Abstract
The GroEL/GroES reaction cycle involves steps of ATP and polypeptide binding to an open GroEL ring before the GroES encapsulation step that triggers productive folding in a sequestered chamber. The physiological order of addition of ATP and nonnative polypeptide, typically to the open trans ring of an asymmetrical GroEL/GroES/ADP complex, has been unknown, although there have been assumptions that polypeptide binds first, allowing subsequent ATP-mediated movement of the GroEL apical domains to exert an action of forceful unfolding on the nonnative polypeptide. Here, using fluorescence measurements, we show that the physiological order of addition is the opposite, involving rapid binding of ATP, accompanied by nearly as rapid apical domain movements, followed by slower binding of nonnative polypeptide. In order-of-addition experiments, approximately twice as much Rubisco activity was recovered when nonnative substrate protein was added after ATP compared with it being added before ATP, associated with twice as much Rubisco protein recovered with the chaperonin. Furthermore, the rate of Rubisco binding to an ATP-exposed ring was twice that observed in the absence of nucleotide. Finally, when both ATP and Rubisco were added simultaneously to a GroEL ring, simulating the physiological situation, the rate of Rubisco binding corresponded to that observed when ATP had been added first. We conclude that the physiological order, ATP binding before polypeptide, enables more efficient capture of nonnative substrate proteins, and thus allows greater recovery of the native state for any given round of the chaperonin cycle.
Keywords: chaperonin, polypeptide binding, protein folding
The GroEL/GroES chaperonin system provides assistance to folding of a large number of proteins to the native state via 2 principal actions, one involving binding of nonnative protein in an open ring of GroEL through multivalent hydrophobic contacts formed between the nonnative protein and surrounding GroEL apical domains and the other involving folding occurring in the encapsulated hydrophilic chamber formed when ATP and the GroES “lid” are bound to the same ring as polypeptide (1–3). A number of studies have made it clear that the polypeptide binding step can rescue misfolded substrate proteins from kinetically trapped states that occur during folding (e.g., 4, 5), despite the lack of stable secondary structure in such conformations (6, 7). Such rescue has been associated with topological “stretching” of the substrate protein, as observed in a number of FRET studies (5, 8, 9). Substrate protein is released during large ATP/GroES-directed rigid body movements of the GroEL apical domains into the GroES-domed hydrophilic chamber, in which it proceeds to fold (10–12). As shown in a number of studies, the so-called “cis” chamber facilitates folding by preventing multimolecular aggregation that could reduce both the rate of recovery of the native state (in those cases in which aggregation is reversible) and the extent of recovery (in those cases in which aggregation is irreversible), as occurs for substrate proteins folding in free solution (e.g., 13, 14).
Although the general trajectory of the reaction cycle has been understood for some time, there has remained an open question concerning the order of arrival of the ligands, ATP and substrate protein, to the normal acceptor, the open ring of an asymmetrical GroEL/GroES/ADP complex. Does polypeptide arrive first? This has been assumed in a number of studies in vitro, where an order of addition has been programmed in which substrate is first incubated with such asymmetrical complexes, followed by addition of ATP (8, 9). In such studies, there have been observations that ATP addition produced additional stretching of nonnative protein, and it was suggested that this could constitute a necessary ATP-mediated “forced unfolding” step. Such a step would presumably result from the small rigid body elevation and counterclockwise twist of the apical domains of a GroEL ring that attend ATP binding to equatorial sites in the ring (15), preceding the large rigid body elevation and clockwise twist that accompany GroES binding. Yet, could it be that ATP binds first? If so, such forced unfolding would not likely be operative if the apical domains move on the same approximate time scale as ATP binding. Moreover, polypeptide association with apical domains that have been mobilized could potentially affect the efficiency of binding. Such an order of addition with ATP binding before substrate protein seems plausible considering previous rate measurements that indicate, on one hand, rapid binding of ATP to unliganded GroEL (16–18) and, on the other hand, relatively slow binding of such substrate proteins as MDH and Rubisco to unoccupied GroEL or to asymmetrical GroEL/GroES/ADP complexes, respectively (19, 20). Here, we have systematically investigated the relative rates of arrival of ATP and substrate protein ligands to both unliganded GroEL and asymmetrical GroEL/GroES/ADP complexes and find that the physiological order of arrival entails rapid ATP binding, producing nearly as rapid apical domain movement, followed by slower binding of substrate protein. Further, in order-of-addition experiments, the extent of recovery of the substrate protein Rubisco was greater with the physiological order of addition than with the opposite order, a consequence of more rapid substrate protein binding by ATP-mobilized apical domains.
Results
Rapid ATP Binding to GroEL as Revealed by Fluorescence Intensity Changes of GroEL F44C Labeled with Oregon Green 488.
To monitor binding of ATP by GroEL, we relied on a previously produced GroEL variant, F44C, containing a single cysteine in an otherwise “cysteine-zero” version of GroEL in which all 3 natural cysteines in the GroEL subunit (amino acids 138, 458, and 519) had been substituted with alanine (21). As illustrated in Fig. 1A, F44 lies in a loop pointing up into the GroEL central cavity at the level of the equatorial domain, positioned between equatorial antiparallel β-strands near whose ends are residues that form hydrogen-bonding contacts with the terminal phosphate of bound ATP through a water molecule and a potassium ion. The loop and residue 44 are thus disposed to be affected by the absence vs. presence of ATP. An earlier study reported that an F44W substitution could report on ATP binding (16). Here, we used the mutant, F44C, which had previously been observed to be fully functional, as indicated by the ability of its encoding plasmid to rescue growth of a GroEL-deficient mutant and by the ability of the purified protein to carry out efficient ATP/GroES-dependent protein refolding in vitro (21). F44C was labeled with Oregon Green 488 (OG488)-maleimide to a level of ≈70% occupancy. It remained fully functional in mediating refolding of malate dehydrogenase (MDH) in vitro, exhibiting kinetics nearly identical to those of WT GroEL [supporting information (SI) Fig. S1A]. The steady-state rate of ATP turnover by the modified mutant, called EL44-OG, also resembled that of WT GroEL (Fig. S1B).
Fig. 1.
ATP binds rapidly to unliganded GroEL and to the trans ring of the GroEL/GroES/ADP complex. (A) Ribbons diagram of a portion of the equatorial domain of 1 GroEL subunit in the GroEL/GroES/ADP/AlFx complex (ref. 24; PDB ID code: 1PCQ) showing the position of F44. (B) Emission spectra of EL44-OG alone (black trace), mixed with 500 μM ADP (green), and mixed with 500 μM ATP (red). Excitation was at 496 nm. (C) Change in fluorescence of EL44-OG on stopped-flow mixing with ATP. The trace could be fit (white line) as the sum of 2 exponentials with the rate constants shown. The schematic indicates OG (green) in the equatorial domains. (D) Dependence of the faster rate of fluorescence change on ATP concentration. Rate constants from experiments as in C (black) and E (red) at different ATP concentrations are plotted as a function of ATP concentration, giving straight lines with slopes equal to the respective second-order rate constants for ATP binding, as shown. (E) Change in fluorescence of the EL44-OG/GroES/ADP complex on stopped-flow mixing with ATP. D represents ADP in the cis ring, GroES is colored gray, and ATP (red) is shown adjacent to the ring to which it binds. (F) As in E, except with unliganded EL44-OG. (G) As in E, but with MDH bound to the trans ring, represented as a blue line in the schematic. (H) Stopped-flow mixing experiment similar to that in E, except GroEL in the complex was labeled with OG on position 315 on the outside of the apical domains. Here, the trace could be fit as a single exponential with the indicated rate. (I) Stopped-flow experiment using the single-ring version of GroEL, SR1, labeled with OG on a cysteine substituted at position 315 (see Table S1 for a summary of rates and amplitudes.)
Fluorescence emission spectra of EL44-OG (excited at 496 nm) revealed that addition of ATP produced a large drop in steady-state fluorescence intensity (≈65% in 500 μM ATP), accompanied by a small blue shift of the emission maximum (Fig. 1B). By contrast, ADP produced a smaller drop of intensity, supporting that the γ-phosphate of ATP has a specific effect on the conformation of the 44-containing loop.
On stopped-flow mixing of 0.5 mM ATP with EL44-OG, a rapid drop of emission intensity was observed, with a curve that could be fit as the sum of 2 exponentials, one with a rate constant of ≈100 s−1 and another with a rate constant of 4 s−1 (Fig. 1C). The former rate indicates rapid interaction of ATP with unliganded GroEL and corresponds to rates reported earlier both for F44W (ref. 16; 80 ± 5 s−1) and for 2 other tryptophan-substituted versions of GroEL: Y485W, where a tryptophan was substituted elsewhere in the equatorial domain (ref. 18; 123 ± 2 s−1), and R231W (ref. 17; 80 s−1), where a tryptophan was substituted into the cavity-facing aspect of the apical domain (GroEL is otherwise devoid of tryptophan). As expected, the rate of interaction of ATP with EL44-OG was dependent on ATP concentration (Fig. 1D), and a bimolecular rate constant of 2 × 105 M−1 s−1 was determined for the faster phase. The slower kinetic phase, also dependent on ATP concentration, likely corresponds to the third fluorescent phase observed for GroEL's R231W and Y485W on ATP binding (17, 18). The possibility that this phase reflected ATP hydrolysis was excluded by employing a hydrolysis-defective D398A/EL44-OG chaperonin.
Equally Rapid Fluorescence Change on ATP Binding to Open Trans Ring of GroEL/GroES/ADP Asymmetrical Complex.
Under physiological conditions, the normal acceptor state for ATP is the open trans ring of a GroEL/GroES/ADP asymmetrical complex. With stopped-flow mixing, we observed that the rate of ATP binding to the trans ring of EL44-OG/GroES/ADP complexes was similar to that to an unliganded GroEL ring (Fig. 1E, compare with Fig. 1F). Here, there was also ATP concentration dependence resembling that of unliganded EL44-OG (Fig. 1D).
Substrate Polypeptide Bound to the Trans Ring Does Not Affect Rate of ATP Binding.
In a number of in vitro studies, nonnative polypeptide has been first complexed to the open trans ring of an asymmetrical ADP GroEL/GroES complex before addition of ATP and excess GroES (8, 9). Under these conditions, ATP binding followed by binding of GroES is clearly able to encapsulate the nonnative polypeptide and direct productive folding. Is the rate of ATP binding in such a context affected by the bound substrate protein? To test this, nonnative MDH was bound to the asymmetrical EL44-OG/GroES/ADP complexes, and ATP was then added with stopped-flow mixing. Under these conditions, the rate of fluorescence intensity change was virtually identical to that of the EL44-OG/GroES/ADP complex in the absence of polypeptide (compare Fig. 1G with Fig. 1E). Thus, the presence of a nonnative substrate bound to the trans ring apical domains does not have any detectable effect on the rapid entry of ATP into the trans ring equatorial nucleotide binding pockets.
Rapid Apical Domain Movement Accompanies ATP Binding.
When ATP binds rapidly to GroEL, does this cause significant adjustment in the apical polypeptide binding domains on the same rapid time scale, or are the apical changes in response to ATP binding relatively slow, such that the effects of ATP binding must be considered to be occurring at a later time? The earlier study of R231W had already indicated that apical effects occurred rapidly with that mutant (17). In the present study as well, a fluorescent probe on the outside aspect of the apical domains, remote from the ATP binding site on the inside of the cylinder (GroEL E315C in a cysteine-zero background labeled with OG), showed a rapid change in fluorescence intensity, on the same time scale as ATP binding. For example, a rate constant of 20 s−1 was observed for a GroEL315-OG/GroES/ADP asymmetrical complex at an ATP concentration of 150 μM (Fig. 1H, compare with 25 s−1 in Fig. 1E). Such apical changes occurred in the same ring to which ATP is bound, because analysis of an OG-modified single-ring version of E315C, SR315-OG, showed a similar rate of fluorescence intensity change (Fig. 1I). The rate of apical movement was also ATP concentration-dependent, with the rates paralleling those of ATP binding (Fig. S2, compare with Fig. 1D).
Relatively Slow Binding of Substrate Polypeptide as Measured by FRET.
To enable a comparison of the rate of binding nonnative substrate polypeptide with that of binding ATP, we next measured the rate of binding of substrate proteins to asymmetrical GroEL/GroES/ADP complexes using FRET. Three different substrate proteins were studied: MDH (33 kDa), Rubisco (51 kDa), and a double-mutant form of maltose binding protein (DM-MBP; 41 kDa). All 3 proteins behave as stringent substrate proteins at 25 °C, requiring the presence of GroEL, GroES, and ATP to reach native form (3). In their absence, quantitative aggregation ensues. For monitoring binding, FRET was measured between substrate protein labeled with coumarin propyl maleimide (CPM; donor) on a cysteine residue (see Materials and Methods) and EL44-OG (acceptor). Exciting at the excitation maximum for CPM (384 nm), emission spectra were collected for the CPM (donor)-labeled substrates while bound to unlabeled GroEL (Fig. 2A, DM-MBP-CPM as an example, black trace), for EL44-OG complexed with unlabeled substrate (Fig. 2A, acceptor labeled, blue trace), or for complexes with CPM-labeled substrate bound to EL44-OG (Fig. 2A, red trace). For both DM-MBP-CPM and MDH-CPM, there was strong donor quenching on association with EL44-OG; at the same time, there was the appearance of a substantial acceptor signal.
Fig. 2.
Relatively slow binding of a nonnative substrate, DM-MBP, to unliganded GroEL and to the trans ring of the GroEL/GroES/ADP complex. (A) Emission spectrum of DM-MBP labeled with CPM (donor) while bound to GroEL (D, black trace), emission spectrum of EL44-OG (acceptor) while complexed with nonnative DM-MBP (A, blue trace), and emission spectrum of the complex of DM-MBP-CPM with EL44-OG (D-A, red trace), all excited at 384 nm, the excitation maximum for CPM. (B) Change in donor channel fluorescence on stopped-flow mixing of nonnative DM-MBP-CPM with EL44-OG. Blue line represents nonnative DM-MBP, and D in the yellow circle represents the CPM label. (C) Dependence of the faster rate constant for DM-MBP binding on GroEL concentration. Rate constants from experiments as in B at different concentrations of GroEL (black) or from similar experiments using the GroEL/GroES/ADP complex (red) are plotted vs. GroEL concentration, yielding straight lines with slopes (second-order rate constants) of 6.11 × 106 M−1s−1 and 5.36 × 106 M−1s−1, respectively. (D) Change in donor fluorescence on stopped-flow mixing of nonnative DM-MBP-CPM with the EL44-OG/GroES/ADP complex and 500 μM ATP. In multiple experiments (n = 6), the rate of the faster phase was consistently slightly faster for the trans ring in the presence of ATP than in its absence: 2.08 ± 0.11 vs. 1.36 ± 0.22 (Table S2).
On stopped-flow mixing of 125 nM DM-MBP-CPM with 125 nM EL44-OG, a drop of donor emission intensity was observed, with a curve that could be fit as the sum of 2 exponentials, one with a rate constant of 1.33 s−1 and the other with a rate constant of 0.65 s−1 (Fig. 2B). The faster rate (k1) was dependent on the concentration of chaperonin (Fig. 2C), but the slower one was not. At a GroEL concentration of 125 nM (Fig. 2B), the rate of substrate polypeptide binding (k1 = 1.33 s−1) is 60-fold slower than the rate of ATP binding (100 s−1 at 500 μM ATP; Fig. 1C). Although the rate of substrate binding would be predicted to be severalfold greater at the physiological GroEL concentration of 1–2 μM (Fig. 2C), the rate of ATP binding would also likely be somewhat greater, considering that physiological ATP concentration is several millimolar (Fig. 1D). Thus, the rate of ATP arrival under physiological conditions is likely to be at least 10-fold greater than substrate arrival.
In a further test, the addition of ATP at the same time as fluorescently labeled DM-MBP had a reproducible effect of increasing the rate of DM-MBP binding (Fig. 2D and Table S2). In sum, the physiological order of addition to a trans ring appears to comprise rapid arrival of ATP followed by slower arrival of substrate protein. This order is opposite to that programmed in recent studies, where polypeptide was initially bound to the trans ring and ATP was then added (8, 9). Does the order of addition have any measurable effect on the refolding of the substrate protein? To test this, we carried out order-of-addition experiments and measured recovery of native enzyme under essentially single-turnover conditions.
More Extensive Recovery of the Native State When ATP Is Added First, Followed by Polypeptide, Compared with the Opposite Order.
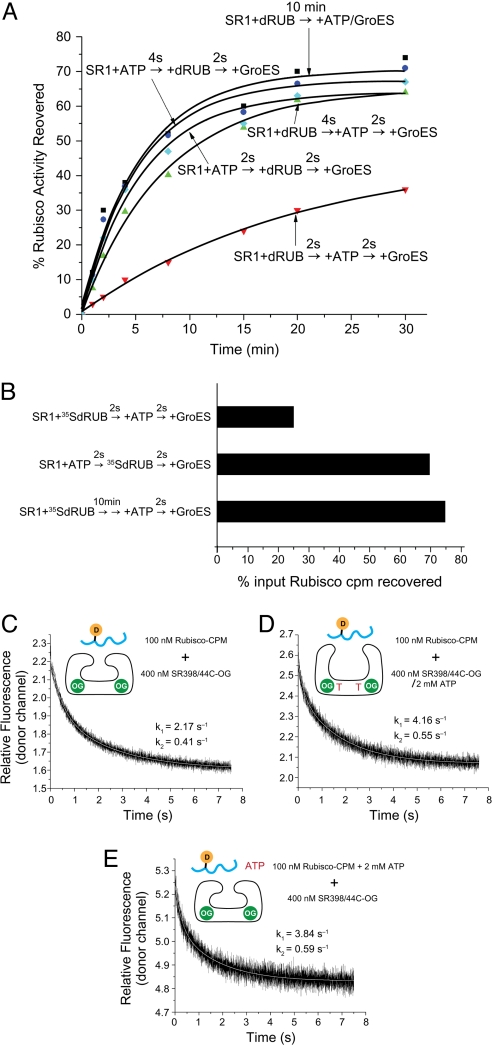
An order-of-addition experiment was carried out using the single-ring version of GroEL, SR1, enabling “single-round” analysis. Polypeptide captured by SR1 is nearly quantitatively folded to native form, after binding of GroES, inside the long-lived cis cavity of the stable SR1/GroES complex (10, 11). Thus, SR1 could be incubated with ATP and nonnative polypeptide in either order, followed by addition of GroES, and the extent of recovery of native protein could be measured in relation both to the order of addition and to the interval between the additions (Fig. 3A). For these tests, GroES was added 2 sec after ATP/polypeptide to allow completion of the initial interactions. In an initial experiment, we reproducibly observed that when nonnative Rubisco was added first followed 2 sec later by ATP, the extent of Rubisco recovery in native form was only ≈30%. This compared with >60% recovery when ATP was added first and with the ≈70% recovery observed when ATP and GroES were added to a preformed Rubisco-SR1 binary complex (produced by a 10-min incubation of nonnative Rubisco with SR1). This suggests that in the context of a cycling reaction in which the trans ring of an acceptor GroEL/GroES/ADP complex is open and available for binding polypeptide for only ≈1–2 s before GroES binds, the mobilization of its apical domains by rapid ATP binding favors the binding of nonnative polypeptide. The opposite order, addition of polypeptide 2 s before ATP, appeared to be less favorable for binding, even though ATP-mobilized apical domains were subsequently available for 2 s before GroES addition. Notably, when the interval between the additions of Rubisco and ATP was increased to 4 s (Fig. 3A), the order-of-addition effect was relieved, with the kinetics and extent of recovery now equal to those of adding ATP first, presumably a function of more extensive polypeptide binding during the interval before ATP addition. Although the extent of recovery of Rubisco in the order-of-addition studies with a 2-s interval was significantly affected, the kinetics of recovery of the native state inside the stable SR1/GroES complexes were similar, consistent with an effect on substrate protein binding and not on the rate of folding in the encapsulated chamber.
Fig. 3.
Binding of ATP before nonnative Rubisco improves folding yield by increasing the extent and rate of binding. (A) Recovery of Rubisco activity with different orders of addition. SR1 was incubated with ATP for 2 or 4 s before nonnative Rubisco (dRUB) was added; alternatively, nonnative Rubisco was added to SR1 first, followed by ATP 2 or 4 s later. For both orders, GroES was added 2 s later, and aliquots were removed at the indicated times for assay of Rubisco activity [cyan (2 s) and blue (4 s) for ATP first, and red (2 s) and green (4 s) for Rubisco first]. For comparison, a “standard” Rubisco refolding assay was carried out by incubating SR1 with nonnative Rubisco for 10 min before ATP and GroES were added together to start refolding (black symbols). (B) Extent of binding 35S-Rubisco to SR1 with different orders of addition. Experiments were carried out with the different orders of addition as in A, except that after adding GroES and then ADP-AlFx (to stabilize the ternary complex), the mixtures were chromatographed on a Superose 6 column and radioactivity of fractions was determined. Total radioactivity recovered at the elution position of the SR1/GroES/ADP-AlFx/Rubisco complex is reported as a percentage of the radioactivity loaded on the column. In identical analyses with 4-s intervals, both orders of addition produced ≈70% recovery, corresponding to the recovery of activity in A (not shown). (C) Rate of Rubisco binding to an SR1 ring in the absence of ATP, measured by FRET. Donor fluorescence of Rubisco-CPM after stopped-flow mixing with a hydrolysis-defective SR1 D398A molecule carrying the OG fluorophore on a substituted Cys-44. (D) Rate of Rubisco binding to SR398A in the presence of ATP (added before loading into the stopped-flow syringe). (E) Rate of Rubisco binding to an SR398A ring when added simultaneously with ATP.
More Extensive Binding of Rubisco with ATP Added First.
To address the effect of the order of addition on substrate protein binding, we used 35S-labeled Rubisco and measured the amount that became bound by SR1 during the order-of-addition incubations using gel filtration of the final product mixtures to separate the stable SR1/GroES/Rubisco complexes from unbound polypeptide. This revealed that ≈10,000 cpm 35S-Rubisco had bound when 40,000 cpm 35S-Rubisco (125 nM) was added 2 sec before ATP and that ≈30,000 cpm had bound when ATP was added 2 sec before Rubisco (Fig. 3B). The latter extent of binding was also achieved when SR1-Rubisco binary complexes formed over a period of 10 min were then incubated with ATP and GroES together (Fig. 3B). These measurements of the extent of substrate binding thus parallel the extent of recovery of enzymatic activity (compare Fig. 3B with Fig. 3A) and support the conclusion that in the context of a 2-s interval between the addition of ATP/polypeptide, the addition of ATP initially favors more efficient substrate protein binding.
Greater Rate of Rubisco Binding to an ATP-Exposed Chaperonin Ring.
The foregoing observation of greater extent of Rubisco binding in an order-of-addition experiment in which ATP is added initially suggests that the rate of Rubisco association with ATP-mobilized apical domains might be greater. To test this, an ATP hydrolysis-defective D398A version of SR1 with OG attached to a cysteine substituted at position 44 (in a C138A background) was used to report by FRET on the rate of binding of CPM-labeled Rubisco (Fig. 3 C–E). We observed that the rate of Rubisco binding, carried out with the same conditions as the foregoing order-of-addition experiments, was 2-fold greater in the presence vs. absence of ATP (compare Fig. 3D with Fig. 3C). When ATP and nonnative Rubisco were added simultaneously, reflecting the physiological situation, the rate of Rubisco binding resembled that when ATP had been added before Rubisco (Fig. 3E, compare with Fig. 3D). These data support that Rubisco associates more rapidly with a ring whose apical domains have been ATP-mobilized and that, under physiological conditions in which both ATP and nonnative substrate are present, it is the ATP-mobilized apical domains that are operative in substrate binding.
Discussion
Physiological Order of Binding to an Open Acceptor Ring Involves Rapid ATP Binding Followed by Nonnative Substrate Protein, Producing More Efficient Capture of Substrate Protein.
The kinetic studies presented here with both unliganded GroEL and asymmetrical GroEL/GroES/ADP acceptor complexes indicate that ATP binds rapidly to an open ring, at rates of >100 s−1 at physiological concentrations, whereas polypeptide binds relatively slowly, at a rate of ≈5–10 s−1. Because the rate of ATP-directed apical domain movement, as reported both elsewhere and in the present study, corresponds closely to that of ATP binding, we conclude that the small apical domain elevation and twist produced by ATP alone, as observed in EM studies (15), is likely to be completed before substrate protein binds to the apical domain; that is, under normal conditions, the substrate collides with already mobilized apical domains. Nevertheless, in order-of-addition experiments carried out here (Fig. 3A), there was no absolute requirement for this to occur; that is, productive GroES encapsulation and folding could occur with either order of addition. At least for the substrate protein Rubisco, however, both the extent of substrate protein binding and ultimate recovery of the native state in a single-round experiment were approximately 2-fold greater when ATP was added 2 s before Rubisco vs. the opposite order. Consistently, the rate of Rubisco binding to an ATP-mobilized ring was 2-fold greater than to an unliganded ring, and approximately the same rate was measured when both ATP and Rubisco were offered simultaneously to a ring. These data would suggest that under physiologic conditions, the apical domains are mobilized by rapid ATP binding, followed by binding of substrate protein. Substrate binding to mobilized apical domains may be more efficient, perhaps as the result of the increased mobility of the apical domains themselves, released by ATP binding from salt bridges (e.g., between amino acids E386 and R197) that hold neighboring subunits together in the nucleotide-free state (15). Freely ATP-mobilized apical domains may be better able to complement the surfaces of nonnative protein, binding more rapidly or with greater affinity.
Effects on Conformation of Nonnative Substrate.
The foregoing physiological order of binding might be expected to have implications for effects on substrate conformation. In this regard, we examined DM-MBP using intramolecular FRET (9) (Fig. S3). On binding nonnative DM-MBP to an unliganded GroEL ring, we observed the same loss of FRET previously reported, consistent with stretching of substrate protein during binding to an open ring. Yet, no further stretching was observed on subsequent addition of ATP (Fig. S3B). With the physiological order of addition of ATP first followed by substrate protein, using the ATP hydrolysis-defective mutant GroEL, D398A or a single-ring version, we observed a similar rate of FRET decrease but with smaller amplitude (Fig. S3 C and D), potentially corresponding to a lesser degree of stretching than that observed with binding to an open ring. On addition of ATP and GroES to the binary complex of GroEL/DM-MBP, we observed the same rise of FRET as reported earlier, corresponding to compaction of substrate protein within the encapsulated chamber at the point of release from the apical domains (8, 9, 22). At the level of substrate protein, then, it seems that binding either to an unliganded open GroEL ring or to ATP-mobilized apical domains is associated with conformational stretching, whereas neither ATP nor ATP/GroES addition to an already formed substrate-GroEL binary complex produces any appreciable stretching that would constitute an ATP-mediated forced unfolding.
Two Distinct ATP Actions During Production of a Folding-Active Ring.
In summary, the present observations indicate that the physiological sequence of the chaperonin reaction cycle involves ATP action at 2 steps in the formation of a folding-active complex. Initially, on rapid ATP binding, fast mobilization of the apical domains occurs (≈100 s−1), entailing their release from each other and producing a small degree of elevation and counterclockwise twist. Next, substrate protein binds (≈5–10 s−1), with, as shown here, the mobilized state of the apical domains accelerating association. Then, in a step that must be slower than substrate binding (ref. 23; 1–2 s−1), GroES collides with the mobilized apical domains via its mobile loops and triggers the large subsequent apical domain movements (60° elevation and 120° clockwise twist) that release the bound substrate protein into the domed folding chamber. These latter movements require the presence of ATP so that its binding energy can provide the force necessary for releasing substrate protein (23). Thus, in effect, ATP binding to the open trans ring of an asymmetrical complex supports 2 distinct events in this ring, separated by the binding of nonnative polypeptide, which lead to the formation of a folding-active complex and the initiation of folding.
Materials and Methods
Proteins and Assays.
Proteins were expressed, purified, and assayed as in SI Materials and Methods.
Formation of Asymmetrical GroEL/GroES Complexes.
In all experiments, asymmetrical GroEL/GroES/ADP complexes were formed by incubating 7 μM EL44-OG with 8 μM GroES and 250 μM ATP in 50 mM Tris-HCl (pH 7.5), 50 mM KCl, 10 mM MgCl2, and 1 mM DTT for 10 min at 25 °C. Before loading into the stopped-flow syringe, the complex was diluted 25-fold in the same buffer, giving a residual ADP concentration of 10 μM.
Fluorescence Labeling.
The cysteine-substituted GroEL and SR1 complexes were labeled with OG488, and MDH and Rubisco were labeled with 7-diethylamino-3-(4′-maleimidylphenyl)-4-methyl coumarin (CPM) using standard labeling conditions as described in SI Materials and Methods.
Stopped-Flow Fluorescence.
Labeled DM-MBP was unfolded in 3 M guanidine-HCl, and labeled Rubisco was unfolded in 5 M urea and 10 mM HCl, before loading into the stopped-flow syringe. Both substrates were diluted 25-fold on mixing with the various GroEL complexes. Concentrations given are the final ones after mixing. For all fluorescence experiments using OG488, the fluorophore was excited at 496 nm and emission was measured using a color separation filter (510–570 nm). For substrate binding FRET experiments, the CPM donor was excited at 384 nm and the emission was recorded by using a cutoff filter (410 nm) and a band-pass filter (360–500 nm). Traces are the sum of the data from 10 individual mixes. Fitting of experimental data was performed with Origin (OriginLab).
Supplementary Material
Acknowledgments.
We thank George Farr and other members of the Horwich lab for helpful discussion. This work was supported by the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0911556106/DCSupplemental.
References
- 1.Thirumalai D, Lorimer GH. Chaperonin-mediated protein folding. Annu Rev Biophys Biomol Struct. 2001;30:245–269. doi: 10.1146/annurev.biophys.30.1.245. [DOI] [PubMed] [Google Scholar]
- 2.Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: From nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- 3.Horwich AL, Fenton WA. Chaperonin-mediated protein folding: Using a central cavity to kinetically assist polypeptide chain folding. Q Rev Biophys. 2009;42:83–116. doi: 10.1017/S0033583509004764. [DOI] [PubMed] [Google Scholar]
- 4.Todd MJ, Lorimer GH, Thirumalai D. Chaperonin-facilitated protein folding: Optimization of rate and yield by an iterative annealing mechanism. Proc Natl Acad Sci USA. 1996:4030–4035. doi: 10.1073/pnas.93.9.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin Z, Rye HS. Expansion and compression of a protein folding intermediate by GroEL. Mol Cell. 2004;16:23–34. doi: 10.1016/j.molcel.2004.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park ES, Fenton WA, Horwich AL. No evidence for a forced-unfolding mechanism during ATP/GroES binding to substrate-bound GroEL: No observable protection of metastable Rubisco intermediate, or GroEL-bound Rubisco from tritium exchange. FEBS Lett. 2005;579:1183–1186. doi: 10.1016/j.febslet.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 7.Horst R, et al. Direct NMR observation of a substrate protein bound to the chaperonin GroEL. Proc Natl Acad Sci USA. 2005:12748–12753. doi: 10.1073/pnas.0505642102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin Z, Madan Z, Rye HS. GroEL stimulates protein folding through forced unfolding. Nat Struct Mol Biol. 2008;15:303–311. doi: 10.1038/nsmb.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma S, et al. Monitoring protein conformation along the pathway of chaperonin-assisted folding. Cell. 2008;133:142–153. doi: 10.1016/j.cell.2008.01.048. [DOI] [PubMed] [Google Scholar]
- 10.Weissman JS, Rye HS, Fenton WA, Beechem JM, Horwich AL. Characterization of the active intermediate of a GroEL-GroES-mediated folding reaction. Cell. 1996;84:481–490. doi: 10.1016/s0092-8674(00)81293-3. [DOI] [PubMed] [Google Scholar]
- 11.Rye HS, et al. Distinct actions of cis and trans ATP within the double ring of the chaperonin GroEL. Nature. 1997;388:792–798. doi: 10.1038/42047. [DOI] [PubMed] [Google Scholar]
- 12.Xu Z, Horwich AL, Sigler PB. The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex. Nature. 1997;388:741–751. doi: 10.1038/41944. [DOI] [PubMed] [Google Scholar]
- 13.Horst R, Fenton WA, Englander SW, Wüthrich K, Horwich AL. Folding trajectories of human dihydrofolate reductase inside the GroEL-GroES chaperonin cavity and free in solution. Proc Natl Acad Sci USA. 2007:20788–20792. doi: 10.1073/pnas.0710042105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Apetri AC, Horwich AL. Chaperonin chamber accelerates protein folding through passive action of preventing aggregation. Proc Natl Acad Sci USA. 2008:17351–17355. doi: 10.1073/pnas.0809794105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ranson NA, et al. ATP-bound states of GroEL captured by cryo-electron microscopy. Cell. 2001;107:869–879. doi: 10.1016/s0092-8674(01)00617-1. [DOI] [PubMed] [Google Scholar]
- 16.Yifrach O, Horovitz A. Transient kinetic analysis of adenosine 5′ triphosphate binding-induced conformational changes in the allosteric chaperonin GroEL. Biochemistry. 1998;37:7083–7088. doi: 10.1021/bi980370o. [DOI] [PubMed] [Google Scholar]
- 17.Taniguchi M, Yoshimi T, Hongo K, Mizobata T, Kawata Y. Stopped-flow fluorescent analysis of the conformational changes in the GroEL apical domain. J Biol Chem. 2004;279:16368–16376. doi: 10.1074/jbc.M311806200. [DOI] [PubMed] [Google Scholar]
- 18.Cliff MJ, Limpkin C, Cameron A, Burston SG, Clarke AR. Elucidation of steps in the capture of a protein substrate for efficient encapsulation by GroE. J Biol Chem. 2006;281:21266–21275. doi: 10.1074/jbc.M601605200. [DOI] [PubMed] [Google Scholar]
- 19.Ranson NA, Burston SG, Clarke AR. Binding, encapsulation and ejection: Substrate dynamics during a chaperonin-assisted folding reaction. J Mol Biol. 1997;266:656–664. doi: 10.1006/jmbi.1996.0815. [DOI] [PubMed] [Google Scholar]
- 20.Rye HS, et al. GroEL-GroES cycling: ATP and non-native polypeptide direct alternation of folding-active rings. Cell. 1999;97:325–338. doi: 10.1016/s0092-8674(00)80742-4. [DOI] [PubMed] [Google Scholar]
- 21.Elad N, et al. Topologies of a substrate protein bound to the chaperonin GroEL. Mol Cell. 2007;26:415–426. doi: 10.1016/j.molcel.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hillger F, et al. Probing protein-chaperone interactions with single-molecule fluorescence spectroscopy. Angew Chem. 2008;47:6184–6188. doi: 10.1002/anie.200800298. [DOI] [PubMed] [Google Scholar]
- 23.Motojima F, Chaudhry C, Fenton WA, Farr GW, Horwich AL. Substrate polypeptide presents a load on the apical domains of the chaperonin GroEL. Proc Natl Acad Sci USA. 2004:15005–15012. doi: 10.1073/pnas.0406132101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chaudhry C, et al. Role of the γ-phosphate of ATP in triggering protein folding by GroEL-GroES: Function, structure, and energetics. EMBO J. 2003;22:4877–4887. doi: 10.1093/emboj/cdg477. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.