1040 or AZD6244 potently inhibited ERK phosphorylation (p-ERK) in both parental A375 cells and those expressing MEK-WT or MEK-DD, but this effect was markedly attenuated in cells expressing each resistant allele (Fig. 2C and Fig. S3A). The presence of MEK1 mutations strongly diminished both the magnitude and durability of MEK inhibition (Fig. S3B). In several instances, levels of MEK phosphorylation (p-MEK) increased at higher doses of MEK inhibitor (e.g., A375, MEK-WT, and MEK(V211D); Fig. 2C), consistent with modulation of feedback inhibition (19). Together, these results validated the functional relevance of MEK1 mutations identified by random mutagenesis.
An Acquired MEK1 Mutation in AZD6244-Resistant Melanoma.
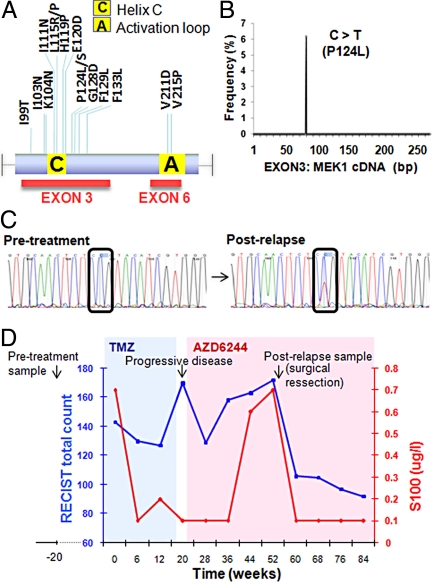
To determine whether any aforementioned resistance mechanisms might influence the clinical response to MEK inhibition, we used targeted deep sequencing of tumors from melanoma patients treated with MEK inhibitors. We reasoned that MEK1 exons 3 and 6, which encompass the validated resistant alleles described above (Fig. 3A), might represent promising candidate loci for the clinical emergence of resistance to MEK inhibition. Accordingly, we characterized the MEK1 locus in tumor genomic DNA from advanced melanoma patients enrolled in a phase II clinical trial of AZD6244 (20). Table S3 summarizes the pertinent clinical characteristics of the patients examined. Seven patients were identified who experienced transient disease stabilization followed by relapse on AZD6244 (mean stable disease duration = 55 ± 18 days in patients 1–6). Five patients contained lymph node or dermal metastases amenable to both pretreatment and postrelapse biopsy, and three harbored BRAFV600E melanomas. One patient with BRAFV600E melanoma (patient 7) experienced prolonged disease stabilization on AZD6244.
Fig. 3.
An acquired MEK1 mutation in AZD6244-resistant metastatic melanoma. PCR products corresponding to MEK1 exons 3 and 6 (A) were generated from relapsed melanomas treated with AZD6244; products were pooled and interrogated by massively parallel sequencing (a single Illumina lane). (B) A single mutation, MEK1P124L, was recovered by this analysis. (C) Sanger sequencing chromatograms from MEK1 exon 3 corresponding to pretreatment and postrelapse melanoma DNA from patient 7 are shown. A C > T transition, indicative of MEK1P124L, is evident only in the relapsed sample. (D) Changes in RECIST total count (blue graph) and serum S100 levels (red graph) are plotted over the clinical course of patient 7. The times of pretreatment and postrelapse biopsies are indicated. The intervals of temozolomide treatment (shaded blue) and AZD6244 treatment (shaded pink) are shown.
MEK1 exons 3 and 6 were amplified by PCR using tumor genomic DNA from five patients who relapsed following MEK inhibitor treatment. All PCR products were pooled and subjected to massively parallel sequencing (see Experimental Procedures). This analysis identified a C → T transition in MEK1 exon 3 that encodes a P124L substitution (Fig. 3B). MEK1 codon 124 was implicated as a secondary resistance allele during the random mutagenesis screens, although alternative amino acid substitutions (P124S or P124Q) were selected. Sanger sequencing of individual pretreatment and postrelapse melanoma samples revealed that MEK1P124L occurred in the postrelapse tumor DNA from patient 7 but was absent in the pretreatment tumor sample (Fig. 3C). This finding suggested that mutations within the drug target might provide a clinically relevant means of resistance to MEK inhibitors.
The MEK1P124L allele was identified in a 55-year-old male with metastatic melanoma, including pulmonary and skeletal involvement at the time of clinical trial enrollment. The patient was treated with oral temozolomide (435 mg per day) but developed progressive disease characterized by mediastinal lymph node enlargement and pulmonary infiltration (Fig. 3D). In accordance with protocol specifications, the patient was switched to the AZD6244 treatment arm. This resulted in regression or stabilization of all disease sites except for a left axillary lymph node metastasis, which expanded over a period of 16 weeks with a concomitant rise in the serum S100 tumor marker. Surgical removal of this metastasis led to a marked decrease in S100 levels; MEK1P124L was detected in this specimen. Following resection, the patient continued on AZD6244 and showed prolonged disease stabilization (>44 weeks), including a steadily declining RECIST score and a 28% overall reduction in tumor burden (Fig. 3D).
Ex Vivo and Functional Analysis of MEK1(P124L).
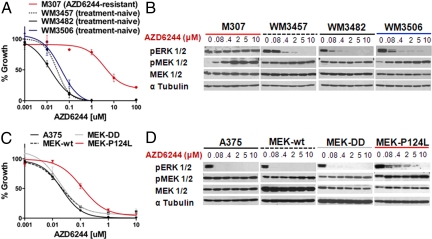
To confirm the role of the P124L allele in resistance to MEK inhibition, we examined the effects of MEK inhibition on ex vivo melanoma cultures derived from the AZD6244-resistant metastasis of patient 7 (M307 cells). M307 cells were obtained from the left axillary lymph node at the same time as the postrelapse resection described above (see SI Text). The presence of MEK1P124L in M307 was verified by Sanger sequencing (Fig. S4). The GI50 for AZD6244 was 10–50 nM in treatment-naïve BRAFV600E melanoma cultures (WM3482, WM3457, and WM3506) but exceeded 2 μM in cells derived from the AZD6244-resistant metastasis (M307; Fig. 4A). Likewise, marked biochemical inhibition of p-ERK was achieved following a 16-hour exposure to ≈400 nM AZD6244 in treatment-naïve lines, but p-ERK remained robust in the resistant cells even at 10 μM AZD6244 (Fig. 4B).
Fig. 4.
Ex vivo and functional characterization of MEK1(P124L). (A) AZD6244-mediated growth inhibition ex vivo of treatment-naïve BRAFV600E melanoma cells (black and blue) or cells cultured from an AZD6244-resistant metastatic focus (red). (B) ERK phosphorylation (p-ERK) and MEK phosphorylation (p-MEK) are shown following treatment with increasing concentrations of AZD6244 in treatment-naïve or AZD6244-resistant melanoma cells cultured ex vivo. The tubulin loading control (α-tubulin) is also shown. (C) AZD6244 growth inhibition curves of parental A375 (solid black), A375 cells expressing MEK-DD (grey), wild-type MEK1 (hatched black), or MEK1(P124L) (red) are shown. In each instance n = 6 and ± error = standard deviation. (D) p-ERK and p-MEK are shown following treatment with increasing AZD6244 concentrations in the cell lines described in C, Above. The α-tubulin control is also shown.
Next, we introduced recombinant MEK1(P124L) into parental A375 cells and examined pharmacological resistance to MEK inhibition. As observed with the MEK1(P124S) allele above, MEK1(P124L) expression resulted in an ≈5-fold increase in AZD6244 GI50 when compared to A375, MEK-WT, or MEK-DD (Fig. 4C). Similarly, MEK1(P124L) conferred sustained p-ERK expression following exposure to varying concentrations of MEK inhibitor, with measureable p-ERK even at 2 μM of AZD6244 (Fig. 4D). In contrast, parental A375 cells and those expressing MEK-WT and MEK-DD showed diminished p-ERK levels even at the lowest drug concentration (80 nM; Fig. 4D).
Secondary MEK1 Mutations Confer Cross-Resistance to B-RAF Inhibition.
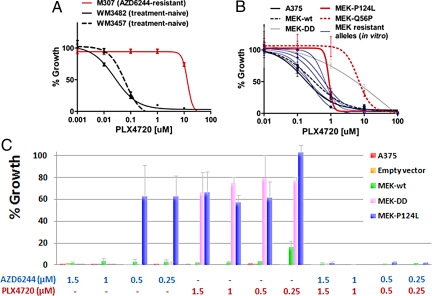
Given the high prevalence of oncogenic BRAF mutations in melanoma, several RAF inhibitors have entered clinical trials (8, 9, 21). To determine whether MEK1 mutations confer cross-resistance to RAF inhibition, we examined the effects of the selective B-RAF inhibitor PLX4720 (21) in the cultured melanoma lines described above. AZD6244-resistant primary melanoma cells (M307) demonstrated profound cross-resistance to PLX4720, with a GI50 value of >10 μM compared to 5–10 nM in treatment-naïve lines (Fig. 5A). These findings were reflected in biochemical studies of p-MEK following B-RAF inhibition (Fig. S5A).
Fig. 5.
Effects of B-RAF and combined MEK/BRAF inhibition on MEK1 resistance alleles. (A) PLX4720-mediated growth inhibition curves are shown for ex vivo cultures of treatment-naïve B-RAFV600E melanoma cells (black) or cells cultured from an AZD6244-resistant metastatic focus (red). (B) PLX4720 growth inhibition curves are shown for parental A375 cells (solid black) and A375 cells expressing selected primary MEK1 resistance alleles (blue), MEK-DD (grey), wild-type MEK1 (hatched black), MEK1(Q56P), or MEK1(P124L) (red). In each instance, n = 6 and ± error = standard deviation. (C) Colony formation assays are shown for parental A375 (red bars) and cells expressing empty vector (orange bars), MEK-WT (green bars), MEK(DD) (pink bars), or MEK(P124L) (blue bars), as indicated in the legend Inset. Concentrations of AZD6244 and PLX4720 (μM) are indicated (Below). y-axis indicates the number of dense colonies formed compared to the vehicle control for each case. n = 3 and ± error = standard deviation.
In general, expression of primary MEK1 resistance alleles identified in vitro only modestly affected the PLX4720 GI50 in A375 cells (Fig. 5B and Table S2). However, the P124L and P124S mutations conferred a two- to threefold resistance relative to wild-type MEK1 (approximately three- to fourfold resistance compared to parental A375), and the Q56P mutation conferred robust resistance (>50-fold) to PLX4720, comparable to the MEK(DD) allele (Fig. 5B and Table S2). Levels of p-MEK following PLX4720 treatment showed comparable reduction across all MEK1 resistance alleles (Fig. S5B). Overall, these results indicated that clinically relevant MEK1 resistance mutations may confer cross-resistance to B-RAF inhibition.
Combined B-RAF and MEK Inhibition Prevents the Emergence of Resistant Clones.
Finally, we tested whether the stringency of MAP kinase pathway inhibition might influence the emergence of on-target resistance variants. A375 melanoma cells expressing empty vector or various derivatives were cultured for up to 4 weeks in the presence of varying doses of AZD6244 or PLX4720 singly or in combination, and colony formation was monitored (see Materials and Methods). Exposure to AZD6244 completely suppressed the growth of parental A375 cells and those expressing empty vector or MEK(DD) at all concentrations examined (Fig. 5C). Expression of wild-type MEK1 resulted in low-level breakthrough growth under these conditions (Fig. S6; see SI Text), but only marginally affected MEK inhibitor GI50 values compared to parental A375 (Table S2). In contrast, expression of MEK1(P124L) resulted in numerous resistant colonies at 0.5 μM and 0.25 μM of AZD6244 (Fig. 5C). As expected, cells expressing the mutagenized MEK1-cDNA library that was used in the primary resistance screens formed some AZD6244-resistant colonies at all concentrations tested (Fig. S6).
As with AZD6244, the B-RAF inhibitor PLX4720 also suppressed the growth of wild-type and empty vector-expressing A375 melanoma cells. On the other hand, MEK1(P124L) expression yielded PLX4720-resistant colonies at all concentrations tested (Fig. 5C and Fig. S6). In this assay, the magnitude of MEK1(P124L)-mediated resistance to B-RAF inhibition approached that seen following MEK(DD) expression (Fig. 5C and Fig. S6). These results complemented the MEK1(P124L) growth inhibition studies above. Strikingly, however, combined exposure to both AZD6244 and PLX4720 potently suppressed the emergence of resistant variants at 0.5–1.5 μM of each compound (Fig. 5C and Fig. S6). These results raised the possibility that combined RAF and MEK inhibition might circumvent acquired resistance to targeted therapeutics directed against the MAP kinase pathway.
Discussion
Tumors harboring a “druggable” oncogene dependency often develop on-target clinical resistance through point mutation or genomic amplification of the target locus (22–24). Elaboration of such resistance mechanisms has enabled the design of second-generation inhibitors with enhanced potency against a range of resistant variants (25, 26). In this study, we used a systematic approach that combined random mutagenesis and massively parallel sequencing to characterize on-target resistance to MEK inhibition in melanoma.
Most MEK1 resistance alleles that emerged from in vitro screens populate the allosteric drug binding pocket adjacent to the ATP binding motif or the second protein shell situated within or near helix C. Arylamine MEK inhibitors function by locking the kinase into a “closed” inactive conformation, in which the activation loop causes helix C to become externally rotated and displaced (27). Thus, primary MEK1 mutations may introduce resistance through direct interference of drug binding or by forcing helix C toward a closed conformation that disrupts the binding pocket. Conceivably, the design of ATP-competitive inhibitors, which have proved successful in tyrosine kinase-driven cancers, may offer one approach to override resistance to non-ATP-competitive kinase inhibition.
Numerous candidate MEK1 resistance alleles identified in vitro reside outside of the drug binding pocket, localizing to regions such as the C-terminal kinase domain or the interface between helix A and the core kinase domain (15). Notably, several such secondary mutations correspond closely to missense germline variants that are observed in CFC syndrome (28). This disorder is characterized by mental retardation as well as cardiac and facial abnormalities and results from aberrant MAP kinase signaling during development. Consistent with the fact that several CFC alleles are known to enhance MEK kinase activity, the Q56P and P124S/L mutations demonstrated a substantial resistance phenotype as measured by pharmacologic growth inhibition studies. These mutations may modulate intrinsic MEK kinase activity in a manner that becomes expressly manifest in vivo during prolonged exposure to arylamine MEK inhibition.
Guided by resistance alleles identified in vitro, we analyzed the MEK1 locus in melanoma patients who relapsed during treatment with the small-molecule MEK inhibitor AZD6244. These studies identified the MEK1P124L mutation in a metastatic focus that progressed in the context of otherwise stable disease on AZD6244. The proline residue at codon 124 is uniquely positioned such that it may exert an indirect influence on helix C conformation while also interfacing directly with helix A, a negative regulatory motif whose crystal structure was recently solved (15). We speculate that mutation of this proline may disrupt a key regulatory interaction between helix A and the rest of the kinase, while simultaneously altering helix C indirectly through loss of a turn motif proximal to this segment. These results illuminate clinical mechanisms of resistance to both MEK and B-RAF inhibition and may inform rational therapeutic approaches to target this prominent tumor dependency. More generally, this work also highlights the power of systematic tumor tissue procurement both before treatment and following relapse in clinical trials of targeted anticancer agents.
The clinical emergence of a resistant MEK1 mutation in metastatic BRAFV600E melanoma suggests that the prior failure of first-generation RAF or MEK inhibitors to elicit meaningful tumor responses in many BRAFV600E melanomas may have resulted at least in part from suboptimal drug potency or pharmacodynamics in vivo. This notion is supported by recent phase I clinical trial results using selective RAF inhibitors, in which favorable drug pharmacodynamics correlated strongly with clinical response (8, 9). Whether the MEK dependency observed in BRAF-mutant melanomas is manifest in other MAP kinase-driven contexts remains an open question. Preclinical studies suggest that some NRAS-mutant melanomas may also exhibit sensitivity to RAF or MEK inhibition (4, 29), whereas KRAS mutations have conferred only marginal sensitivity (30). These findings may point to the future need to target multiple cellular pathways simultaneously (e.g., combined MAP kinase and PI3 kinase pathway blockade). On the other hand, combined pharmacologic blockade within the MAPK pathway may suppress the emergence of on-target resistance in tumors harboring the BRAFV600E mutation.
Finally, these findings highlight the increasing importance of tumor genomic profiling to guide patient selection in clinical trials of targeted therapeutics (31). The presence of BRAF mutations is expected to denote patient subpopulations whose tumors are enriched for RAF/MEK dependency. In the future, profiling BRAF-mutant melanomas for genetic alterations affecting MEK1 (and conceivably MEK2) in cases where resistance emerges may be required both to determine the mechanism of resistance and to specify optimal salvage therapy. Robust diagnostic approaches to stratify patients on the basis of tumor genotype and to identify clinically pertinent resistance mechanisms should speed the advent of “personalized” cancer treatment.
Experimental Procedures
Cell Lines and Primary Melanoma Cultures.
The origins and growth conditions of all cell lines used are described in the SI Text.
MEK1 Random Mutagenesis Screen.
Generation of mutagenized libraries was accomplished using a modification of published methods (10), and is described in the SI Text.
Sequencing of MEK1 DNA.
MEK1 cDNA or exons 3 and 6 of MEK1 genomic DNA were characterized by second generation sequencing or Sanger sequencing as described in the SI Text.
Retroviral Infections.
293T cells (70% confluent) were transfected with pWZL-Blast-MEK1 and pCL-Ampho packaging vector using Lipofectamine 2000 (Invitrogen). Supernatants containing virus were passed through a 0.45-μm syringe. The A375 cells were infected for 16 h with virus together with polybrene (4 μg/mL, Sigma). The selective marker blasticidin (3 μg/mL) was introduced 48 h postinfection.
In Vitro Pharmacologic Studies.
CI-1040 (13) was purchased from Shanghai Lechen International Trading Company; AZD6244 (12) was purchased from Selleck Chemicals, and PLX4720 (21) was purchased from Symansis. Cell growth inhibition assays were performed as described in the SI Text.
Colony Formation Assays.
For each cell line, 40,000 cells where seeded into 15-cm dishes in triplicate, and colony formation was characterized as described in the SI Text.
Patients and Samples.
Pretreatment and postrelapse metastatic melanoma samples were obtained from patients enrolled in a phase II clinical trial of AZD6244 (20). Consent was received from all patients according to the approved biobanking IRB protocol (University of Zürich, no. 647) before biopsy.
Supplementary Material
Acknowledgments.
This work was supported by grants from the Swiss National Foundation (grant 310040–103671), the Gottfried and Julia Bangerter Rhyner Stiftung, the Burroughs-Wellcome Fund, the Robert Wood Johnson Foundation, the Melanoma Research Alliance, The Starr Cancer Consortium, and the Novartis Institute for Biomedical Research, grants K08CA115927, P50CA093683, and DP2OD002750.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0905833106/DCSupplemental.
References
- 1.Martin GS. Cell signaling and cancer. Cancer Cell. 2003;4:167–174. doi: 10.1016/s1535-6108(03)00216-2. [DOI] [PubMed] [Google Scholar]
- 2.Weinstein IB. Cancer. Addiction to oncogenes—the Achilles heal of cancer. Science. 2002;297:63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 3.Davies H, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 4.Solit DB, et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006;439:358–362. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McDermott U, et al. Identification of genotype-correlated sensitivity to selective kinase inhibitors by using high-throughput tumor cell line profiling. Proc Natl Acad Sci USA. 2007;104:19936–19941. doi: 10.1073/pnas.0707498104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McDermott DF, et al. Double-blind randomized phase II study of the combination of sorafenib and dacarbazine in patients with advanced melanoma: A report from the 11715 Study Group. J Clin Oncol. 2008;26:2178–2185. doi: 10.1200/JCO.2007.14.8288. [DOI] [PubMed] [Google Scholar]
- 7.Rinehart J, et al. Multicenter phase II study of the oral MEK inhibitor, CI-1040, in patients with advanced non-small-cell lung, breast, colon, and pancreatic cancer. J Clin Oncol. 2004;22:4456–4462. doi: 10.1200/JCO.2004.01.185. [DOI] [PubMed] [Google Scholar]
- 8.Flaherty K, et al. Phase I study of PLX4032: Proof of concept for V600E BRAF mutation as a therapeutic target in human cancer. ASCO Meeting Abstracts May 20. 2009;2009:9000. [Google Scholar]
- 9.Schwartz GK, et al. A phase I study of XL281, a selective oral RAF kinase inhibitor, in patients (Pts) with advanced solid tumors. ASCO Meeting Abstracts May 20. 2009;2009:3513. [Google Scholar]
- 10.Azam M, Latek RR, Daley GQ. Mechanisms of autoinhibition and STI-571/imatinib resistance revealed by mutagenesis of BCR-ABL. Cell. 2003;112:831–843. doi: 10.1016/s0092-8674(03)00190-9. [DOI] [PubMed] [Google Scholar]
- 11.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davies BR, et al. AZD6244 (ARRY-142886), a potent inhibitor of mitogen-activated protein kinase/extracellular signal-regulated kinase kinase 1/2 kinases: Mechanism of action in vivo, pharmacokinetic/pharmacodynamic relationship, and potential for combination in preclinical models. Mol Cancer Ther. 2007;6:2209–2219. doi: 10.1158/1535-7163.MCT-07-0231. [DOI] [PubMed] [Google Scholar]
- 13.Allen LF, Sebolt-Leopold J, Meyer MB. CI-1040 (PD184352), a targeted signal transduction inhibitor of MEK (MAPKK) Semin Oncol. 2003;30:105–116. doi: 10.1053/j.seminoncol.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 14.Delaney AM, Printen JA, Chen H, Fauman EB, Dudley DT. Identification of a novel mitogen-activated protein kinase kinase activation domain recognized by the inhibitor PD 184352. Mol Cell Biol. 2002;22:7593–7602. doi: 10.1128/MCB.22.21.7593-7602.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischmann T, et al. Crystal structures of MEK1 binary and ternary complexes with nucleotides and inhibitors. Biochemistry. 2009;48:2661–2674. doi: 10.1021/bi801898e. [DOI] [PubMed] [Google Scholar]
- 16.Rodriguez-Viciana P, Rauen KA. Biochemical characterization of novel germline BRAF and MEK mutations in cardio-facio-cutaneous syndrome. Methods Enzymol. 2008;438:277–289. doi: 10.1016/S0076-6879(07)38019-1. [DOI] [PubMed] [Google Scholar]
- 17.Nava C, et al. Cardio-facio-cutaneous and Noonan syndromes due to mutations in the RAS/MAPK signalling pathway: Genotype-phenotype relationships and overlap with Costello syndrome. J Med Genet. 2007;44:763–771. doi: 10.1136/jmg.2007.050450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Estep AL, Palmer C, McCormick F, Rauen KA. Mutation analysis of BRAF, MEK1 and MEK2 in 15 ovarian cancer cell lines: Implications for therapy. PLoS ONE. 2007;2:e1279. doi: 10.1371/journal.pone.0001279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pratilas CA, et al. (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc Natl Acad Sci USA. 2009;106:4519–4524. doi: 10.1073/pnas.0900780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dummer R, et al. AZD6244 (ARRY-142886) vs temozolomide (TMZ) in patients (pts) with advanced melanoma: An open-label, randomized, multicenter, phase II study. ASCO Meeting Abstracts May 20. 2008;2008:9033. [Google Scholar]
- 21.Tsai J, et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc Natl Acad Sci USA. 2008;105:3041–3046. doi: 10.1073/pnas.0711741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorre ME, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 23.Heinrich MC, et al. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J Clin Oncol. 2003;21:4342–4349. doi: 10.1200/JCO.2003.04.190. [DOI] [PubMed] [Google Scholar]
- 24.Kobayashi S, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 25.Talpaz M, et al. Dasatinib in imatinib-resistant Philadelphia chromosome-positive leukemias. N Engl J Med. 2006;354:2531–2541. doi: 10.1056/NEJMoa055229. [DOI] [PubMed] [Google Scholar]
- 26.Kantarjian H, et al. Nilotinib in imatinib-resistant CML and Philadelphia chromosome-positive ALL. N Engl J Med. 2006;354:2542–2551. doi: 10.1056/NEJMoa055104. [DOI] [PubMed] [Google Scholar]
- 27.Ohren JF, et al. Structures of human MAP kinase kinase 1 (MEK1) and MEK2 describe novel noncompetitive kinase inhibition. Nat Struct Mol Biol. 2004;11:1192–1197. doi: 10.1038/nsmb859. [DOI] [PubMed] [Google Scholar]
- 28.Dentici ML, et al. Spectrum of MEK1 and MEK2 gene mutations in cardio-facio-cutaneous syndrome and genotype-phenotype correlations. Eur J Hum Genet. 2009;17:733–740. doi: 10.1038/ejhg.2008.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin WM, et al. Modeling genomic diversity and tumor dependency in malignant melanoma. Cancer Res. 2008;68:664–673. doi: 10.1158/0008-5472.CAN-07-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haigis KM, et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat Genet. 2008;40:600–608. doi: 10.1038/ngXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomas RK, et al. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39:347–351. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.