Abstract
Nucleotide-binding site (NBS)-encoding resistance genes are key plant disease-resistance genes and are abundant in plant genomes, comprising up to 2% of all genes. The availability of genome sequences from several plant models enables the identification and cloning of NBS-encoding genes from closely related species based on a comparative genomics approach. In this study, we used the genome sequence of Brassica rapa to identify NBS-encoding genes in the Brassica genome. We identified 92 non-redundant NBS-encoding genes [30 CC-NBS-LRR (CNL) and 62 TIR-NBS-LRR (TNL) genes] in approximately 100 Mbp of B. rapa euchromatic genome sequence. Despite the fact that B. rapa has a significantly larger genome than Arabidopsis thaliana due to a recent whole genome triplication event after speciation, B. rapa contains relatively small number of NBS-encoding genes compared to A. thaliana, presumably because of deletion of redundant genes related to genome diploidization. Phylogenetic and evolutionary analyses suggest that relatively higher relaxation of selective constraints on the TNL group after the old duplication event resulted in greater accumulation of TNLs than CNLs in both Arabidopsis and Brassica genomes. Recent tandem duplication and ectopic deletion are likely to have played a role in the generation of novel Brassica lineage-specific resistance genes.
Electronic supplementary material
The online version of this article (doi:10.1007/s00438-009-0492-0) contains supplementary material, which is available to authorized users.
Keywords: Brassica rapa, NBS-encoding gene, Disease resistance, Genome diploidization, Evolution
Introduction
Plants have developed a variety of defense systems to protect themselves from environmental pathogens and pests. One well-characterized defense mechanism is recognition of specific pathogens mediated by disease-resistance (R) proteins and initiation of defense responses, including localized cell death and systemic acquired resistance (Dangl and Jones 2001). The largest class of known R genes includes those that encode the nucleotide-binding site (NBS) domain showing similarity to eukaryotic cell death effectors and the leucine-rich repeat (LRR) domains. Most characterized R genes can be subdivided into two distinct types based on the structure of the N-terminal domain of the protein: the N-terminal domain is either a coiled-coil (CC) motif or a Drosophila Toll/mammalian Interleukin-1 Receptor (TIR) domain. Variant types with different domains or truncated structures also exist, but generally they can be classified into the CC-NBS-LRR (CNL) or TIR-NBS-LRR (TNL) types based on the NBS-coding sequences (Ameline-Torregrosa et al. 2008; Meyers et al. 2003).
NBS-encoding genes belong to one of the largest gene families in plant genomes, and have been identified in non-vascular plants to angiosperms. Sequencing of the genome of model plant species has enabled genome-level investigation of the NBS-encoding gene family in monocot and dicot species, including rice (Monosi et al. 2004; Zhou et al. 2004), sorghum (Paterson et al. 2009), Arabidopsis thaliana (Meyers et al. 2003; Tan et al. 2007), Medicago truncatula (Ameline-Torregrosa et al. 2008), poplar (Kohler et al. 2008), grape (Yang et al. 2008b), and papaya (Porter et al. 2009) (Table 1). According to these studies, approximately 0.6–1.8% of genes encoded by plant genomes are NBS-encoding genes that occur at a density of 0.3–1.6 genes per megabase, with the exception of papaya. In the papaya genome, which did not have its whole genome duplicated recently in contrast to other plants, only 0.2% of genes encode NBS domains at a density of 0.1 gene per megabase (Porter et al. 2009). Most NBS-encoding genes are unevenly distributed and clustered in specific chromosomal regions (Ameline-Torregrosa et al. 2008; Meyers et al. 2003; Monosi et al. 2004). These observations suggest that whole genome duplication (WGD) along with tandem and segmental gene duplication have generated the high number of NBS-encoding genes in the plant genome.
Table 1.
The number of predicted NBS-encoding R genes identified in sequenced plant genomes
| Br | At | Mt | Pt | Vv | Cp | Os | |
|---|---|---|---|---|---|---|---|
| NBS-encoding genes | |||||||
| CC-NBS-LRR | 17 | 58 | 152 | 278 | 203 | 4 | 392 |
| TIR-NBS-LRR | 42 | 110 | 94 | 121 | 97 | 6 | 0 |
| Others | 33 | 39 | 87 | 3 | 159 | 44 | 81 |
| Total | 92 | 207 | 333 | 402 | 459 | 54 | 473 |
| Genome size (Mbp) | ≈100 (529)a | 125 | ≈186 (500)a | 485 | 487 | 372 | 389 |
| Source | Meyers et al. (2003) | Ameline-Torregrosa et al. (2008) | Kohler et al. (2008) | Yang et al. (2008b) | Porter et al. (2009) | Monosi et al. (2004) | |
Br, Brassica rapa; At, Arabidopsis thaliana; Mt, Medicago truncatula; Pt, Populus trichocarpa; Vv, Vitis vinifera; Cp, Carica papaya; Os, Oryza sativa
Others represent NBS-encoding genes with unusual structures or unknown motifs
aThe NBS-encoding genes of Br and Mt were identified based on partial genome sequences. Overall genome sizes are indicated in parentheses
Many of the NBS-encoding genes are ancient in origin and orthologous relationships are difficult to determine owing to lineage-specific gene amplifications and deletions. Therefore, the spectrum of NBS-encoding genes present in one species is not characteristic of the diversity of NBS-encoding genes in other plant families (McHale et al. 2006). In general, CNL types are found widely in both monocot and dicot genomes, whereas no TNL type genes have been identified in monocot genomes, although a small number of TNL genes have been found in gymnosperms (Bai et al. 2002; Cannon et al. 2002; Liu and Ekramoddoullah 2003). Previous phylogenetic analyses suggested that the CNL group originated before the divergence of the monocots and dicots, whereas the TNL group expanded recently after the divergence of the two groups (McHale et al. 2006; Yang et al. 2008b). Consequently, the TNL group proteins are thought to be involved in recognizing “species-specific” pathogens while the CNL group proteins are predicted to have basic defense roles as a result of regional adaptation to the biotic environment (Yang et al. 2008b).
Brassica rapa is one of the most valuable vegetable crops worldwide. However, susceptibility to multiple pathogens, including Plasmodiophora brassicae, a clubroot fungus, Pectobacterium carotovorum, a bacterial pathogen of soft rot decay, turnip mosaic virus (TuMV), and Meloidogyne root knot nematodes, hinders cultivation and reduces produce yield and quality. One of the significant challenges in B. rapa breeding has been to introduce durable disease resistance into agronomically important cultivars. One approach that can be used to understand the fundamental basis of Brassica resistance and susceptibility is genomic evaluation of disease-resistance gene homologs. In an initial study to identify candidate disease-resistance genes in Brassica species, a comparative mapping approach was taken using the closely related A. thaliana genome sequence as a source (Sillito et al. 2000). However, the polyploid nature of Brassica genomes along with frequent genome rearrangements and high sequence similarity between homologs has hampered the identification of orthologous genes. Genome-wide, sequence-level comparative analysis of conserved sequence blocks between A. thaliana and B. rapa reveals that they shared three rounds of WGD events (hereafter referred to as 1R, 2R, and 3R, which are equivalent to the γ, β, and α duplication events, respectively) before diverging from a common ancestor approximately 13–17 million years ago (MYA). Moreover, Brassica genomes contain triplicated homologous counterparts of corresponding segments of the A. thaliana genome due to a whole genome triplication (WGT) event (4R) approximately 11–12 MYA after speciation (Mun et al. 2009). Furthermore, an additional natural allopolyploidization event, which happened during the last 10,000 years and resulted in a change in chromosome number and genome size, has shaped the diversification of Brassica crops (Johnston et al. 2005; UN 1935). An alternative approach that can be used to identify Brassica NBS-encoding genes is mining of expressed sequence tag (EST) databases. However, although approximately 600K B. napus and 150K B. rapa ESTs are available in the dbEST database of the National Center for Biotechnology Information (NCBI), only a few R gene candidates are represented, indicating that not all R gene families are present in the current EST datasets. Recent progress in genome sequencing of B. rapa, however, allows genome-level identification of defense-related NBS-encoding genes.
Approximately 70 Mbp seed BAC sequences were recently made available on public databases and an additional 75 Mbp BAC sequences are accessible through the sequencing pipeline of the Korea Brassica rapa Genome Project (KBGP). The available genome sequences cover approximately 64% of the gene-rich euchromatic space of B. rapa based on estimation of the euchromatin amount as ≈220 Mbp (Mun et al. 2009). The main objective of this study was to use the available genome sequences to identify putative NBS-encoding gene candidates of Brassica. We determined the sequence characteristics of the NBS-encoding genes we identified, and examined the distribution of NBS-encoding genes across linkage groups. In addition, we performed phylogenetic analysis and calculated the number of substitutions per synonymous (K s) and non-synonymous site (K a) in B. rapa and A. thaliana sequences to gain insight into the evolution of NBS-encoding genes in B. rapa. Understanding of the genomic organization of NBS-encoding genes in B. rapa will allow genome-level insights into R gene evolution and inform genetic and breeding strategies to engineer disease resistance in Brassica crops.
Materials and methods
Identification of sequences that encode NBS motifs in the B. rapa genome
We used ≈145 Mbp phase 2 or 3 genome sequences from 1,199 BAC clones of B. rapa ssp. pekinensis cv. Chiifu, of which 568 BAC clones (≈70 Mbp) were deposited in the HTGS database of NCBI and 631 BAC clones (≈75 Mbp) were in the genome sequencing pipeline of KBGP. The 568 BAC clones were selected from the gene-rich euchromatic region of B. rapa based on BAC-end sequence comparison with the A. thaliana counterpart regions and assembled into approximately 70 Mbp of non-redundant sequence contigs (Mun et al. 2009). In addition, we also downloaded 75 Mbp of 631 BAC clones, generated mainly from sequencing of chromosomes A3 and A9, from the KBGP database (The Korea Brassica rapa Genome Project, http://www.brassica-rapa.org/BRGP/index.jsp). Because these sequences are individual BAC sequences that have not been assembled, several of them contain overlapping redundant sequences. In general, minimum tiled BAC clones in the sequencing pipeline of A3 and A9 are allowed approximately 30% overlap. Therefore, using the same proportion of sequence redundancy, we estimate that the sequences we used contained ≈100–145 Mbp of non-redundant sequence covering approximately 19–27% of the entire B. rapa genome (529 Mbp) and capturing almost half (47–66%) of the euchromatic gene space (220 Mbp).
To make a list of NBS-encoding genes from the BAC sequences, protein-coding genes were predicted ab initio using the FGENESH program with the parameters trained using the B. rapa matrix. To reduce sequence redundancy, self-BLAST comparison of gene models was conducted and the redundant gene models were collapsed into a single gene model if BLAST showed perfect matches and source BAC sequences overlapped with one another at the positions that the gene models predicted using Pipmaker (Schwartz et al. 2000). Candidate genes containing NBS domains were identified based on a BLASTP search (Altschul et al. 1997) at a cut-off value of <E −20 using 207 A. thaliana NBS-LRR-encoding genes downloaded from the NIBLRRS server (NIBLRRS database, http://niblrrs.ucdavis.edu/data_links.php) as a query. To ensure that there were no additional related genes missing from the gene prediction, a TBLASTN search was also used against the nucleotide sequences at a cut-off value of <E −10. Matches were considered to be genes encoding NBS motifs if the TBLASTN translation was not interrupted by stop codons in the open reading frame. Sequences found multiple times in the output were identified and manually removed based on BLASTN self comparisons of the initial candidates and sequence comparisons of BAC clones. The sequences used in this study can be found in the GenBank database under the accession numbers listed in Online Resource 1. To identify EST matches of the candidate NBS-encoding genes of B. rapa, 26,416 unigenes and 127,144 EST sequences of B. rapa ssp. pekinensis cv. Chiifu were downloaded from the KBGP database (The Korea Brassica rapa Genome Project). All unigenes and ESTs were compared with the coding sequences of candidate NBS-encoding genes. We considered unigenes and ESTs to have a genome match if more than 90% of sequences matched with at least 95% identity in a BLASTN search (Altschul et al. 1997). If one unigene or EST matched with multiple candidate genes, only the gene with the best alignment score was considered.
Analysis of conserved domains and motif searches
Conserved domains and motifs were identified using hmmpfam comparison to Pfam v23 (Finn et al. 2008). Protein sequences of candidate NBS-encoding genes were compared to the Pfam v23 HMMs using HMMER 2.3.2. (Eddy 2003). To classify the candidate NBS-encoding R genes based on domain structure, each sequence was examined for N-terminal regions, the NBS domains, and the LRR domains plus C-terminal regions using the Multiple Expectation Maximization for Motif Elicitation (MEME) program (Bailey and Elkan 1995). Because Pfam and MEME analyses could not identify the CC motif in the N-terminal region, the region from the N-terminal to the beginning of the P-loop of the NBS domain was also examined using the MARCOIL program with a threshold probability of 90 (Delorenzi and Speed 2002).
Alignment and phylogenetic analysis of sequences
Nucleotide-binding site-encoding genes are known to be subdivided into CNL and TNL groups. However, we did not separate the groups in the phylogenetic analysis as we were interested in examining the divergence between groups based on phylogeny. Amino acid sequences of the NBS domain spanning the P-loop/Kin1 motif to the RNBS-D motif were used. The selected sequences were aligned using ClustalW (Thompson et al. 1994) with default options, and the alignment was manually corrected using the alignment editor Jalview (Waterhouse et al. 2009). Aligned sequences were trimmed at both ends to eliminate regions of poor alignment. A phylogenetic tree was constructed using the Bootstrap neighbor-joining (NJ) method in MEGA4 (Tamura et al. 2007). The stability of tree nodes was tested by bootstrap analysis with 1,000 replicates.
Chromosome assignment of NBS-encoding genes
The source BAC clones which contain the candidate NBS-encoding R genes were anchored on the 10 B. rapa chromosomes using sequence-tagged site markers (Kim et al. 2006) with additional information of assembled sequence contigs (The Korea Brassica rapa Genome Project), fluorescent in situ hybridization (FISH) of BAC clones, and physical map information (Mun et al. 2008).
Ks and Ka analyses of homologous sequences
The timing of a duplication event or divergence of homologous segments and selective pressure on duplicated genes were estimated by calculating the synonymous (K s) and non-synonymous substitution (K a) per site between homologous genes. To calculate the K s and K a value, the protein sequences of the gene models were aligned using the all-against-all alignment and the resulting alignment was used as a reference to align the nucleotide sequences. After the removal of gaps, the K s and K a values from pairwise alignments of homologous sequences were determined using the maximum likelihood method implemented in the CODEML (Goldman and Yang 1994) module of the PAML (Yang 2007) package using the F3 × 4 model, similar to the analysis described by Blanc et al. (2003). We compared the mode rather than the means of K s distributions because the mode is not affected by bias due to incorrectly defined homolog pairs, which is partly responsible for overestimation of K s values. Only gene pairs with a K s estimate of <5 were considered for further evaluation and their K s age distribution was represented at an interval of 0.1. The calculation of divergence time was based on the neutral substitution rate of 1.5 × 10−8 substitutions per site per year for chalcone synthase (Chs) (Koch et al. 2000).
Results
Identification of NBS-encoding genes and analysis of domains and motifs
The B. rapa NBS-encoding genes were identified based on BLAST searches of non-redundant gene models against the A. thaliana R gene family followed by a hmmpfam search. A total of 92 non-redundant NBS-encoding R gene candidates were identified from the genomic sequences we collected (Table 1; Online Resource 1). Among the candidate genes, 17 genes were identified as CNLs and 42 genes were identified as TNLs. In addition, 33 different predicted protein sequences that lack specific motifs or domains were classified into three distinct groups namely CN (2), NL (21), and TN (10); some of these genes are probably pseudogenes. TIR-only and TIR-X sequences were not indentified in this study.
Because typical NBS domains often contain conserved motifs that allow CNL and TNL sequences to be distinguished, the 33 genes lacking the full complement of domains were further divided into CNL or TNL according to their NBS signature. A previous study in A. thaliana identified eight major NBS motifs that differed in sequence between CNL and TNL proteins (Meyers et al. 2003). As expected, MEME analyses identified consensus sequence motifs for the P-loop, kinase-2, NBS-A to -D, GLPL, and MHDV regions; these consensus sequence motifs are similar to those of A. thaliana with some slight modifications (Table 2). These motifs were relatively well conserved between CNL members. In contrast, the NBS-A and kinase-2 motifs were frequently deleted in the TNL members (Online Resource 1). In particular, the MHDV motif in TNL proteins was often modified into an MHSL motif, and several TNs (BrTN2, BrTN7, and BrTN10) also contained a modified MHDV motif that is not present in A. thaliana TNs, indicating two different origins of TNs in Brassica. We also examined proteins with multiple NBS domains and found three proteins (BrTNL14, BrTNL26, and BrTNL35) that have two domains within a single protein (TNLN). The NBS domains were similar in motif structure, but the first NBS domain was truncated. This domain type has only ever been detected in a single poplar R gene (Kohler et al. 2008).
Table 2.
Consensus sequences of major NBS domain motifs found within the predicted B. rapa CNL and TNL proteins
| Class | Motif | Consensus sequencea |
|---|---|---|
| TIR-NBS-LRR | TN linker | TPSKDFDDLVGIEAHIEKLKS |
| TNL P-loop/Kin1 | VRMVGIWGPAGIGKTTIARALYNQLSSRF | |
| TNL RNBS-A | DDYGLKLRLQENFLSQIFNQK | |
| TNL Kin2 | DIEVRHLGRAQEMLSDKKVLVVLDEVDNWWQLE | |
| TNL RNBS-B | LKDKKVLVVLDDVDDLEQLEALAGETQWFGPGSRIIITTRD | |
| TNL RNBS-C | IDHIYEVxFPSEDEALQIFCQYAFGQKSPPDGF | |
| TNL GLPL | LAREVTKLAGNLPLGLRVLGSYLRGMSKEEWIEALPRLRTS | |
| TNL RNBS-D | LDGEIESTLRFSYDALDDNEKTLFLHIACFFNGEKVDYVKS | |
| TNL MHDV | MHSLLQQMGREIVR | |
| CC-NBS-LRR | CNL P-loop/Kin1 | VIGIYGMGGIGKTTLARQIYN |
| CNL RNBS-A | VIWVVVSQDLTIHKIQDDIAQ | |
| CNL Kin2 | WDKKEExEKAxDIHNVLRRKKFVLLLDDIWEKVDLSEIGVP | |
| CNL RNBS-B | FPTKENGCKVVFTTRSKEVCGRMGVDDPM | |
| CNL RNBS-C | VKCLDTDKAWDLFKKKVGENTLKSHPDIPELARKVAGKCRG | |
| CNL GLPL | VAEKCRGLPLALNVIGETMSCKRTVQEWRHAVDVLTSSAAEFSGMEDKILPILKYSYD | |
| CNL RNBS-D | KSCFLYCSLFPEDYKIDKEELIEYWICEG | |
| CNL MHDV | MHDVVREMALWIAS |
aConsensus amino acid sequence was derived from MEME. Underlined residues indicate consensus sequences identical to the motifs detected in the Arabidopsis NBS domains by Meyers et al. (2003). x indicates a non-conserved residue
The LRR regions were characterized by LRRs that were variable in sequence, size, and occurrence per gene. The average number of LRR domains in TNL (5.2 ± 2.4 SD, range 2–12) was more than twofold higher than in CNL (2.4 ± 0.9 SD, range 1–4). Pfam and MEME analyses identified three conserved LRR motifs and two of them showed group specificity (Online Resource 2). LRR motif 1 (FNPxxLVELNxxNSKLEKLWEGxxPLTxL), which is similar to A. thaliana TNL-LRR motif 4, was common in TNL proteins. In contrast, LRR motif 3 (SLxYLxLSGxSIxxLPSxLKx) was frequently identified in CNL proteins. However, the LRR motif 2 (SNxTxLExLDLSxCxSLVELPSSxxNLKK), which corresponds to the A. thaliana LDL-LRR motif, showed relatively low group specificity. Furthermore, duplicated repeated motif patterns were frequently observed in TNLs, suggesting that duplication of the LRRs of TNL accounts for their higher length variation in Brassica. A putative nuclear localization signal (NLS) has been described in the C-terminal domains of the A. thaliana RRS1 (Deslandes et al. 2003) and RPS4 (Wirthmueller et al. 2007) proteins and the N-terminal of five poplar TNL proteins (Kohler et al. 2008). We identified two proteins containing a NLS at the N-terminal (BrNL16) and C-terminal (BrCNL16), respectively. The C-terminal NLS of BrCNL16 (PRCRRKKRK) was similar to that of the A. thaliana RRS1 protein (KKRKRS), suggesting that this signal may target the protein to the nucleus. However, the putative N-terminal NLS of BrNL16 (RRRRLEFKLRFINSPRPVVFTYSNIKAK) appears to be a unique variant or a spurious domain, because it does not have sequence similarity to any R protein N-terminal NLS sequence.
In summary, we found 30 CNL and 62 TNL genes in B. rapa showing 1:2 ratio of CNL to TNL, which is similar to what is found in A. thaliana (Table 1) (Meyers et al. 2003). The average number of exons in NBS-encoding genes was 4.2 in B. rapa (Online Resource 1). This exon estimate was less than that in all predicted genes (4.7 per gene). In addition, the average number of exons per CNL genes (2.3 per gene) was significantly lower than that in TNL genes (5.2 per gene; t test, P < 0.001) and 43% of CNL genes were encoded by a single exon. This result is consistent with what is found in A. thaliana. In A. thaliana, NBS-encoding genes contain 4.2 exons, but CNLs and TNLs have 2.2 and 5.3 exons per gene on average, respectively (Meyers et al. 2003).
Genomic organization of NBS-encoding genes in the B. rapa genome
To determine the distribution of B. rapa NBS-encoding genes in the genome, genetic mapping information, namely the position of source BACs, was used. Most of the source BAC sequences have been anchored on two existing “Jangwon” and “VCS” genetic maps (Kim et al. 2006; The Korea Brassica rapa Genome Project). Seventy-nine B. rapa NBS-encoding genes were distributed across linkage groups except A4, whereas 13 genes did not map to any characterized BACs (Online Resource 3). Of the 79 NBS-encoding R genes located on linkage groups, almost 57% of the genes are encoded on chromosomes A3 and A9 (Fig. 1; Online Resource 3), primarily due to the higher representation of sequences from these two chromosomes in this study. Additionally, 48% (44/92) of NBS-encoding genes form physically homogenous clusters and are unevenly distributed in the genome as tandem arrays. There were more TNL tandem duplicates (53%) than CNL tandem duplicates (37%). Uneven distribution of NBS-encoding genes on chromosomes appears to be common in plants (Kohler et al. 2008; Meyers et al. 2003; Porter et al. 2009; Yang et al. 2008b; Zhou et al. 2004). Interestingly, TNL genes are generally widely distributed across the B. rapa chromosomes except for chromosomes A4 and A5, whereas 60% of CNL genes are located on chromosome A9.
Fig. 1.
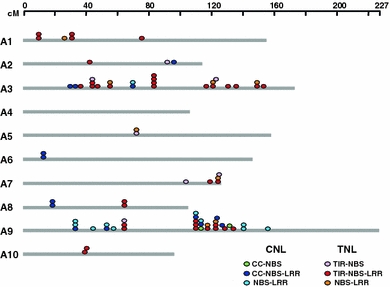
Distribution of predicted NBS-encoding genes on the ten linkage groups of B. rapa. Color ovals above each linkage group (gray bars) indicate the approximate locations of each gene. For more details, including the BAC clone identities and associated markers, see Online Resource 3
As the B. rapa genome has not been fully sequenced, we attempted to estimate how many NBS-encoding R genes exist in the B. rapa genome using the B. rapa unigene sets (The Korea Brassica rapa Genome Project) with a BLASTN similarity E-value cut-off of E −20 to B. rapa and A. thaliana NBS-encoding genes. Applying this criterion, we found 98 NBS-like expressed sequences in the 26,416 unigenes. We identified 34 B. rapa NBS-encoding genes that showed a nearly identical match to the unigene sequences. Therefore, we estimate that the current genome sequence contains roughly 35% (34/98) of B. rapa NBS-encoding genes. Taken together with the approximately 47% euchromatic genome coverage of sequences used in this study, we estimate that the overall number of NBS-encoding genes in the B. rapa genome is at least ≈200–270, which is relatively lower than other sequenced model species with similar genome sizes (Ameline-Torregrosa et al. 2008; Kohler et al. 2008; Monosi et al. 2004; Yang et al. 2008b). Because B. rapa NBS-encoding genes are not evenly distributed in the genome, it is possible that major NBS-encoding gene clusters lie in the regions of genome that is not analyzed.
Phylogeny construction and in silico analysis of gene expression
A phylogenetic analysis of 90 NBS-encoding sequences was performed to classify subgroups and identify relationships between genes. Two genes (BrCNL1 and BrNL7) were excluded due to more than 50% deletion of the NBS domain. Because a previous study showed that phylogenetic analyses of NBS domains allow subgroups of R genes to be distinguished (Meyers et al. 1999), we constructed a composite phylogenic tree for all candidate genes using NBS consensus sequences. Both distance and parsimony algorithms produced similar trees (data not shown). Figure 2 shows a phylogeny of B. rapa NBS-encoding genes with the chromosomal origin of genes and expression support indicated. Six subgroups, comprising three CNL subgroups and three TNL subgroups, are present in the B. rapa genome. There was a clear separation between the CNL and TNL groups, reflecting ancient differentiation of NBS-encoding genes into two major groups in the B. rapa genome. Among the CNL and TNL subgroups, TNL-3 was paraphyletic; phylogenetic and motif analyses of the NBS domain suggested that the TNL-3 subgroup was derived from the other two TNL subgroups by deletion or recombination of motifs (Fig. 2; Online Resource 1). Most of the CNL genes clustered in the two distinct major subgroups, CNL-1 and CNL-2, with the exception of two genes, BrCNL8 and BrCNL15, which clustered in the CNL-3 subgroup due to deletion of the kinase-2, NBS-B, and NBS-C motifs in the NBS domain. NBS sequences from the truncated R genes (TN or NL) were dispersed among the different subgroups, indicating a diverse origin rather than a monophyletic origin due to domain loss. Local expansion of specific types was also apparent. For example, CNLs on chromosome A9 comprised 65 and 71% of CNL-1 and CNL-2 subgroups, respectively. Similarly, TNLs on chromosome A3 comprised approximately 40% of TNL-1 and TNL-2 subgroups. In silico analysis of gene expression indicated that only 25 predicted genes (28%) have EST support with an average of 2.4 ESTs per expressed NBS-encoding gene. These genes were expressed in a wide range of cDNA libraries, including those constructed from various development stages, tissue types, and abiotic stress-treated tissues. There was no difference in the ratio of expressed genes between CNL (29%) and TNL (27%) groups. However, expression patterns vary between subgroups and even between highly similar genes within the same subgroup. For example, CNL-3 is not expressed and the TNL-2 subgroup has only 18% of the expression. Moreover, among the four highly similar genes in the TNL-1 subgroup (BrNL21, BrTNL42, BrTNL43, and BrTNL45), BrTNL45 displayed seven ESTs while the other genes had no ESTs.
Fig. 2.
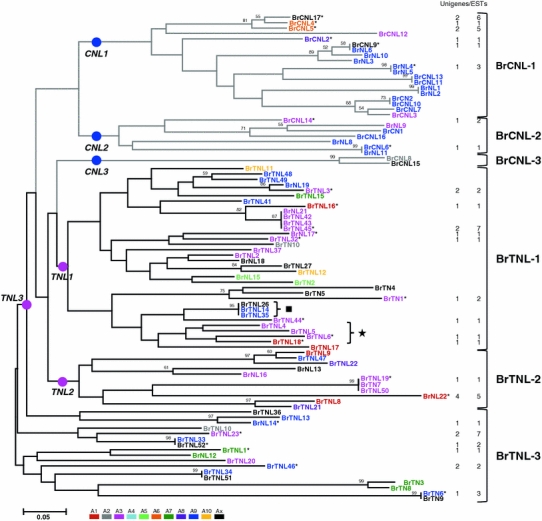
A phylogenetic tree of 90 NBS-encoding genes of B. rapa based on neighbor-joining analysis. CNL and TNL groups are represented as gray and black bars, respectively. Bootstrap values are indicated on the branches and the branch length reflects the estimated number of substitutions per 100 sites; bootstrap results were not reported if the support was <50%. To visualize genomic location better, the names of genes located on the same chromosome are indicated in the same color. Gene names are intended to represent domain configurations. Asterisks indicate the gene models with expression support. The number of matched unigenes and ESTs for each gene is indicated on the right margin. Brackets on the right of the tree indicate subgroup names. A brace with square indicates genes containing two NBS domains (TNLN); the second complete NBS was used for phylogeny construction. A brace with asterisk denotes TNL members that were generated by recent lineage-specific events, as shown in Figs. 3 and 4. For all other relevant information, please see Online Resource 1
Relationship between NBS-encoding genes of B. rapa and A. thaliana
Further phylogenetic reconstruction using the conserved NBS domains of A. thaliana and B. rapa NBS-encoding genes was performed to assess the support for R gene subgroups and to investigate the evolution of B. rapa-specific NBS-encoding genes (Fig. 3). Because A. thaliana CNL and TNL groups have been categorized (Meyers et al. 2003), we constructed a composite phylogenetic tree for the two groups. The analysis separated CNL and TNL type sequences, and each group was divided into three major clades: clades I–III for TNL, and V–VII for CNL. Clade IV was a paraphyletic group that contained TN genes; four from B. rapa (BrTN1, BrTN4, BrTN6, and BrTN9) and two from A. thaliana (At1G72850 and At5G48780). These proteins were characterized by a missing MHDV motif and are considered to have an ancient origin based on a previous report that TN proteins are encoded in the genomes of conifers and grasses, and that two A. thaliana TN proteins are extremely well conserved in rice (Meyers et al. 2002). Although bootstrap values were low for the branches, the topology of the composite tree for A. thaliana-B. rapa NBS-encoding genes was consistent with that of the B. rapa NBS-encoding gene tree. All B. rapa subgroups present in Fig. 2 were present in the composite phylogenetic tree in Fig. 3, which supports the categorization of B. rapa NBS-encoding genes into six subgroups. In addition, the tree indicates syntenic relationships between A. thaliana and B. rapa subgroups. B. rapa subgroups were clustered together with one or two distinct A. thaliana subgroups except clade I in which BrTNL-1 were paraphyletic with AtTNL-E to -H, suggesting possible further division of BrTNL-1 members. Further identification of NBS-encoding genes from the B. rapa genome will enable the reconstruction of a more robust phylogeny.
Fig. 3.
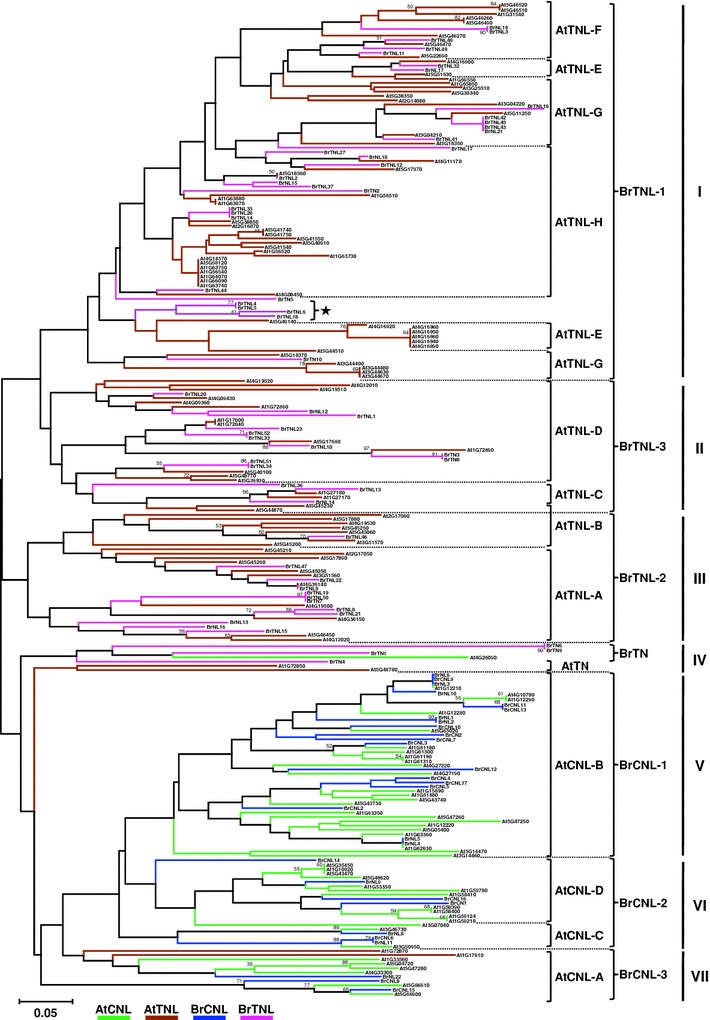
Phylogenetic relationships between NBS-encoding genes in the B. rapa and A. thaliana genomes. The neighbor-joining tree was constructed using the NBS domains of 90 B. rapa and 140 Arabidopsis NBS-encoding proteins reported by Meyers et al. (2003) using MEGA4 software. CNL and TNL groups of each species are represented by color bars. Bootstrap values are indicated on the branches and the branch length reflects the estimated number of substitution per 100 sites; bootstrap results were not reported if the support was <50%. Brackets and braces in the tree indicate the subgroup names of Arabidopsis as reported by Meyers et al. (2003) and the B. rapa groups identified in Fig. 2, respectively. On the right margin, clades I–III represent the TNL group; clade IV represents a mixed TN group; and clades V–VII represent the CNL group. An example of local expansion in BrTNL-1 is indicated with an asterisk
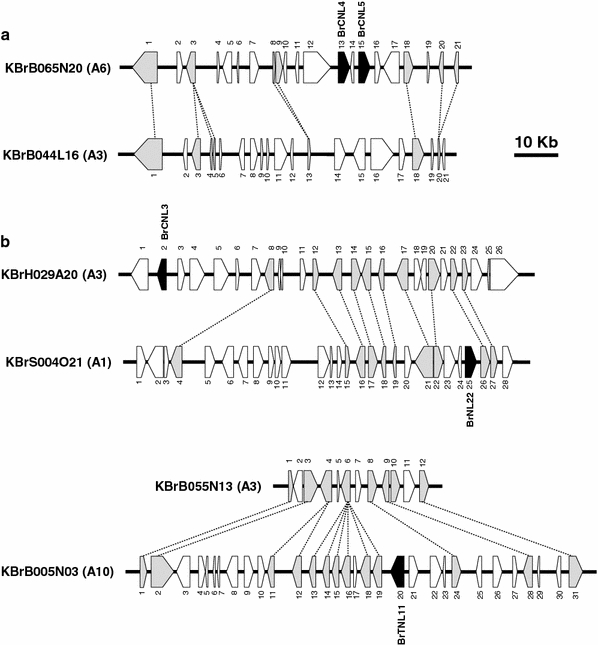
The phylogenetic distribution of A. thaliana and B. rapa NBS-encoding genes does not indicate a major B. rapa-specific expansion. Rather, the local expansion of particular types was detected. Four genes in the BrTNL-1 subgroup located on chromosome A1 (BrTNL18) and A3 (BrTNL4, BrTNL5, and BrTNL6) formed a local cluster separated from the AtTNL-E subgroup, suggesting that these genes might have been generated recently after the divergence of A. thaliana and B. rapa. We compared the microsyntenic structure of B. rapa source BAC clones containing the four genes and syntenic counterpart regions of A. thaliana (Fig. 4a; Online Resource 4). This comparison revealed that the B. rapa region is shrunken in size compared to the syntenic A. thaliana region as reported by Mun et al. (2009) and that BrTNL4, BrTNL5, and BrTNL6 are tandem duplicates without homologous A. thaliana counterparts. Estimation of K s in these genes indicated that they are recent duplicates that occurred after the split of A. thaliana and B. rapa (Fig. 4c). Because BrTNL17 and BrTNL18 have homologous counterparts of ancient origin in the AtTNL-G subgroup (At5G18350, At5G18360, and At5G18370) and frequent ectopic translocation of NBS-encoding genes among different chromosome does not occur in A. thaliana (Baumgarten et al. 2003), loss of synteny for BrTNL4, BrTNL5, and BrTNL6 suggests gene deletion in the A. thaliana genome. This result suggests that recent tandem duplications and ectopic deletions may play a role in the generation of novel lineage-specific Brassica R genes.
Fig. 4.
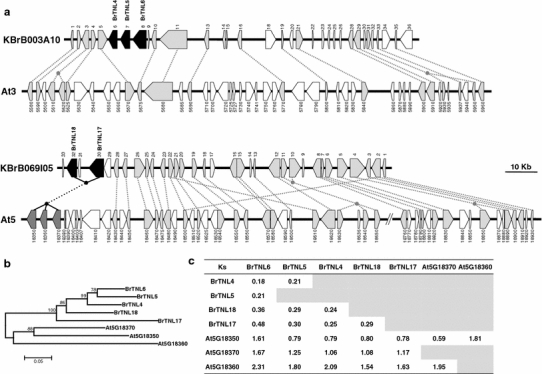
An example of lineage-specific expansion of B. rapa TNL genes. a Microsynteny between syntenic regions of B. rapa and Arabidopsis showing unique amplification of B. rapa BrTNL4, BrTNL5, and BrTNL6 due to tandem duplication and/or deletion of counterparts. Lines indicate homologous pairs. Supporting data and gene annotations are provided in Online Resource 4. b A neighbor-joining tree showing phylogenetic relationship among the NBS-encoding genes in the syntenic regions. c K s estimates for the NBS-encoding genes in the syntenic regions
Duplication and differential selection of the B. rapa NBS-encoding gene family
Similar to Arabidopsis, TNL genes also out-number CNL genes in B. rapa, indicating either amplification of TNL genes or loss of CNL genes. To investigate the evolution of NBS-encoding genes and to estimate the approximate timing of duplication, we compared the distribution of synonymous substitutions (K s) between CNL and TNL genes in A. thaliana and B. rapa. As shown in Fig. 5a–c, CNL and TNL groups in both the A. thaliana and B. rapa genomes appear to have diverged after a single ancient duplication event 115–125 MYA based on K s modes of 2.4–2.6, which might be resulted from the oldest WGD (1R). The ancient duplication of NBS-encoding genes in the A. thaliana and B. rapa genomes is supported by the K s distribution of each CNL and TNL group (Fig. 5d–i). The age distributions of CNL and TNL indicate that each group has been generated by several rounds of duplication events that may be related to the WGDs. The K s distributions for the CNL and TNL groups between A. thaliana and B. rapa are characterized by three major peaks at K s = 2.5–2.8, 1.3–1.7, and 0.5–0.8, which roughly correspond to three WGD events (1R, 2R, and 3R, respectively) in the common ancestor of Arabidopsis and Brassica (Fig. 5d, g). In particular, note that the TNL group has a reduced K s peak for the ancient duplication event (1R) compared to the CNL group, suggesting that the relatively recent duplication in the TNL group resulted from the old duplication event (2R) or that more TNL genes were deleted after the ancient duplication. A comparison of the K s mode for the paralogs of each group in both A. thaliana and B. rapa identified differences in the duplicated genes retained in the two genomes. For CNL group, two peaks representing 1R and 2R are evident, but 3R peak has reduced (Fig. 5e, f). However, for TNL group, the peak representing 2R are evident, but the 1R and 3R peaks have collapsed (Fig. 5h, i). Surprisingly, in the B. rapa genome, a peak (mode K s = 0.2–0.3) for WGT event (4R) has also been underestimated in both types (Fig. 5f, i). Microsynteny comparison of selected paralogous regions showed none of NBS-encoding genes conserved in the duplicated or triplicated regions (Fig. 6; Online Resource 5). Taken together, these findings suggest that duplicated R genes produced by the 3R or 4R events were widely lost in the triplicated B. rapa genome.
Fig. 5.
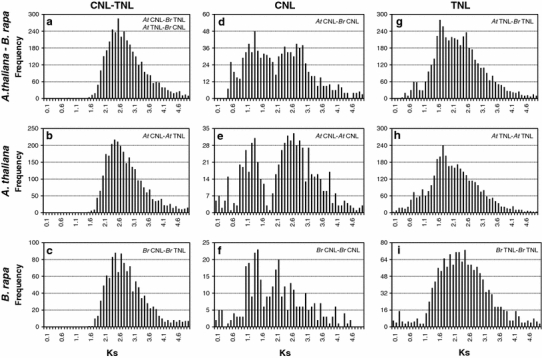
Polyploidy events in the A. thaliana and B. rapa genomes viewed through the frequency distribution of relative K s modes for NBS-encoding genes. Distributions of K s values were obtained from homologous CNL and TNL genes (a–c), paralogous CNL genes (d–f), and paralogous TNL genes sets (g–i) in the Arabidopsis and B. rapa genomes. The vertical axes indicate the frequency of paired sequences and the horizontal axes denote K s values at 0.1 intervals. The black bars depict the positions of the modes of the K s distribution obtained from homologous or paralogous gene pairs. At, A. thaliana; Br, B. rapa
Fig. 6.
An example of microsynteny relationship between paralogous BAC pairs of B. rapa showing loss of redundant NBS-encoding genes in the duplicated (a) or triplicated regions (b). Paralogous BAC pairs containing NBS-encoding genes were selected based on the report of Mun et al. (2009). Lines indicate paralogous pairs. Assigned chromosome of BAC clone was indicated in parenthesis. Supporting data and gene annotations are provided in Online Resource 5
To test whether different selection pressures can explain the differential accumulation of the two groups in the Brassicaceae genome, K a/K s ratios were calculated. In both A. thaliana and B. rapa genomes, the overall K a/K s ratios for CNL (0.404 and 0.387, respectively) were higher than those for TNL (0.356 and 0.353, respectively). The difference between CNL and TNL was highly significant in a two-tailed t test (P < 0.001). This result is consistent with previous studies (Cannon et al. 2002; Yang et al. 2008b), suggesting higher selective constraints and purifying selection in the TNL group. Intriguingly, the distribution of K a/K s ratios according to K s value for most homologs in each group indicated a significant change in selection pressure on the CNL and TNL groups at a specific time point. As shown in Fig. 7, the CNL group had a higher average K a/K s than the TNL group after the ancient duplication event (1R), suggesting that CNL genes were positively selected after the ancient WGD event in both the A. thaliana and B. rapa genomes. However, soon after the old duplication (2R), around K s = 1.6, the TNL group genes had a higher K a/K s ratio than recent duplicates in the CNL group. This result suggests relaxation of selective constraint in the TNL group than the CNL group, which might have resulted in recent amplification of TNL genes in both the Arabidopsis and Brassica genomes. The difference in K a/K s ratios between the CNL and TNL groups in the B. rapa genome is smaller than that seen in A. thaliana; this may be because the whole genome sequence of B. rapa is not yet available for analysis.
Fig. 7.
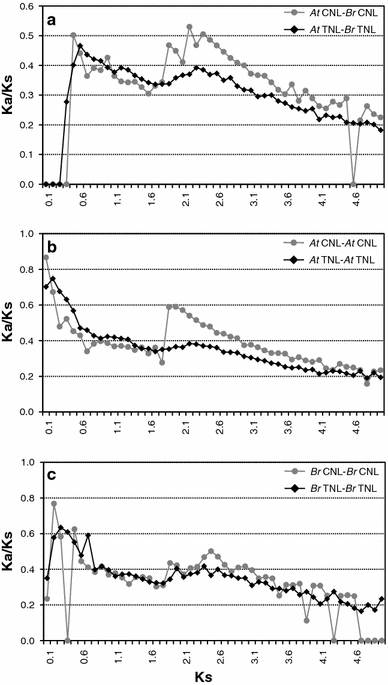
The relationship between K a/K s and K s of paralogous NBS-encoding gene sets between the A. thaliana and B. rapa genomes (a) or in each individual genome (b, c). The vertical axes indicate the average K a/K s ratio in CNL (gray line) and TNL (black line) groups. The horizontal axes denote a K s value at 0.1 intervals
Discussion
Brassica crop species have long been cultivated worldwide as vegetables, oilseed crops, forages, and condiments. The past couple of years have seen the development of abundant genetic and genomic resources for these crop species. In particular, various genomics tools for B. rapa have greatly facilitated our understanding of the structure and evolution of the Brassica genome. The value of this species has been elevated by its close relationship not only with A. thaliana but also with its diploid sibling species (B. oleracea and B. nigra) and allopolyploid hybrids (B. napus, B. carinata, and B. juncea), which is reflected by similar polyploidy genome structures and conserved phenotypes such as enlarged vegetative or floral meristems. Because Brassica species have a similar genome content and genome structure, B. rapa can serve as an alternative to A. thaliana for cloning the orthologs of many economically important genes in other Brassica crops. Likewise, because most B. rapa pathogens also infect other vegetable-type Brassica crop cultivars, B. rapa can be used as a source to identify R genes for common pathogens of Brassica crops. Furthermore, the long cultivation history of Brassica crops in a wide diversity of environmental conditions suggests that R gene products for diverse disease phenotypes can be discovered in the B. rapa genome. In such cases, R genes with novel specificities that are discovered in B. rapa could be directly applied to other cultivated Brassica crops using simple transformation techniques. A successful example of a translational strategy to introduce disease resistance from a model organism into a crop species is the introduction of the RCT1 gene from M. truncatula into alfalfa cultivars for anthracnose resistance (Yang et al. 2008a). Therefore, a better understanding of the genomic organization of NBS-encoding genes in the B. rapa genome and characterization of their genetic and molecular mechanisms is highly desirable.
This study is the first report of genome-wide discovery of NBS-encoding R genes in the Brassica genome. Although the complete genome of B. rapa has not yet been fully sequenced, the availability of BAC sequences covering approximately 50% of the euchromatic genome of B. rapa enabled identification of NBS-encoding R genes in the Brassica genome. We estimated that B. rapa possesses smaller number of NBS-encoding genes recorded than sequenced crop species with similar genome sizes (≈400–500 Mbp) with the exception of papaya. This finding is surprising, because the B. rapa genome has evolved as a triplicated hexaploid from the common ancestor of A. thaliana, and thus is expected to have high levels of genomic redundancy. However, the estimated number of NBS-encoding genes in the B. rapa genome is slightly higher than that in A. thaliana. This result suggests that roughly half of the NBS-encoding genes generated as a result of WGT approximately 11–12 MYA were lost in the current B. rapa genome, demonstrating a rapid birth and death dynamic in the Brassica R gene family.
One possible explanation for the low number of NBS-encoding genes despite the WGT event is the overall downsizing of the euchromatic gene space of the B. rapa genome, related to the diploidization process after the WGT event. A synteny comparison of B. rapa seed BAC sequence contigs with their homologous counterparts in the A. thaliana genome indicated that the B. rapa genome shrunk by ≈30% due mainly to the deletion of roughly one-third of the redundant proteome as well as transposable elements in the euchromatic regions (Mun et al. 2009). In addition, as shown in Fig. 6, loss of redundant genes in the paralogous regions generated by WGD or WGT was frequently observed in the B. rapa genome. Similar genome-wide deletion of redundant NBS-encoding gene components has also been reported in other plant genomes. None or only a small number of NBS-encoding genes have been identified in the WGD blocks in rice (Yu et al. 2005), grape (Yang et al. 2008b), A. thaliana (Nobuta et al. 2005), and poplar (Yang et al. 2008b), indicating that most duplicated NBS-encoding genes were lost soon after WGD. Functional redundancy or artificial selection during domestication could accelerate the deletion or silencing of duplicated NBS-encoding genes that have not functionally diversified in the polyploid Brassica genome. The remaining copies are likely to retain their original function, thus B. rapa may contain sufficient NBS-encoding genes to defend against pathogens. Moreover, gene loss in polyploid genomes is known to be non-random (Blanc and Wolfe 2004; Seoighe and Gehring 2004), and polyploidy followed by the genome-wide removal of some but not all redundant genomic material can result in species-specific differences in the function of homologous genes (Adams and Wendel 2005). Therefore, some of the NBS-encoding genes of B. rapa that survived may have become more specific or gained novel disease resistance. Furthermore, almost 50% of NBS family members were detected as tandem arrays within homogenous clusters, and this percentage is expected to increase when complete sequence scaffolds are analyzed. This result is consistent with previous reports from other plants (Ameline-Torregrosa et al. 2008; Nobuta et al. 2005; Yang et al. 2008b; Yu et al. 2005), suggesting that tandem duplication in combination with polyploidy played an important role in the expansion of NBS-encoding genes in the Brassica genome. The tandem-clustered R gene family could provide a source of lineage-specific R genes with taxonomically restricted functionality, though identification of the entire NBS-encoding gene family in the B. rapa genome is required to evaluate if this hypothesis is correct.
The availability of B. rapa genome sequences enabled genome-wide evolutionary analysis of NBS-encoding genes in the Arabidopsis–Brassica lineage. We found that the CNL and TNL groups in the Arabidopsis and Brassica genomes shared a single ancient duplication event (Fig. 5a–c). NBS-encoding genes in the A. thaliana and B. rapa genomes showed unique patterns of CNL and TNL expansion based on K s analysis; these patterns were related to several WGD events. In addition, genes of each group that were retained in individual genomes had a specific K s distribution pattern, suggesting that the two groups experienced different selection pressures. The relatively high K a/K s ratio found in ancient duplicates in the CNL group implies that relaxation of purifying selection on the ancient duplicated CNL genes might have resulted in ancient diversification (accumulation of degenerative mutations) of this group before the second WGD event (2R), which is estimated to have occurred ≈53 MYA. In contrast, the K s distributions for TNL genes in the A. thaliana and B. rapa genomes have relatively high frequencies for the old duplication event (2R) and some skew toward ancient (1R) and recent duplication events (3R). This pattern of K s distributions indicates that the 2R event contributed significantly to the evolution of the TNL group. Analysis of K a/K s ratio indicates that the relatively higher relaxation of selective constraints on the TNL genes after the 2R event was sustained until separation of the Arabidopsis and Brassica lineages, resulting in amplification of the TNL group genes in the common ancestor of Arabidopsis and Brassica. These data are in good agreement with the previous finding that TNL group genes are more homogenous and have been amplified more recently in the A. thaliana genome than CNL group genes (Meyers et al. 2003). Interestingly, ancient tandem duplicates of CNL are very rare compared to those of TNL genes in the B. rapa genome; tandem duplicates of TNL genes appear to have originated over a long period of time (Online Resource 6). These results collectively indicate that greater deletion of ancient duplicate CNL genes and relatively strong relaxation of selective constraints of TNL genes after the 2R event may be responsible for greater amplification of TNL group genes than CNL group genes in the Brassica genome. Duplications to distant positions were also observed. Some of these rearrangements could be associated with segmental duplications of entire chromosomal regions, which are frequently observed in the Brassica genomes (Town et al. 2006; Yang et al. 2006).
Lineage-specific amplification of NBS-encoding genes in the Brassica genome is an ongoing process as emphasized by the local expansion of B. rapa TNL genes. The detection of R genes not present in A. thaliana (TNLN) indicates that domain acquisition has occurred in the Brassica lineage, which may result in biological innovation. The uneven and clustered distribution of NBS-encoding genes and the discovery of recently duplicated redundant genes provide insight into how novel pathogen-resistant genes can evolve via additional gene duplication events, ectopic recombination, unequal crossing over, and diversifying selection (Michelmore and Meyers 1998). We anticipate that further analysis of complete genome sequences with accurate annotation supported by manual verification will provide more insight into the evolution of NBS-encoding genes in the B. rapa genome.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank Young-Joo Seol and Mina Jin of the National Academy of Agricultural Science, RDA for sequence and genetic marker information, and Seung-Chan Lee of the National Institute of Horticultural and Herbal Science, RDA and Dae-Soo Kim of the Korean Bioinformation Center, KRIBB, for bioinformatics assistance. This work was supported by grants from the National Academy of Agricultural Science (200901FHT020710397) and the National Institute of Horticultural and Herbal Science (2009FHT020508395), Rural Development Administration, Korea.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
J.-H. Mun and H.-J. Yu contributed equally to this work.
Contributor Information
Jeong-Hwan Mun, Phone: +82-31-2991635, FAX: 82-31-2991636, Email: munjh@rda.go.kr.
Hee-Ju Yu, Phone: +82-31-2906209, FAX: +82-31-2906219, Email: yuheeju@rda.go.kr.
References
- Adams KL, Wendel JF. Novel patterns of gene expression in polyploid plants. Trends Genet. 2005;21:539–543. doi: 10.1016/j.tig.2005.07.009. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acid Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameline-Torregrosa C, Wang BB, O’Bleness MS, Deshpande S, Zhu H, Roe B, Young ND, Cannon SB. Identification and characterization of nucleotide-binding site-leucine-rich repeat genes in the model plant Medicago truncatula. Plant Physiol. 2008;146:5–21. doi: 10.1104/pp.107.104588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai J, Pennill LA, Ning J, Lee SW, Ramalingam J, Webb CA, Zhao B, Sun Q, Nelson JC, Leach JE, Hulbert SH. Diversity in nucleotide binding site-leucine-rich repeat genes in cereals. Genome Res. 2002;12:1871–1884. doi: 10.1101/gr.454902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. The value of prior knowledge in discovering motifs with MEME. Proc Int Conf Intell Syst Mol Biol. 1995;3:21–29. [PubMed] [Google Scholar]
- Baumgarten A, Cannon S, Spangler R, May G. Genome-level evolution of resistance genes in Arabidopsis thaliana. Genetics. 2003;165:309–319. doi: 10.1093/genetics/165.1.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell. 2004;16:1679–1691. doi: 10.1105/tpc.021410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Hokamp K, Wolfe KH. A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 2003;13:137–144. doi: 10.1101/gr.751803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon SB, Zhu H, Baumgarten AM, Spangler R, May G, Cook DR, Young ND. Diversity, distribution, and ancient taxonomic relationships within the TIR and non-TIR NBS-LRR resistance gene subfamilies. J Mol Evol. 2002;54:548–562. doi: 10.1007/s00239-001-0057-2. [DOI] [PubMed] [Google Scholar]
- Dangl JL, Jones JD. Plant pathogens and integrated defense responses to infection. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- Delorenzi M, Speed T. An HMM model for coiled-coil domains and a comparison with PSSM-based predictions. Bioinformatics. 2002;18:617–625. doi: 10.1093/bioinformatics/18.4.617. [DOI] [PubMed] [Google Scholar]
- Deslandes L, Olivier J, Peeters N, Feng DX, Khounlotham M, Boucher C, Somssich I, Genin S, Marco Y. Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc Natl Acad Sci USA. 2003;100:8024–8029. doi: 10.1073/pnas.1230660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR (2003) HMMER user’s guide. ftp://selab.janelia.org/pub/software/hmmer/CURRENT/Userguide.pdf
- Finn RD, Tate J, Mistry J, Coggill PC, Sammut JS, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, Bateman A. The Pfam protein families database. Nucleic Acid Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman N, Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol. 1994;11:725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- Johnston JS, Pepper AE, Hall AE, Chen ZJ, Hodnett G, Drabek J, Lopez R, Price HJ. Evolution of genome size in Brassicaceae. Ann Bot. 2005;95:229–235. doi: 10.1093/aob/mci016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Chung TY, King GJ, Jin M, Yang TJ, Jin YM, Kim HI, Park BS. A sequence-tagged linkage map of Brassica rapa. Genetics. 2006;174:29–39. doi: 10.1534/genetics.106.060152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch MA, Haubold B, Mitchell-Olds T. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae) Mol Biol Evol. 2000;17:1483–1498. doi: 10.1093/oxfordjournals.molbev.a026248. [DOI] [PubMed] [Google Scholar]
- Kohler A, Rinaldi C, Duplessis S, Baucher M, Geelen D, Duchaussoy F, Meyers BC, Boerjan W, Martin F. Genome-wide identification of NBS resistance genes in Populus trichocarpa. Plant Mol Biol. 2008;66:619–636. doi: 10.1007/s11103-008-9293-9. [DOI] [PubMed] [Google Scholar]
- Liu JJ, Ekramoddoullah AK. Isolation, genetic variation and expression of TIR-NBS-LRR resistance gene analogs from western white pine (Pinus monticola Dougl. ex. D. Don.) Mol Genet Genomics. 2003;270:432–441. doi: 10.1007/s00438-003-0940-1. [DOI] [PubMed] [Google Scholar]
- McHale L, Tan X, Koehl P, Michelmore RW. Plant NBS-LRR proteins: adaptable guards. Genome Biol. 2006;7:212. doi: 10.1186/gb-2006-7-4-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Dickerman AW, Michelmore RW, Sivaramakrishnan S, Sobral BW, Young ND. Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J. 1999;20:317–332. doi: 10.1046/j.1365-313X.1999.t01-1-00606.x. [DOI] [PubMed] [Google Scholar]
- Meyers BC, Morgante M, Michelmore RW. TIR-X and TIR-NBS proteins: two new families related to disease resistance TIR-NBS-LRR proteins encoded in Arabidopsis and other plant genomes. Plant J. 2002;32:77–92. doi: 10.1046/j.1365-313X.2002.01404.x. [DOI] [PubMed] [Google Scholar]
- Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell. 2003;15:809–834. doi: 10.1105/tpc.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore RW, Meyers BC. Clusters of resistance genes in plant evolve by divergent selection and a birth-and-death process. Genome Res. 1998;8:1113–1130. doi: 10.1101/gr.8.11.1113. [DOI] [PubMed] [Google Scholar]
- Monosi B, Wisser RJ, Pennill L, Hulbert SH. Full-genome analysis of resistance gene homologues in rice. Theor Appl Genet. 2004;109:1434–1447. doi: 10.1007/s00122-004-1758-x. [DOI] [PubMed] [Google Scholar]
- Mun JH, Kwon SJ, Yang TJ, Kim HS, Choi BS, Baek S, Kim JS, Jin M, Kim JA, Lim MH, Lee SI, Kim HI, Kim H, Lim YP, Park BS. The first generation of a BAC-based physical map of Brassica rapa. BMC Genomics. 2008;9:280. doi: 10.1186/1471-2164-9-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mun JH, Kwon SJ, Yang TJ, Seol YJ, Jin M, Kim JA, Lim MH, Kim JS, Baek S, Choi BS, Yu HJ, Kim DS, Kim N, Lim KB, Lee SI, Hahn JH, Lim YP, Bancroft I, Park BS. Genome-wide comparative analysis of the Brassica rapa gene space reveals genome shrinkage and differential loss of duplicated genes after whole genome triplication. Genome Biol. 2009;10:R111. doi: 10.1186/gb-2009-10-10-r111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobuta K, Ashfield T, Kim S, Innes RW. Diversification of non-TIR class NB-LRR genes in relation to whole-genome duplication events in Arabidopsis. Mol Plant Microbe Interact. 2005;18:103–109. doi: 10.1094/MPMI-18-0103. [DOI] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, Schmutz J, Spannagl M, Tang H, Wang X, Wicker T, Bharti AK, Chapman J, Feltus FA, Gowik U, Grigoriev IV, Lyons E, Maher CA, Martis M, Narechania A, Otillar RP, Penning BW, Salamov AA, Wang Y, Zhang L, Carpita NC, Freeling M, Gingle AR, Hash CT, Keller B, Klein P, Kresovich S, McCann MC, Ming R, Peterson DG, Mehboob-ur-Rahman, Ware D, Westhoff P, Mayer KF, Messing J, Rokhsar DS. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457:551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- Porter BW, Paidi M, Ming R, Alam M, Nishijima WT, Zhu YJ. Genome-wide analysis of Carica papaya reveals a small NBS resistance gene family. Mol Genet Genomics. 2009;281:609–626. doi: 10.1007/s00438-009-0434-x. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Zhang Z, Frazer KA, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W. PipMaker-a web server for aligning two genomic DNA sequences. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoighe C, Gehring C. Genome duplication led to highly selective expansion of the Arabidopsis thaliana proteome. Trends Genet. 2004;20:461–464. doi: 10.1016/j.tig.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Sillito D, Parkin I, Mayerhofer R, Lydiate D, Good A. Arabidopsis thaliana: a source of candidate disease-resistance genes for Brassica napus. Genome. 2000;43:452–460. doi: 10.1139/gen-43-3-452. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tan X, Meyers BC, Kozik A, West MA, Morgante M, St Clair DA, Bent AF, Michelmore RW. Global expression analysis of nucleotide binding site-leucine rich repeat-encoding and related genes in Arabidopsis. BMC Plant Biol. 2007;7:56. doi: 10.1186/1471-2229-7-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acid Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Town CD, Cheung F, Maiti R, Crabtree J, Haas BJ, Wortman JR, Hine EE, Althoff R, Arbogast TS, Tallon LJ, Vigouroux M, Trick M, Bancroft I. Comparative genomics of Brassica oleracea and Arabidopsis thaliana reveal gene loss, fragmentation, and dispersal after polyploidy. Plant Cell. 2006;18:1348–1359. doi: 10.1105/tpc.106.041665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UN Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn J Bot. 1935;7:389–452. [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ (2009) Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. doi:10.1093/bioinformatics/btp1033 [DOI] [PMC free article] [PubMed]
- Wirthmueller L, Zhang Y, Jones JD, Parker JE. Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr Biol. 2007;17:2023–2029. doi: 10.1016/j.cub.2007.10.042. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang TJ, Kim JS, Kwon SJ, Lim KB, Choi BS, Kim JA, Jin M, Park JY, Lim MH, Kim HI, Lim YP, Kang JJ, Hong JH, Kim CB, Bhak J, Bancroft I, Park BS. Sequence-level analysis of the diploidization process in the triplicated FLOWERING LOCUS C region of Brassica rapa. Plant Cell. 2006;18:1339–1347. doi: 10.1105/tpc.105.040535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Gao M, Xu C, Gao J, Deshpande S, Lin S, Roe BA, Zhu H. Alfalfa benefits from Medicago truncatula: the RCT1 gene from M. truncatula confers broad-spectrum resistance to anthracnose in alfalfa. Proc Natl Acad Sci USA. 2008;105:12164–12169. doi: 10.1073/pnas.0802518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Zhang X, Yue JX, Tian D, Chen JQ. Recent duplications dominate NBS-encoding gene expansion in two woody species. Mol Genet Genomics. 2008;280:187–198. doi: 10.1007/s00438-008-0355-0. [DOI] [PubMed] [Google Scholar]
- Yu J, Wang J, Lin W, Li S, Li H, Zhou J, Ni P, Dong W, Hu S, Zeng C, Zhang J, Zhang Y, Li R, Xu Z, Li S, Li X, Zheng H, Cong L, Lin L, Yin J, Geng J, Li G, Shi J, Liu J, Lv H, Li J, Wang J, Deng Y, Ran L, Shi X, Wang X, Wu Q, Li C, Ren X, Wang J, Wang X, Li D, Liu D, Zhang X, Ji Z, Zhao W, Sun Y, Zhang Z, Bao J, Han Y, Dong L, Ji J, Chen P, Wu S, Liu J, Xiao Y, Bu D, Tan J, Yang L, Ye C, Zhang J, Xu J, Zhou Y, Yu Y, Zhang B, Zhuang S, Wei H, Liu B, Lei M, Yu H, Li Y, Xu H, Wei S, He X, Fang L, Zhang Z, Zhang Y, Huang X, Su Z, Tong W, Li J, Tong Z, Li S, Ye J, Wang L, Fang L, Lei T, Chen C, Chen H, Xu Z, Li H, Huang H, Zhang F, Xu H, Li N, Zhao C, Li S, Dong L, Huang Y, Li L, Xi Y, Qi Q, Li W, Zhang B, Hu W, Zhang Y, Tian X, Jiao Y, Liang X, Jin J, Gao L, Zheng W, Hao B, Liu S, Wang W, Yuan L, Cao M, McDermott J, Samudrala R, Wang J, Wong G, Yang H. The Genomes of Oryza sativa: a history of duplications. PLoS Biol. 2005;3:e38. doi: 10.1371/journal.pbio.0030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Wang Y, Chen J-Q, Araki H, Jing Z, Jiang K, Shen J, Tian D. Genome-wide identification of NBS genes in japonica rice reveals significant expression of divergent non-TIR NBS-LRR genes. Mol Genet Genomics. 2004;271:402–415. doi: 10.1007/s00438-004-0990-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


