Abstract
RNA is a key molecule in life, and comprehending its structure/function relationships is a crucial step towards a more complete understanding of molecular biology. Even though most of the information required for their correct folding is contained in their primary sequences, we are as yet unable to accurately predict both the folding pathways and active tertiary structures of RNA species. Ribozymes are interesting molecules to study when addressing these questions because any modifications in their structures are often reflected in their catalytic properties. The recent progress in the study of the structures, the folding pathways and the modulation of the small ribozymes derived from natural, self-cleaving, RNA motifs have significantly contributed to today’s knowledge in the field.
Keywords: Catalytic RNA, Modulation, RNA folding, Riboswitch, Ribozyme, Structure
Introduction
RNA is a key molecule in life, and the understanding of its structure/function relationships is crucial in molecular biology and therefore has important implications in terms of human health [1]. However, we are as yet unable to accurately predict either the folding pathway or the active tertiary structure of RNA molecules from their primary sequences. Clearly, these two fundamental questions need to be addressed. A long-term goal is to be able to interpret an RNA sequence in terms of its functionally folded three-dimensional form. RNA molecules possess a hierarchical structure: the primary sequence determines the formation of the secondary structure elements, and the secondary structure in turn determines the tertiary folding [2–4]. RNA molecules fold sequentially from 5′ to 3′ using recurring stable submotifs [5, 6], and the folding intermediates tend to become increasingly stable as the tertiary interaction network progresses [7–9]. RNA can fold into multiple structures; however, only a single structure is usually functional. In order to fold correctly, the RNA must avoid the problem of folding into alternative, non-functional structures and kinetic traps [10–13].
Ribozymes are interesting molecules to study when addressing these RNA questions because modifications in their structures are reflected in their catalytic properties. More precisely, small ribozymes are suitable for this task since considerable progress has been made in the determination of their structures [14, 15], and a versatile toolbox is available for their study. Here we present a review of the recent progress in the study of the tertiary structures, the folding pathways and the modulation of the small ribozymes derived from natural, self-cleaving, RNA motifs.
Small self-cleaving ribozymes
This group of natural ribozymes includes five RNA species that have been derived from self-cleaving sequences ranging from ~40 to 200 nucleotides in length and possessing various secondary structures. One subgroup includes three self-catalytic RNA motifs that are part of species belonging to the brotherhood of small, circular, self-replicating RNAs and are essential components of the rolling circle replication mechanism of these infectious RNAs. According to this mechanism, the circular monomer strands are replicated into linear multimeric strands of complementary polarity that then are self-cleaved, on the basis of the RNA motifs, into monomers that, following circularization, participate in the next round of replication [16]. More specifically, the hammerhead and hairpin RNA motifs were obtained from viroids and viroid-like satellite RNAs of plant origin [17–22], while the HDV self-cleaving RNA motif was obtained from the hepatitis delta virus (HDV) that infects humans [23–25]. The hammerhead motif has also been detected in eukaryote satellite DNA transcripts obtained from newt, schistosome and cricket [26–28], while the HDV motif has also been detected in the human genome [29].
The members of the second subgroup of natural self-cleaving RNA motifs are found within the bodies of larger transcripts. For example, the VS self-cleaving motif was identified by accident in the Varkud satellite RNA, an abundant transcript from the circular mitochondrial Varkud satellite DNA found in numerous Neurospora species and in other simple eukaryotes [30]. In common with the above ribozymes, this self-catalytic motif, in vivo, processes a multimeric RNA into monomers [31]. Another example is the glmS self-cleaving motif that was identified, by computer analysis of the 5′ untranslated region (5′UTR) of the glucosamine-6-phosphate synthase (glmS) mRNA of certain gram-positive bacteria, to be a putative riboswitch [32]. In fact, upon the binding of glucosamine-6-phosphate (GlcN6P), the glmS motif has been shown to undergo a site-specific self-cleavage reaction making it the first, and as yet the only, natural allosteric ribozyme [33]. This cleavage dramatically decreases the half-life of glmS mRNA, and thus this ribozyme acts as a regulatory element controlling the glucosamine-6-phosphate synthase level [33].
All of these self-cleaving RNA motifs catalyze the cleavage of the RNA phosphodiester backbone through a transesterification reaction involving the attack of the vicinal 2′-hydroxyl group (2′-OH) on the scissile phosphate, yielding a 2′-3′-cyclic phosphate and a 5′-hydroxyl termini as products [34]. The mechanism involved is a bimolecular nucleophilic substitution (SN2) that is general acid–base catalyzed and requires in-line geometry between the 2′ oxygen, the phosphate and the 5′ oxygen. The catalytic strategies and active site organization at the atomic level for each of these ribozymes have been extensively characterized and do not fall under the scope of this work since these aspects have been recently been reviewed [1, 15]. Finally, the presence of divalent metal cations is important in RNA folding in order to obtain well-defined structures, as well as in the catalysis, by functioning either as a general acid or as a general base when coordinated with water. These self-cleaving RNA motifs are metalloenzymes under physiological conditions, but are also active in the absence of divalent metals under certain conditions such as in the presence of high monovalent metal ion concentrations or at low pH levels [35, 36].
HDV ribozyme
The self-cleaving RNA motif derived from HDV varies from 85 to 95 nucleotides in length depending on the sequence variant and the polarity. It has been possible to separate this self-catalytic sequence into two molecules, thereby creating a trans-acting system where one molecule, the ribozyme, possesses the catalytic properties required to cleave multiple copies of the other molecule, the substrate. This separation can be achieved in several ways, the most frequent being the removal of the junction between stems I and II (Fig. 1a) [25]. The development of trans-acting ribozymes, for HDV as well as all other self-catalytic RNA motifs, has permitted experiments leading to the identification of the important structural features of these motifs [24, 25, 37]. For example, direct mutagenesis experiments led to the identification of the crucial nucleotides in the catalytic core, as well as identifying essential base-paired positions [for examples see Refs. 37, 38]. It should be noted, however, that an unbiased in vitro selection revealed that, with the exception of the catalytic nucleotide (position 76), none of the bases were absolutely required for cleavage [39]. This work clearly revealed that the ribozyme supports more variability than was originally thought to be the case based on the isolation of natural sequences. In addition, it also revealed that higher cleavage levels were observed for sequences closely related to the natural ones, suggesting that the combination of all of the structural features is important for optimal cleavage activity.
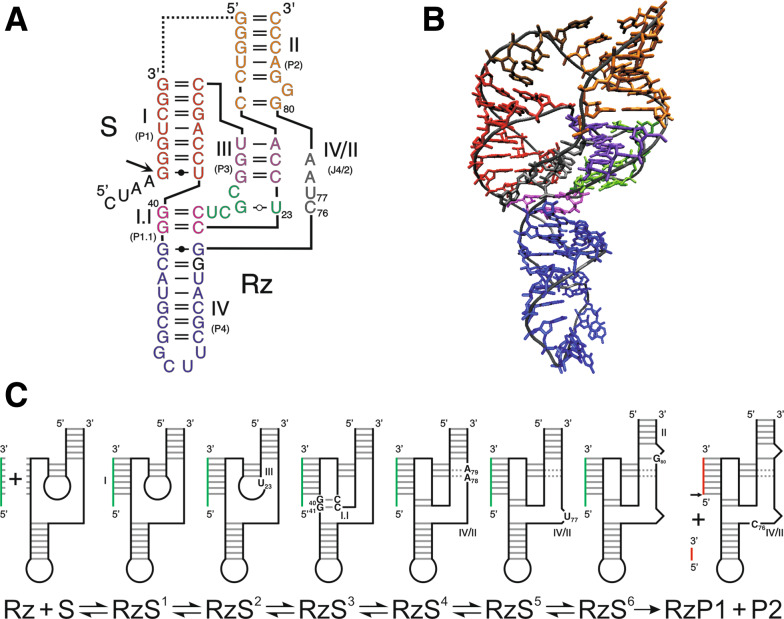
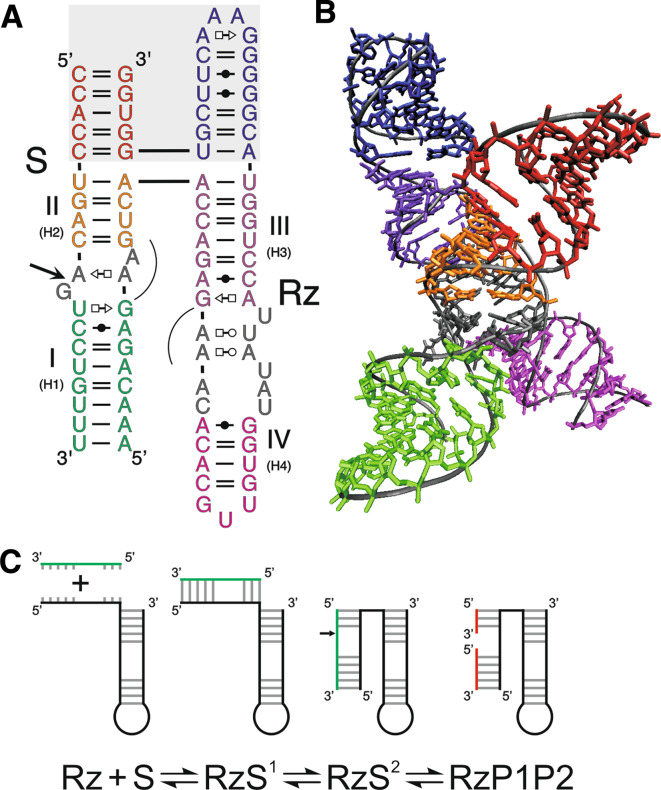
Fig. 1.
Structure and folding pathway of HDV ribozyme. The ribozyme and substrate are denoted by Rz and S, respectively. The cleavage site is indicated by an arrow. a Secondary structure of a shortened trans-acting version of the wild-type antigenomic HDV ribozyme according to the notation of Leontis and Westhof [145]. The harmonized nomenclature using roman numbering for the stems is indicated as well as the previous one in parenthesis. The dotted line represents the junction that has been removed in order to generate a trans-acting version. b Crystal structure of the genomic version of HDV ribozyme [46]. The colors of the various domains are the same in both panels A and B. c Proposed folding pathway of HDV ribozyme according to Reymond et al. [8]. The substrate is shown in green and the products in red. Important nucleotides are indicated in the various intermediates
According to the experimentally well-supported pseudoknot model, the HDV ribozyme is composed of one stem (I or P1), one pseudoknot (II or P2), two stem-loops (stems III or P3 and IV or P4) and three single-stranded junctions (I/II, I/IV and IV/II). The structure depicted in Fig. 1a is derived from the HDV of antigenomic polarity. Previous nomenclature of the various regions and the harmonized one using roman numbers for the stems are indicated. It is composed of a 57-nucleotide ribozyme and an 11-nucleotide substrate. By using this design, the stem I is composed of one Wobble base pair followed by six Watson–Crick base pairs formed between the ribozyme and the substrate and becomes the recognition domain. Both the junction I/IV and the loop III are single-stranded in the initial folding steps, but are eventually involved in the formation of a second pseudoknot (I.I) [37, 40, 41]. This structure requires the presence of divalent cations such as calcium or magnesium in order to be formed [42]. Both X-ray diffraction analysis and nuclear magnetic resonance studies have provided high-resolution tertiary structures of the genomic HDV ribozyme [40, 43, 44]. Overall, these approaches have shown that the global shape of the ribozyme is dominated by two coaxial helices formed by the stacking of the stems I–I.I–IV and the stems II–III (Fig. 1b). The stems II and IV have a structural role and are located above and below the catalytic core, which is formed by a network of interactions located at the interface between the two helical stacks. This network relies heavily on the nucleotides’ identities, as well as on several of the 2′-OH groups of the ribose moieties [38, 45].
The crystal structure of the genomic HDV ribozyme also revealed that this catalytic RNA adopts a highly ordered structure [40, 43, 46], in agreement with the previously reported unusual properties of this motif. For example, the self-catalytic motif retains activity at temperatures as high as 80°C and in solutions containing up to 5 M urea [47, 48]. Moreover, it was shown that for some trans-acting HDV ribozyme variants the cleavage level was near 100%, suggesting that HDV ribozyme forms a homogeneous population of structures and is not prone to alternative structure formation [8]. This important characteristic has been fully exploited in order to decipher the folding pathway of the HDV ribozyme, by far the most complex folding pathway elucidated to date for a small ribozyme. Extensive work has been performed using a number of approaches, including in vitro selection and photo-induced cross linking, in order to identify a specific set of mutants able to halt the folding pathway at each stable intermediate [39, 49]. Briefly, after the recognition of the substrate by the ribozyme leading to the formation of the stem I, five subsequent conformational steps are required in order to form the catalytically active structure (Fig. 1c). The first conformational transition is the docking of the stem I within the catalytic core. A specific interaction between the substrate in the middle of the stem I, which is slightly unfolded at the position +4 from the cleavage site, and both the C22 and the U23 residues of the loop III are most likely responsible for this docking [50]. This conclusion is supported by the fact that replacing a weak base pair, such as UA, in the middle of the stem I by a stronger base pair, such as GC, is detrimental to the ribozyme’s activity [51]. The substitution of an A for a U in position 23 blocks stem I docking to the catalytic core [50]. The second conformational transition is the formation of the pseudoknot I.I, which includes two GC base pairs [37, 41]. Any mutation of the pseudoknot I.I reducing either its stability, or the coaxial stacking, is detrimental to the cleavage activity [52]. The third conformational step is the formation of the A-minor motif between the two consecutive adenosines of the junction IV/II and the minor groove of the stem III, more specifically with the two GC base pairs of the latter [40]. This is a key interaction for the positioning of the junction IV/II, which is otherwise quite flexible. Moreover, this conformational transition initiates the positioning of the catalytic cytosine (C76), which is also located within the junction IV/II. The replacement of the adenosines in positions 78 and 79 by uridines prevents any further progression along the folding pathway [8, 40]. The fourth and fifth conformational steps are the formation of the trefoil motif and a base pair switch at the bottom of the stem II, respectively [40, 53, 54]. These two steps most likely occur simultaneously, although an order has been suggested based on physico-chemical analyses [8]. The trefoil turn is a particular motif identified by X-ray crystallography involving the catalytically active cytosine and its two flanking nucleotides [40]. This motif is induced by the formation of the A-minor motif and is proposed to contribute relaxing the phosphodiester backbone of the junction IV/II, thereby positioning the cytosine deep inside the catalytic centre [54]. Altering the trefoil turn motif by deleting the uridine in position 77 results in a ribozyme completely deprived of catalytic activity [53]. The other conformation transition, the base pair switch, takes place at the bottom of the stem II. It consists of switching the C19–G81 base pair of the stem II to a new C19–G80 base pair and also bulging out the G81 residue [53]. The replacement of the guanosine in position 80 by a cytosine prevents the adoption of the ribozyme–substrate transition complex. This base pair switch is also induced by the formation of the A-minor motif. Upon completion of the base pair switch, the geometry at the catalytic site is correct, and the chemical reaction occurs. The nucleotide involved in this chemical step is the catalytically active cytosine C76, which plays the role of a general base [55, 56]. Any mutation of this cytosine results in the complete loss of cleavage activity. When the transesterification reaction occurs, it is simultaneously accompanied by the products’ release. However, under certain conditions, it has been shown that the 3′-product might remain associated with the ribozyme, thereby causing product inhibition.
In summary, each of these conformational steps is separated by the formation of specific interactions. Several mutants have been designed in order to block the folding pathway at stable intermediates by disrupting these interactions. These mutants have been used in an isothermal titration calorimetry study that yielded the complete thermodynamic characterization of the HDV folding pathway [8]. This folding pathway is enthalpy driven, and the formation of additional interactions, such as the stacking in the coaxial helices, is responsible for the stability of this complex structure. The formation of the pseudoknot I.I has been shown to be the limiting step in the molecular mechanism of the HDV ribozyme [8, 46]. At the beginning of the folding pathway, the loop III and junction I/IV are located relatively far from each other and have a significant amount of freedom. The formation of the pseudoknot I.I is required in order to bring together these two single-stranded regions, thereby trapping the substrate within the catalytic core [49]. This requires an important entropic loss, and the driving force of this step is most likely the formation of two GC base pairs and the coaxial stacking of the helices I–I.I–IV.
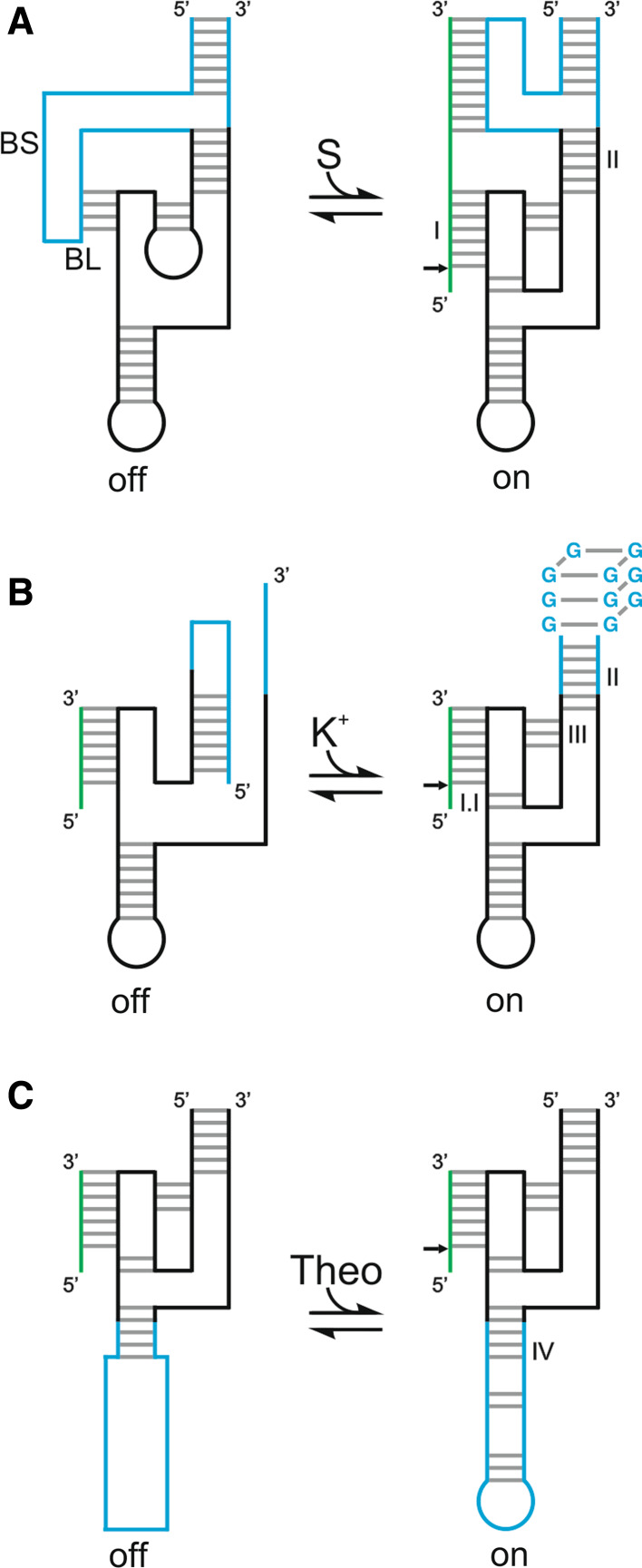
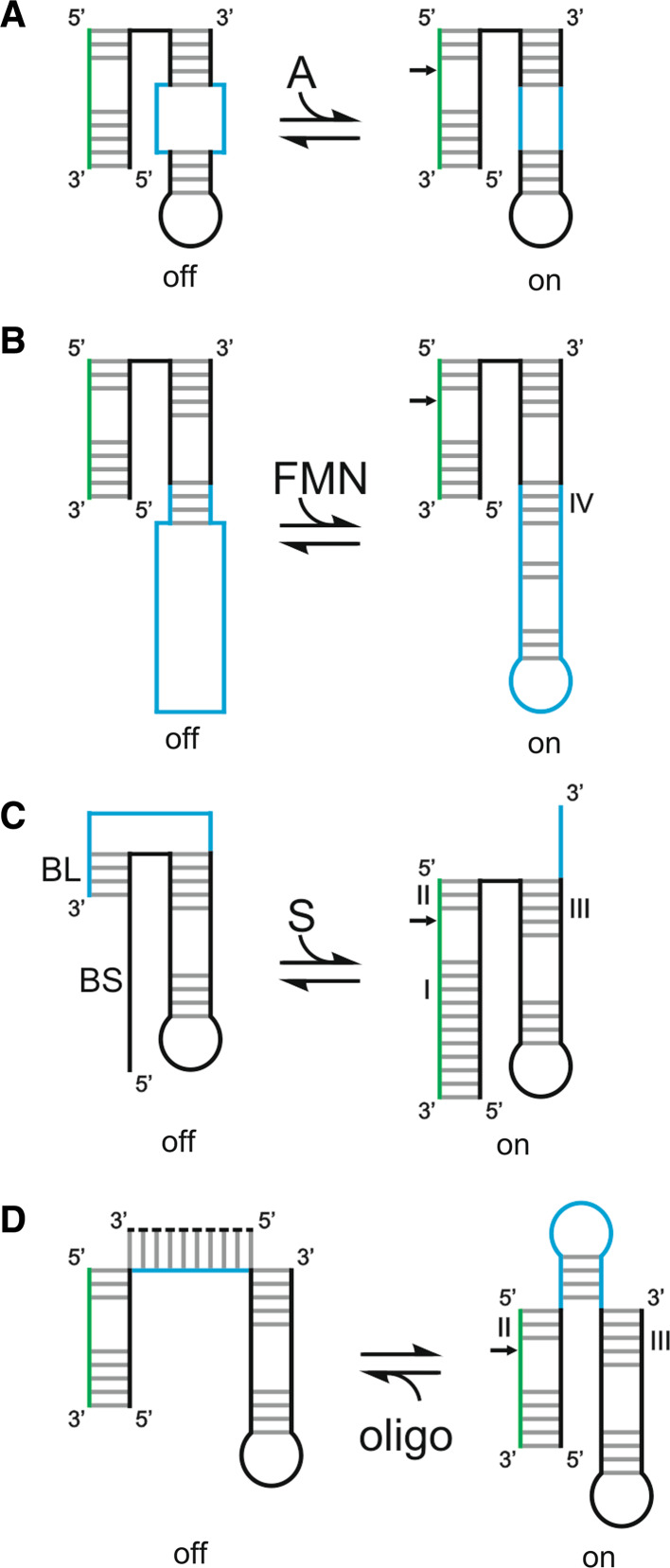
Three different strategies have been used to modify activity of the HDV ribozyme. The first one is based on the fact that, due to its self-cleaving origin, the HDV ribozyme suffers from a lack of substrate specificity when used in trans as a molecular tool. Its substrate specificity depends on the formation of stem I that contains only seven base pairs (Fig. 1a), while a total of 15–16 base pairs has been estimated to be required in order to ensure the targeting of a unique RNA species from the human transcriptome [57]. This is the reason why the past interest in this ribozyme was only moderate. In order to overcome this hurdle a module named the SOFA (Specific On/ofF Adaptor) was engineered for the HDV ribozyme (Fig. 2a) [58]. The SOFA adaptor switches the cleavage activity from “off” to “on” state solely in the presence of its cognate substrate. This modulation acts at two different levels. The substrate binding site forms a short duplex with an inserted sequence element (the blocker); this increases the energetic barrier for non-specific base-pairing interactions with the substrate binding site, thus reducing the potential for off-target cleavages. A second inserted sequence element (the biosensor) extends base-pairing with the substrate to favor binding of the genuine substrate, and formation of this duplex concomitantly results in disruption of the short duplex involving the blocker sequence. Analysis of the cleavage activity using a large collection of substrates and SOFA-ribozyme mutants provided evidence as to the roles of each domain and gave hints for design optimization [59]. The proof-of-concept of this adaptor was demonstrated both in vitro and in vivo using several SOFA-ribozymes that cleaved various RNA transcripts [58, 60, 61]. Several alternative versions of the SOFA module have been reported for the HDV ribozyme, and it has been adapted to various ribozymes including both the hammerhead and the hairpin ribozymes [62].
Fig. 2.
Various modulated versions of the HDV ribozyme. The inactive and active states are indicated by “off” and “on”, respectively. The substrate is shown in green, and the portion of the ribozyme responsible for the modulation in blue. a The SOFA module [58]. BL and BS indicate blocker and biosensor, respectively. b The G-quartzyme [64]. c The theophylline aptamer attached by a communication module [65]
Two other ways by which the HDV ribozyme’s activity can be modified involve the use of allosteric modules adapted to the HDV ribozyme. The first involves using an aptamer known to fold into an unstable hairpin structure in the absence of potassium and into a G-quadruplex structure upon the addition of potassium [63]. A rationally designed HDV ribozyme in which most of the stem II was replaced by this aptamer lost its cleavage activity in the absence of potassium (Fig. 2b) [64]. Further structural characterization led to the proposal that the 5′-strand of the aptamer was interacting with loop III, inactivating the HDV ribozyme. The addition of potassium promotes the formation of the G-quadruplex structure involving both strands of the aptamer, releasing the catalytic core of the ribozyme and stabilizing it by the formation of the stem II. The resulting RNA species represents a new class of ribozyme that exhibits a monovalent cation-dependent activity. The second method involves replacing most of the stem IV by a randomized region followed by a theophylline aptamer previously developed by in vitro selection (Fig. 2c) [65]. A negative selection was performed so as to remove all the sequences that cleave in the absence of theophylline. A positive selection was then performed in order to enrich the pool of sequences that are active in the presence of theophylline, thereby selecting a theophylline-dependent allosteric HDV ribozyme.
Hammerhead ribozyme
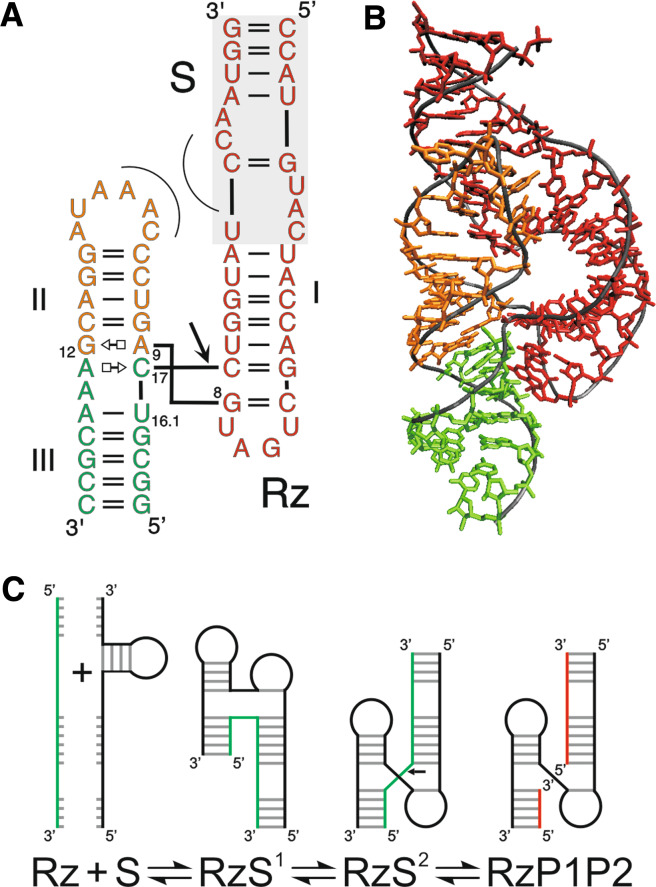
The hammerhead ribozyme is undoubtedly the most studied catalytic RNA so far. It was the first to be discovered [18, 66] and to have its crystal structure determined [67, 68]. It is famous for the dilemma surrounding the poor correlation that exists between the biochemical data and the crystal structures of minimal versions [69–71]. The hammerhead ribozyme adopts a “Y-shape” consisting of three helical stems of variable sequence and length named stems I–III (Fig. 3a). In the center of the three-way junction of the Y, 11 highly conserved nucleotides form the catalytic core that includes the cleavage site located 3′ of an important single-stranded cytosine residue [17]. A recent X-ray crystal structure of a larger version derived from Schistosoma mansoni underlines the importance of two peripheral elements, specifically a hairpin loop located at the end of stem II and a bulge located in the longer stem I (Fig. 3b) [72]. Interaction of these two motifs in a tertiary manner increases the cleavage rate by at least 50-fold [73]. Most, if not all, natural hammerhead self-cleaving motifs appear to contain similar interactions between loops or bulges located in stems I and II [74, 75]. Moreover, this new crystal structure reconciled older chemical data and provided a basis to hypothesize how the transition state might be achieved [76]. In this transition state an interaction network implicating key nucleotides (G8, A9, G12, U16.1 and C17) positions the 2′-hydroxyl nucleophile, the scissile phosphate and the 5′-oxygen leaving group at an angle very close to 180° [72].
Fig. 3.
Structure and folding pathway of the hammerhead ribozyme. The ribozyme and substrate are denoted by Rz and S, respectively. The cleavage site is indicated by an arrow. a Secondary structure of a trans-acting version of the hammerhead ribozyme [72] according to the notation of Leontis and Westhof [145]. The corresponding minimal version is obtained by removing the shaded region at the top. The tertiary interactions between the loop of stem II and the bulge of stem I are illustrated by the thin curved lines. b Crystal structure of the Schistosome version of hammerhead ribozyme [72]. The colors of the various domains are the same in panels A and B. c Hammerhead ribozyme’s folding pathway. The substrate is shown in green, and the products in red
Trans-acting hammerhead ribozymes have been extensively used to develop gene-inactivation systems either for functional genomic applications or for therapeutic aims [77, 78]. Several trans-acting versions have been produced by cleaving the RNA strand at various locations within the loops, with the most common involving strand breaks in loops I and III. In such trans-acting versions, the initial step of the folding pathway is the formation of the ribozyme–substrate complex via assembly of both stems I and III. Then the tertiary interaction network is established, which enables formation of the transition state for cleavage (Fig. 3c). Depending on the length and the base composition of the stems formed between the substrate and the ribozyme, product release inhibition has been observed.
The hammerhead ribozyme is also by far the most studied RNA molecule in terms of the modulation of the catalytic activity, either directly or by the addition of motifs leading to the production of allosteric versions. Several compounds were reported to either positively or negatively modulate the hammerhead cleavage activity [79–81]. For example, in the case of antibiotics, usually inhibitory effects were observed. This is nicely exemplified by the effect of neomycin on hammerhead activity [79]. In this case, it has been proposed that neomycin interacts directly with the ribozyme–substrate complex, stabilizing the ground state over the transition state and thereby inhibiting cleavage. Other antibiotics, such as chlorotetracycline, either stimulate or inhibit depending on the reaction conditions [80]. However, in general the mechanisms by which antibiotics affect the cleavage activity remain elusive [79]. Another interesting mechanism has been reported in a study of metal ion inhibition of the hammerhead cleavage. For example, terbium ions tightly bind to the ribozyme and subsequently displace an adjacent magnesium cation important for the catalytic activity [82]. In the case of cobalt hexammine, a study revealed that it binds to the hammerhead ribozyme and provokes a conformational change that produces an inactive structure [83]. Finally, several proteins and peptides with RNA chaperone activities have been shown to positively modulate hammerhead ribozyme activity [84–89]. For example, the p7 nucleocapsid protein of HIV has been demonstrated to resolve misfolded ribozyme–substrate complexes [84]. Other proteins, such as hnRNP A1, enhance the binding rate of the substrate and significantly accelerate product release, thereby increasing ribozyme turnover [85].
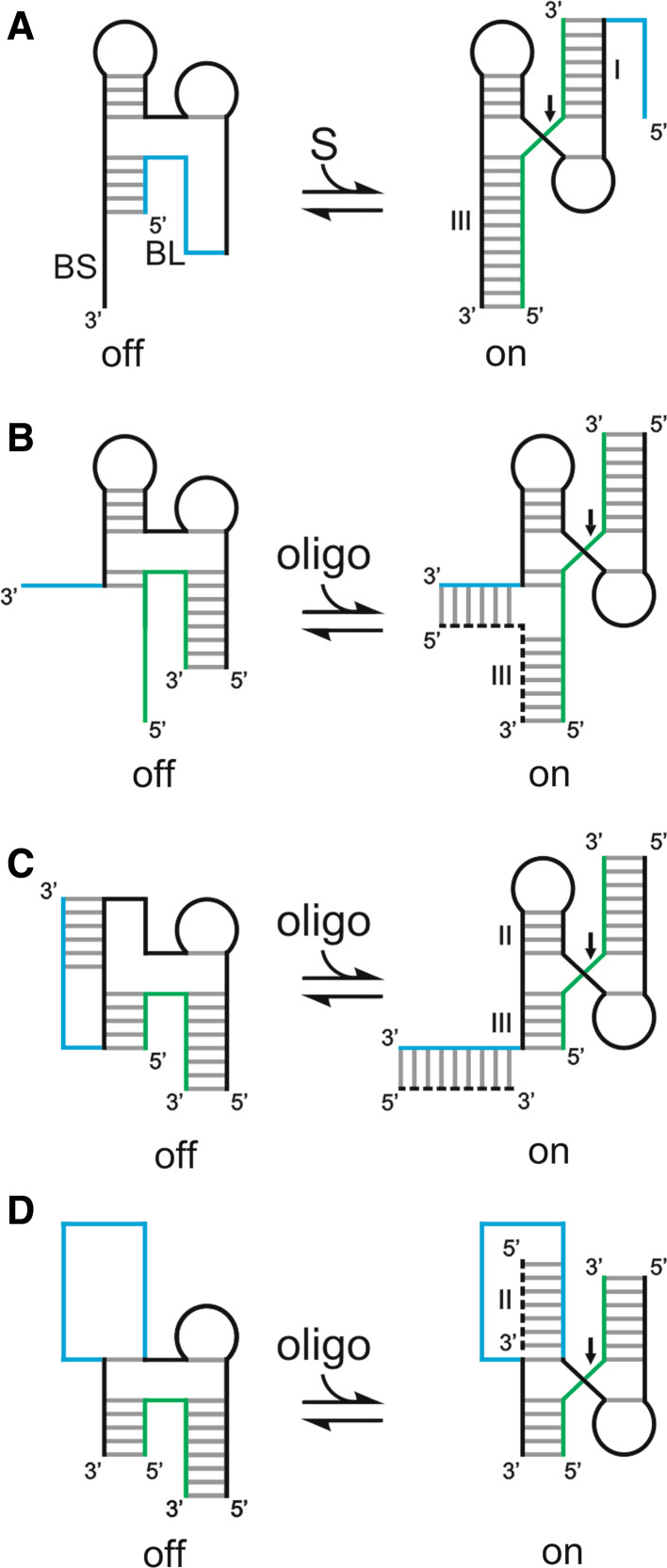
Since Watson–Crick base pairs are predictable, several strategies directed towards controlling hammerhead activity were based on the use of either additional sequences or complementary oligonucleotides. For example, in the case of modulation through additional sequences, the SOFA module developed for the HDV ribozyme was adapted to the hammerhead ribozyme [62] (Fig. 4a). The blocker was inserted at the 5′ end of stem I and interacts with the 3′ arm of stem III, thereby blocking the ribozyme in an “off” state. The biosensor complementary to the 5′ region of the substrate was added to the 3′ end of stem III. Substrate annealing to the biosensor then promotes the displacement of the blocker, yielding a ribozyme in an “on” state. In the case of modulation through complementary oligonucleotides, facilitator oligonucleotides that interact with regions of the substrate outside of the binding site of the ribozyme were shown to modify the ribozyme’s kinetic parameters. Positive effects are seen when the facilitator binds the substrate 3′ of the ribozyme, while negative ones are observed when it binds 5′ of the ribozyme [90]. Alternatively, other strategies involving trans-acting oligonucleotides that interact with the ribozyme were designed in order to modulate the cleavage activity (Fig. 4b–d) [91–94].
Fig. 4.
Various modulation strategies for the hammerhead ribozyme based on complementary sequences. The inactive and active states are indicated by “off” and “on”, respectively. The substrate is shown in green and the portion of the ribozyme responsible for the modulation in blue. The trans-acting oligonucleotides are represented by the dotted lines. a The SOFA module [62]. BL and BS indicate blocker and biosensor, respectively. b Three-strand ribozyme strategy [91]. c Blocker release strategy [93]. d Stem II stabilization strategy [92]
Over the years, rational designs and in vitro selection strategies have been used for the development of a wide repertoire of allosteric hammerhead ribozymes (also named aptazymes). The ribozyme activity is switched from inactive to active, or vice versa, by the binding of an effector to an adjacent aptamer domain (Table 1, upper portion) [95–106]. The general strategy involves using aptamers previously reported to be specific for a compound and then inserting them in one of the structural stems of the hammerhead ribozyme. Insertion in either stem I or III generally regulates the substrate binding step, while insertion in stem II usually either impairs the formation or decreases the stability of this helical region critical for the catalytic activity. Aptamers can be specific to various small components and proteins, including adenosine-5′-triphosphate (ATP), theophylline, flavin mononucleotide (FMN) and the HIV Tat protein, to name only a few examples, and have been reported to regulate hammerhead catalytic activity [95–100, 104]. Interestingly, two allosteric ribozymes using the insertion of the same ATP aptamer within stem II have been demonstrated to regulate the catalysis in different ways [95, 96]. The only difference between the two was in the linker domain located between the aptamer domain and the catalytic core. In one construct this communication module is composed of 4 base pairs that form a proper stem II in the absence of ATP, leading to negative modulation [95, 96]. In the second construct, the presence of a linker composed of 14 bases including only 3 base pairs prevents the formation of stem II and yields an inactive ribozyme in the absence of ATP, leading to positive modulation [95]. Clearly, this illustrates the importance of the communication module in the allosteric behavior of a ribozyme. Moreover, cooperativity between the binding of two different effectors has also been reported [100]. In this case, FMN and theophylline aptamers have been juxtaposed within stem II and sequential binding of both effectors is required in order to activate the ribozyme.
Table 1.
Allosteric hammerhead ribozymes
| Effector | Modulation | Region | Mechanism | Ref. |
|---|---|---|---|---|
| Allosteric hammerhead ribozymes developed by rational design | ||||
| ATP | _ | Stem II | Steric interference between bound aptamer and HHRz tertiary structure | [96] |
| FMN | + | Stem II | Stabilization of stem II | [97] |
| ATP and FMN | + | Stem III | Control the accessibility/formation of the substrate binding site | [99] |
| ATP and theophylline | ± | Stem II | Stabilization or disruption of stem II | [95] |
| FMN and theophylline | + | Stem II | Stabilization of stem II | [98] |
| FMN and theophylline | + | Stem II | Stabilization of stem II | [100] |
| Theophylline, tetracycline and xanthine | ± | Stem II | In vivo RNA-based gene-regulatory platform | [106] |
| TMPyP4 | ± | Stem II | Stabilization of stem II | [101] |
| Light and caged theophylline | + | Stem II | Stabilization of stem II | [105] |
| ERK2 phosphorylated or unphosphorylated | + | Stem II | Stabilization of stem II | [102] |
| HCV helicase/replicase | + | Stems I, II, III and I,III | Increase the affinity of the HHRz for its substrate | [103] |
| Tat protein | ± | Stem II | Stabilization of stem II or slip-structure mechanism | [104] |
| Allosteric hammerhead ribozymes developed by in vitro selection | ||||
| Oligonucleotides | + | Stem II | Reorganization and stabilization of stem II | [114] |
| cAMP, cCMP and cGMP | + | Stem II | Slip-structure mechanism | [107] [108] |
| FMN, theophylline and ATP | ± | Stem II | Slip-structure mechanism | [113] |
| Theophylline and 3-methylxanthine | + | Stem II | The level of modulation depend on the communication module selected | [110] |
| Caffeine and aspartame | + | Stem II | n.d. | [109] |
| Mn2+, Fe2+, Co2+, Ni2+, Zn2+ and Cd2+ | + | Stem II | Stabilization of stem II | [112] |
| Light and (BDHP-COOH or BCPD-COOH) | + | Stem II | Stabilization of stem II | [111] |
n.d. Not determined
Several allosteric hammerhead ribozymes have been isolated by in vitro selection (see Table 1, lower portion) [107–114]. Generally, in terms of allosteric ribozymes, the initial rounds are performed using a negative selection process that removes the sequences that do not require the presence of the effector for cleavage to occur, followed by subsequent rounds using a positive selection in the presence of the effector. For most of the reported allosteric hammerhead ribozymes, the region including the randomized positions was introduced in stem II. In vitro selection yielded allosteric domains that exhibit a very high level of specificity for their effectors, even discriminating between closely related compounds such as cAMP, cCMP and cGMP [107, 108]. Alternatively, the reselection of an allosteric hammerhead ribozyme with an improved discrimination power for a given effector after mutagenesis of the original aptamer domain has also been reported [110]. This type of strategy has been used to isolate ribozymes whose activities are modulated by photosensitive compounds [105, 111], opening the way to in vivo light-regulated gene expression through the activity of photochemically modulated hammerhead ribozymes.
Finally, as is the case with the rational design of allosteric ribozymes, the domain linking the aptamer and the catalytic core is very important, in terms of the modulation, for in vitro selected allosteric ribozymes. In vitro selection of a communication module connected to an FMN-aptamer has shown that in some cases the effector causes a positive modulation, while for other sequence variants it causes a negative regulation [113]. In a similar study, several theophylline-dependent allosteric hammerhead ribozymes harboring various sequences as communication sequences have been isolated, illustrating the flexibility of this domain [110]. Furthermore, a communication module is not restricted to one specific aptamer or particular ribozyme. The same activating communication module selected for the FMN aptamer was used in a construction with both an ATP aptamer and a theophylline aptamer, causing a positive modulation of the activity in the presence of the ligand. In the same way, a communication module selected with the HDV ribozyme was also effective when transferred into a hammerhead ribozyme [65]. Clearly, the communication module is a key feature of allostreric ribozymes, permitting the transfer of properties from the aptamer domain to the catalytic core. The molecular mechanism leading to the modulation of in vitro selected hammerhead ribozymes remains largely unknown, mainly because the efforts to elucidate the structures of both the inactive and active conformation were limited. In many cases, the stabilization of stem II has been proposed to be required for the adoption of a catalytically active structure.
Hairpin ribozyme
In its naturally occurring form, the hairpin ribozyme adopts a four-way junction secondary structure [115, 116]. Both two-way and four-way junction trans-acting ribozymes have been derived and extensively studied (Fig. 5a) [117–119]. In the minimal two-way junction version, the 49-nucleotide-long ribozyme is composed of four helices separated into two domains (A and B). Domain A comprises stems I and II (or H1 and H2) and an internal loop of eight nucleotides, while domain B comprises stems III and IV (or H3 and H4) and an internal loop of 16 nucleotides. These two domains dock into a non-coaxial orientation, and the interactions between the two internal loops form the catalytic core of the ribozyme (Fig. 5b) [120]. This catalytic core can be formed with a variety of domain-connecting setups; in fact, it can even be formed if the two domains are separated into two distinct RNA molecules [121, 122]. The cleavage site and almost all of the conserved nucleotides are located in these two internal loops [123]. The presence of a guanosine 3′ of the cleavage site is absolutely required as it is involved in stacking tertiary interactions within the internal loop of domain B, being a key feature of the hairpin catalytic core [124]. In contrast, a variety of mutations can be found in the base-paired regions as long as they support helix formation, the only requirement being the presence of a guanosine as the first nucleotide in the ribozyme strand of stem II [125].
Fig. 5.
Structure and folding pathway of the hairpin ribozyme. The ribozyme and substrate are denoted by Rz and S, respectively. The cleavage site is indicated by an arrow. a Secondary structure of a four-way junction trans-acting version of the hairpin ribozyme [116] according to the notation of Leontis and Westhof [145]. The corresponding minimal two-way version is obtained by removing the shaded region at the top. The tertiary interactions between the internal loops A and B are illustrated by the thin curved lines. The harmonized nomenclature using roman numbers for the stems is indicated as well as the previous one in parenthesis. b Crystal structure of the four-way junction version [116]). The colors of the various domains are the same in panels A and B. c Hairpin ribozyme’s folding pathway. The substrate is shown in green and the products in red
Folding of the hairpin ribozyme is a two-step folding pathway: after the binding of the substrate that results in the formation of domain A, the docking of the two internal loops caused by tertiary interactions forms the catalytic site (Fig. 5c). Once it is formed, the chemical step can then occur using in-line geometry. Interestingly, the ligation reaction is around tenfold more efficient than the cleavage reaction [126]. The hypothesis to explain this observation is that the docking of the two internal loops confers sufficient rigidity to the catalytic core that the optimal geometry is maintained and the highly reactive 2′–3′-cyclic phosphate can open and react with the 5′-hydroxyl [126].
At least four different ways of modulating the activity of the hairpin ribozyme have been succesfully used. First, in vitro selected adenine-dependent hairpin ribozymes have been developed; those require the metabolite adenine as a structural component to be able to form their catalytic cores (Fig. 6a) [127]. Second, the replacement of the short stem IV with a communicator module has permitted the introduction of a redox active aptamer into the ribozyme resulting in allosteric regulation of the ribozyme’s activity (Fig. 6b) [128]. Third, a SOFA version was developed in which a short blocker sequence forming a stem II equivalent was attached to the 3′end of the ribozyme, and a biosensor recognition sequence that is able to bind the substrate was added to the 5′end (Fig. 6c) [62]. As with the hammerhead ribozyme, the binding of the substrate to the biosensor allows the substrate to unfold the blocker and bind in its place, thereby increasing the specificity of the reaction. Finally, a modulation based on the presence of a trans-acting oligonucleotide has been achieved by introducing a short stem loop between stems II and III. When an oligonucleotide composed of either DNA or RNA complementary to this new stem loop is bound, domains A and B can no longer interact together and the catalytic activity is disrupted (Fig. 6d) [129, 130].
Fig. 6.
Various modulations of the activity of the hairpin ribozyme. The inactive and active states are indicated by “off” and “on”, respectively. The substrate is shown in green, and the portion of the ribozyme responsible for the modulation in blue. a The adenosine-dependant hairpin ribozyme [127]. b The FMN aptamer attached by a communication module [128]. c The SOFA module [62]. BL and BS indicate blocker and biosensor, respectively. d The oligonucleotide modulation [129]. The trans-acting oligonucleotide is represented by the dotted line
VS ribozyme
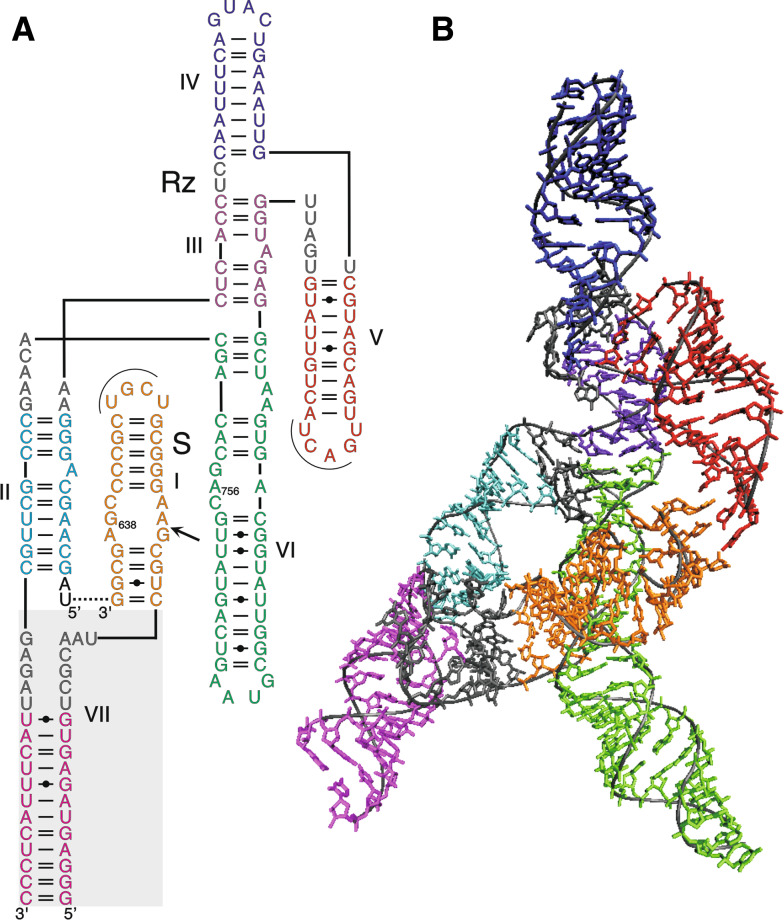
With a length of around 140 nucleotides in its trans-acting version, the VS ribozyme is the largest known small self-cleaving ribozyme. Its crystal structure has not yet been solved, although a recent small-angle X-ray scattering study gave a low-resolution structure, which was used to fit the ribozyme and to confirm its predicted overall shape [131]. The secondary structure of this ribozyme is formed by 5 helices, numbered from II to VI, that form two three-way junctions sharing stem III (Fig. 7a). The substrate is composed of stem I and, in contrast to other ribozymes, the VS ribozyme is able to rely exclusively on tertiary interactions in order to recognize its substrate [132]. The substrate appears to be docked between stems II and VI, in a cleft formed by the three-way junction involving stems II, III and VI, where it makes close interactions with the internal loop of stem VI (Fig. 7b). An adenosine (A756) in this highly conserved internal loop and a guanosine (G638) in the internal loop of the substrate have been shown to be important for catalysis and are probably directly implicated in the chemical reaction, leading to the hypothesis that these internal loops are in fact the ribozyme’s active site [133]. The cleavage occurs in the internal loop of stem I through a general acid–base catalyzed transesterification mechanism. This cleavage has been shown to be a consequence of a one nucleotide shift in the stable secondary structure of stem I [134], a secondary structure rearrangement that is stabilized by the formation of a kissing loop pseudoknot interaction. This pseudoknot is composed of three base pairs formed between the loops of stems I and V, and is required for the cleavage by VS ribozyme [135]. Hydroxyl radical footprinting showed that a solvent-inaccessible core is rapidly formed in this region [136]; the absence of water is supported by a previous kinetic study that reported pH variation has only a small effect on this catalytic RNA [137]. The folding of the VS ribozyme is ion-induced, and the presence of divalent ions is required, while the presence of monovalent ions further enhances the reaction rate [137].
Fig. 7.
Structure of the VS ribozyme. The ribozyme and substrate are denoted by Rz and S, respectively. The cleavage site is indicated by an arrow. a Secondary structure of a trans-acting version of the VS ribozyme according to the notation of Leontis and Westhof [145]. A shortened version can be obtained by removing the non-essential shaded region at the bottom. The tertiary interactions forming the kissing loop between loops I and V are illustrated by the thin curved lines. The dotted line represents the junction that has been removed in order to generate a trans-acting version. b Structure of the VS ribozyme obtained by SAXS [131]. The colors of the various domains are the same in panels A and B
While screening a collection of antibiotics looking for inhibitors of the VS ribozyme, a class of simple cyclic peptides was found to enhance ribozyme cleavage [138]. The presence of tuberactinomycin antibiotics and, especially, viomycin, in a trans-cleavage experiment results in a more extensive reaction and a decrease in the magnesium requirements for VS ribozyme cleavage. These antibiotics are able to interact with RNA and, in the case of the VS ribozyme, to restore the activity of some mutants, although the exact mechanism is not known.
Finally, a longer version of the natural VS ribozyme has a non-essential stem (called stem VII) that increases the stabilization between the 3′ end of the ribozyme and the 5′ end of its substrate (Fig. 7a). This version is particularly interesting in ligation assays since it permits substrate binding through a secondary structure [139].
GlmS ribozyme
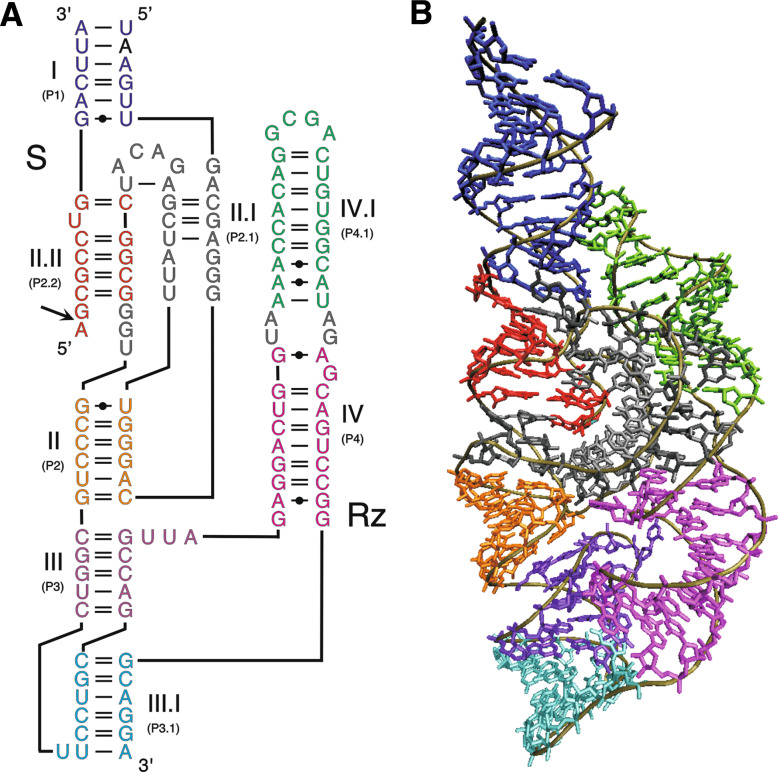
The trans-acting version of the glmS ribozyme is around 120 nucleotides long and is composed of eight small stems, two of which are pseudoknots (Fig. 8a). The available crystal structures have shown that coaxial stacking of RNA helices defines the global tertiary structure of the glmS ribozyme, while the catalytic core is formed by an internal loop involved in the two pseudoknots (Fig. 8b) [140, 141]. The double pseudoknot catalytic core forms a rigid binding pocket that is able to recognize and use a variety of small hydroxylamines, such as ethanolamine or tris-hydroxymehylaminomethane, although only binding of glucosamine results in efficient cleavage [142]. The glmS ribozyme does not appear to undergo important conformational changes upon the binding of its cofactor glucosamine-6-phosphate (GlcN6P). When the cofactor GlcN6P is bound in the catalytic site through the recognition of the sugar moiety and of the phosphate [143], 80% of the solvent-accessible surface of the metabolite is buried, and the primary amine is well positioned for catalysis, either directly as a general acid, or as a general base through water molecules [140]. Metal ions are required for the formation of the precleavage structure, but do not appear to play any catalytic roles in glmS ribozyme activity [143].
Fig. 8.
Structure of the glmS ribozyme. The ribozyme and substrate are denoted by Rz and S, respectively. The cleavage site is indicated by an arrow. a Secondary structure of a trans-acting version of the glmS ribozyme according to the notation of Leontis and Westhof [145]. The harmonized nomenclature using roman numbers for the stems is indicated as well as the previous one in parentheses. b Crystal structure of the glmS ribozyme from Thermoanaerobacter tengcongensis [140]. The colors of the various domains are the same in panels A and B
Even if glmS ribozyme activity does not rely on conformational changes, its unique metabolite dependence implies an extremely interesting and as yet poorly understood folding pathway. The binding site for the GlcN6P metabolite is formed through a complex network of interactions that provides sufficient rigidity to the pocket so that it stays open, with the functional groups ready to accommodate the cofactor, quite a different scenario from the induced fit folding usually seen upon substrate binding with other RNA enzymes. Most of the research to date has focused on understanding the cleavage reaction, but very little is known about the folding steps leading to the cofactor-free structure. This ribozyme has only recently been discovered, and already some progress has been made toward the elucidation of its folding pathway. In addition, both the structural and functional versatility of this ribozyme has been examined by in vitro selection [144].
Concluding remarks
Studies on the natural small ribozymes have made key contributions to our understanding of the folding of RNA molecules, and have helped to identify structural features important for the formation of catalytically active structures. Although the accurate prediction of the secondary and tertiary structures of long RNA species is not yet possible, the current knowledge obtained from these small ribozymes has considerably improved our view of the folding pathways of RNA molecules in general in addition to providing opportunities for the development of a substantial number of methodologies for their study. Moreover, the discovery of other small, natural, self-catalytic ribozymes and riboswitches will widen the repertoire of this class of catalytic ribozyme and will most likely increase our ability to correctly predict their folding. The characterization of the interactions involved, combined with the various components modulating ribozyme activity, has paved the way for many developments in the area of the nanotechnology. Progress in comprehending how the various RNA motifs interplay together in order to control activity should lead to a better understanding of complex biological mechanisms such as the regulation of translation involving several RNA motifs that form the 5′ untranslated region of mRNA. From this point of view, RNA molecules can be considered as combinations of building blocks. The designing of complex ribozymes with several motifs modulating either together, or sequentially, the catalytic activity is expected to propel further progress in this direction.
Acknowledgments
The work in J.P.P.’s laboratory is supported by a grant from the Canadian Institute for Health Research (CIHR: MOP-44022). The RNA group is supported by grants from the CIHR and the Université de Sherbrooke. J.P.P. holds the Canada Research Chair in Genomics and Catalytic RNA and is a member of the Infectious Diseases group of the Centre de Recherche Clinique Étienne-Lebel.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Serganov A, Patel DJ. Ribozymes, riboswitches and beyond: regulation of gene expression without proteins. Nat Rev Genet. 2007;8:776–790. doi: 10.1038/nrg2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Onoa B, Tinoco I., Jr RNA folding and unfolding. Curr Opin Struct Biol. 2004;14:374–379. doi: 10.1016/j.sbi.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Zhuang Z, Jaeger L, Shea JE. Probing the structural hierarchy and energy landscape of an RNA T-loop harpin. Nucleic Acids Res. 2007;35:6995–7002. doi: 10.1093/nar/gkm719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greenleaf WJ, Frieda KL, Foster DA, Woodside MT, Block SM. Direct observation of hierarchical folding in single riboswitch aptamers. Science. 2008;319:630–633. doi: 10.1126/science.1151298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moody EM, Bevilacqua PC. Folding of a stable DNA motif involves a highly cooperative network of interactions. J Am Chem Soc. 2003;125:16285–16293. doi: 10.1021/ja038897y. [DOI] [PubMed] [Google Scholar]
- 6.Jaeger L, Verzemnieks EJ, Geary C. The UA-handle: a versatile submotif in stable RNA architectures. Nucleic Acids Res. 2009;37:215–230. doi: 10.1093/nar/gkn911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Draper DE. RNA folding: thermodynamic and molecular descriptions of the roles of ions. Biophys J. 2008;95:5489–5495. doi: 10.1529/biophysj.108.131813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reymond C, Bisaillon M, Perreault JP. Monitoring of an RNA multistep folding pathway by isothermal titration calorimetry. Biophys J. 2009;96:132–140. doi: 10.1016/j.bpj.2008.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fiore J, Kraemer B, Koberling F, Erdmann R, Nesbitt D. Enthalpy-driven RNA folding: single-molecule thermodynamics of tetraloop receptor tertiary interaction. Biochemistry. 2009;48:2550–2558. doi: 10.1021/bi8019788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pljevaljcic G, Klostermeier D, Millar DP. The tertiary structure of the hairpin ribozyme is formed through a slow conformational search. Biochemistry. 2005;44:4870–4876. doi: 10.1021/bi047772i. [DOI] [PubMed] [Google Scholar]
- 11.Russell R, Das R, Suh H, Travers KJ, Laederach A, Engelhardt MA, Herschlag D. The paradoxical behavior of a highly structured misfolded intermediate in RNA folding. J Mol Biol. 2006;363:531–544. doi: 10.1016/j.jmb.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 12.Bhaskaran H, Russell R. Kinetic redistribution of native and misfolded RNAs by a DEAD-box chaperone. Nature. 2007;449:999–1000. doi: 10.1038/nature06235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alemán EA, Lamichhane R, Rueda D. Exploring RNA folding one molecule at a time. Curr Opin Chem Biol. 2008;12:647–654. doi: 10.1016/j.cbpa.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 14.Lilley DM. Structure, folding and mechanisms of ribozymes. Curr Opin Struct Biol. 2005;15:313–323. doi: 10.1016/j.sbi.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 15.Cochrane JC, Strobel SA. Catalytic strategies of self-cleaving ribozymes. Acc Chem Res. 2008;41:1027–1035. doi: 10.1021/ar800050c. [DOI] [PubMed] [Google Scholar]
- 16.Symons RH. Plant pathogenic RNAs and RNA catalysis. Nucleic Acids Res. 1997;25:2683–2689. doi: 10.1093/nar/25.14.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hutchins CJ, Tathjen PD, Forster AC, Symons RH. Self-cleavage of plus and minus transcripts of avocado sunblotch viroid. Nucleic Acids Res. 1986;14:3627–3640. doi: 10.1093/nar/14.9.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prody GA, Bakos JT, Buzayan JM, Schneider IR, Bruening G. Autolytic processing of dimeric plant virus satellite RNA. Science. 1986;231:1577–1580. doi: 10.1126/science.231.4745.1577. [DOI] [PubMed] [Google Scholar]
- 19.Buzayan JM, Gerlach WL, Bruening G. Non-enzymatic cleavage and ligation of RNAs complementary to a plant virus satellite RNA. Nature. 1986;323:349–353. [Google Scholar]
- 20.Earnshaw DJ, Gait MJ. Progress toward the structure and therapeutic use of the hairpin ribozyme. Antisense Nucleic Acid Drug Dev. 1997;7:403–411. doi: 10.1089/oli.1.1997.7.403. [DOI] [PubMed] [Google Scholar]
- 21.Lilley DM. Folding and catalysis by the hairpin ribozyme. FEBS Lett. 1999;452:26–30. doi: 10.1016/s0014-5793(99)00544-x. [DOI] [PubMed] [Google Scholar]
- 22.Buzayan JM, Gerlach WL, Bruening G. Nonenzymatic cleavage and ligation of RNAs complementary to a plant virus satellite RNA. Nature. 1986;323:349–353. [Google Scholar]
- 23.Shih IH, Been MD. Catalytic strategies of the hepatitis delta virus ribozymes. Annu Rev Biochem. 2002;71:887–917. doi: 10.1146/annurev.biochem.71.110601.135349. [DOI] [PubMed] [Google Scholar]
- 24.Been MD. Cis- and trans-acting ribozymes from a human pathogen, hepatitis delta virus. Trends Biochem Sci. 1994;19:251–256. doi: 10.1016/0968-0004(94)90151-1. [DOI] [PubMed] [Google Scholar]
- 25.Been MD, Wickham GS. Self-cleaving ribozymes of hepatitis delta virus RNA. Eur J Biochem. 1997;247:741–753. doi: 10.1111/j.1432-1033.1997.00741.x. [DOI] [PubMed] [Google Scholar]
- 26.Epstein LM, Gall JG. Self-cleaving transcripts of satellite DNA from the newt. Cell. 1987;48:535–543. doi: 10.1016/0092-8674(87)90204-2. [DOI] [PubMed] [Google Scholar]
- 27.Ferbeyre G, Smith JM, Cedergren R. Schistosome satellite DNA encodes active hammerhead ribozymes. Mol Cell Biol. 1998;18:3880–3888. doi: 10.1128/mcb.18.7.3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rojas AA, Vazquez-Tello A, Ferbeyre G, Venanzetti F, Bachmann L, Paquin B, Sbordoni V, Cedergren R. Hammerhead-mediated processing of satellite pDo500 family transcripts from Dolichopoda cave crickets. Nucleic Acids Res. 2000;28:4037–4043. doi: 10.1093/nar/28.20.4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salehi-Ashtiani K, Lupták A, Litovchick A, Szostak JW. A genomewide search for ribozymes reveals an HDV-like sequence in the human CPEB3 gene. Science. 2006;313:1788–1792. doi: 10.1126/science.1129308. [DOI] [PubMed] [Google Scholar]
- 30.Saville BJ, Collins RA. A site-specific self-cleavage reaction performed by a novel RNA in Neurospora mitochondria. Cell. 1990;61:685–696. doi: 10.1016/0092-8674(90)90480-3. [DOI] [PubMed] [Google Scholar]
- 31.Collins RA. The Neurospora Varkud satellite ribozyme. Biochem Soc Trans. 2002;30:1122–1126. doi: 10.1042/bst0301122. [DOI] [PubMed] [Google Scholar]
- 32.Barrick JE, Corbino KA, Winkler WC, Nahvi A, Mandal M, Collins J, Lee M, Roth A, Sudarsan N, Jona I, et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc Natl Acad Sci USA. 2004;101:6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:263–264. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- 34.Cochrane JC, Strobel SA. Catalytic strategies of self-cleaving ribozymes. Acc Chem Res. 2008;41:102701035. doi: 10.1021/ar800050c. [DOI] [PubMed] [Google Scholar]
- 35.Murray JB, Seyhan AA, Walter NG, Burke JM, Scott WG. The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem Biol. 1998;5:587–595. doi: 10.1016/s1074-5521(98)90116-8. [DOI] [PubMed] [Google Scholar]
- 36.Perrotta AT, Been MD. HDV ribozyme activity in monovalent cations. Biochemistry. 2006;45:11357–11365. doi: 10.1021/bi061215+. [DOI] [PubMed] [Google Scholar]
- 37.Deschênes P, Ouellet J, Perreault J, Perreault JP. Formation of the P1.1 pseudoknot is critical for both the cleavage activity and substrate specificity of an antigenomic trans-acting hepatitis delta ribozyme. Nucleic Acids Res. 2003;31:2087–2096. doi: 10.1093/nar/gkg307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishikawa F, Shirai M, Nishikawa S. Site-specific modification of functional groups in genomic hepatitis delta virus (HDV) ribozyme. Eur J Biochem. 2002;269:5792–5803. doi: 10.1046/j.1432-1033.2002.03280.x. [DOI] [PubMed] [Google Scholar]
- 39.Nehdi A, Perreault JP. Unbiased in vitro selection reveals the unique character of the self-cleaving antigenomic HDV RNA sequence. Nucleic Acids Res. 2006;34:584–592. doi: 10.1093/nar/gkj463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferré-D’Amaré AR, Zhou K, Doudna JA. Crystal structure of a hepatitis delta virus ribozyme. Nature. 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- 41.Wadkins TS, Perrotta AT, Ferré-D’Amaré AR, Doudna JA, Been MD. A nested double pseudoknot is required for self-cleavage activity of both the genomic and antigenomic hepatitis delta virus ribozymes. RNA. 1999;5:720–727. doi: 10.1017/s1355838299990209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suh YA, Kumar PK, Taira K, Nishikawa S. Self-cleavage activity of the genomic HDV ribozyme in the presence of various divalent metal ions. Nucleic Acids Res. 1993;21:3277–3280. doi: 10.1093/nar/21.14.3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferré-D’Amaré AR, Doudna JA. Crystallization and structure determination of a hepatitis delta virus ribozyme: use of the RNA-binding protein U1A as a crystallization module. J Mol Biol. 2000;295:541–556. doi: 10.1006/jmbi.1999.3398. [DOI] [PubMed] [Google Scholar]
- 44.Tanaka Y, Hori T, Tagaya M, Katahira M, Nishikawa F, Sakamoto T, Kurihara Y, Nishikawa S, Uesugi S. NMR analysis of tertiary interactions in HDV ribozymes. Nucleic Acids Symp Ser. 2000;44:285–286. doi: 10.1093/nass/44.1.285. [DOI] [PubMed] [Google Scholar]
- 45.Fiola K, Perreault JP. Kinetic and binding analysis of the catalytic involvement of ribose moieties of a trans-acting delta ribozyme. J Biol Chem. 2002;277:26508–26516. doi: 10.1074/jbc.M203468200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ke A, Zhou K, Ding F, Cate JH, Doudna JA. A conformational switch controls hepatitis delta virus ribozyme catalysis. Nature. 2004;429:201–205. doi: 10.1038/nature02522. [DOI] [PubMed] [Google Scholar]
- 47.Perrotta AT, Been MD. The self-cleaving domain from the genomic RNA of hepatitis delta virus: sequence requirements and the effects of denaturant. Nucleic Acids Res. 1990;18:6821–6827. doi: 10.1093/nar/18.23.6821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duhamel J, Liu DM, Evilia C, Fleysh N, Dinter-Gottlieb G, Lu P. Secondary structure content of the HDV ribozyme in 95% formamide. Nucleic Acids Res. 1996;24:3911–3917. doi: 10.1093/nar/24.20.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reymond C, Ouellet J, Bisaillon M, Perreault JP. Examination of the folding pathway of the antigenomic hepatitis delta virus ribozyme reveals key interactions of the L3 loop. RNA. 2007;13:44–54. doi: 10.1261/rna.263407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ouellet J, Perreault JP. Cross-linking experiments reveal the presence of novel structural features between a hepatitis delta virus ribozyme and its substrate. RNA. 2004;10:1059–1072. doi: 10.1261/rna.7230604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ananvoranich S, Lafontaine DA, Perreault JP. Mutational analysis of the antigenomic trans-acting delta ribozyme: the alterations of the middle nucleotides located on the P1 stem. Nucleic Acids Res. 1999;27:1473–1479. doi: 10.1093/nar/27.6.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nishikawa F, Nishikawa S. Requirement for canonical base pairing in the short pseudoknot structure of genomic hepatitis delta virus ribozyme. Nucleic Acids Res. 2000;28:925–931. doi: 10.1093/nar/28.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nehdi A, Perreault J, Beaudoin JD, Perreault JP. A novel structural rearrangement of hepatitis delta virus antigenomic ribozyme. Nucleic Acids Res. 2007;35:6820–6831. doi: 10.1093/nar/gkm674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harris DA, Rueda D, Walter NG. Local conformational changes in the catalytic core of the trans-acting hepatitis delta virus ribozyme accompany catalysis. Biochemistry. 2002;41:12051–12061. doi: 10.1021/bi026101m. [DOI] [PubMed] [Google Scholar]
- 55.Perrotta AT, Wadkins TS, Been MD. Chemical rescue, multiple ionizable groups, and general acid–base catalysis in the HDV genomic ribozyme. RNA. 2006;12:1282–1291. doi: 10.1261/rna.14106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakano S, Chadalavada DM, Bevilacqua PC. General acid–base catalysis in the mechanism of a hepatitis delta virus ribozyme. Science. 2000;287:1493–1497. doi: 10.1126/science.287.5457.1493. [DOI] [PubMed] [Google Scholar]
- 57.Peracchi A. Prospects for antiviral ribozymes and deoxyribozymes. Rev Med Virol. 2004;14:47–64. doi: 10.1002/rmv.415. [DOI] [PubMed] [Google Scholar]
- 58.Bergeron LJ, Perreault JP. Target-dependent on/off switch increases ribozyme fidelity. Nucleic Acids Res. 2005;33:1240–1248. doi: 10.1093/nar/gki278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bergeron LJ, Reymond C, Perreault JP. Functional characterization of the SOFA delta ribozyme. RNA. 2005;11:1858–1868. doi: 10.1261/rna.2112705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Robichaud GA, Perreault JP, Ouellette RJ. Development of an isoform-specific gene suppression system: the study of the human Pax-5B transcriptional element. Nucleic Acids Res. 2008;36:4609–4620. doi: 10.1093/nar/gkn432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fiola K, Perreault JP, Cousineau B. Gene targeting in the Gram-Positive bacterium Lactococcus lactis, using various delta ribozymes. Appl Env Microbiol. 2006;72:869–879. doi: 10.1128/AEM.72.1.869-879.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lévesque D, Brière FP, Perreault JP. A modern mode of activation for nucleic acid enzymes. PLoS ONE. 2007;2:e673. doi: 10.1371/journal.pone.0000673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lévesque D, Beaudoin JD, Roy S, Perreault JP. In vitro selection and characterization of RNA aptamers binding thyroxine hormone. Biochem J. 2007;403:129–138. doi: 10.1042/BJ20061216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beaudoin JD, Perreault JP. Potassium ions modulate a G-quadruplex-ribozyme’s activity. RNA. 2008;14:1018–1025. doi: 10.1261/rna.963908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kertsburg A, Soukup GA. A versatile communication module for controlling RNA folding and catalysis. Nucleic Acids Res. 2002;30:4599–4606. doi: 10.1093/nar/gkf596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Forster AC, Symons RH. Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell. 1987;49:211–220. doi: 10.1016/0092-8674(87)90562-9. [DOI] [PubMed] [Google Scholar]
- 67.Pley HW, Flaherty KM, McKay DB. Three-dimensional structure of a hammerhead ribozyme. Nature. 1994;372:68–74. doi: 10.1038/372068a0. [DOI] [PubMed] [Google Scholar]
- 68.Scott WG, Finch JT, Klug A. The crystal structure of an all-RNA hammerhead ribozyme: a proposed mechanism for RNA catalytic cleavage. Cell. 1995;81:991–1002. doi: 10.1016/s0092-8674(05)80004-2. [DOI] [PubMed] [Google Scholar]
- 69.McKay DB. Structure and function of the hammerhead ribozyme: an unfinished story. RNA. 1996;2:395–403. [PMC free article] [PubMed] [Google Scholar]
- 70.Verma S, Vaish NK, Eckstein F. Structure–function studies of the hammerhead ribozyme. Curr Opin Chem Biol. 1997;1:532–536. doi: 10.1016/s1367-5931(97)80049-x. [DOI] [PubMed] [Google Scholar]
- 71.Blount KF, Uhlenbeck OC. The structure–function dilemma of the hammerhead ribozyme. Annu Rev Biophys Biomol Struct. 2005;34:415–440. doi: 10.1146/annurev.biophys.34.122004.184428. [DOI] [PubMed] [Google Scholar]
- 72.Martick M, Scott WG. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell. 2006;126:309–320. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Canny MD, Jucker FM, Kellogg E, Khvorova A, Jayasena SD, Pardi A. Fast cleavage kinetics of a natural hammerhead ribozyme. J Am Chem Soc. 2004;126:10848–10849. doi: 10.1021/ja046848v. [DOI] [PubMed] [Google Scholar]
- 74.De la Peña M, Gago S, Flores R. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J. 2003;22:5561–5570. doi: 10.1093/emboj/cdg530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Khvorova A, Lescoute A, Westhof E, Jayasena SD. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat Struct Biol. 2003;10:708–712. doi: 10.1038/nsb959. [DOI] [PubMed] [Google Scholar]
- 76.Nelson JA, Uhlenbeck OC. Hammerhead redux: does the new structure fit the old biochemical data? RNA. 2008;14:605–615. doi: 10.1261/rna.912608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Amarzguioui M, Prudz H. Hammerhead ribozyme design and application. Cell Mol Life Sci. 1998;54:1175–1202. doi: 10.1007/s000180050247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Citti L, Rainaldi G. Synthetic hammerhead ribozymes as therapeutic tools to control disease genes. Curr Gene Ther. 2005;5:11–24. doi: 10.2174/1566523052997541. [DOI] [PubMed] [Google Scholar]
- 79.Stage TK, Hertel KJ, Uhlenbeck OC. Inhibition of the hammerhead ribozyme by neomycin. RNA. 1995;1:95–101. [PMC free article] [PubMed] [Google Scholar]
- 80.Murray JB, Arnold JR. Antibiotics interactions with the hammerhead ribozyme: tetracyclines as a new class of hammerhead inhibitor. Biochem J. 1996;317:860–885. doi: 10.1042/bj3170855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jenne A, Hartig JS, Piganeau N, Tauer A, Samarsky DA, Green MR, Davies J, Fumalok M. Rapid identification and characterization of hammerhead-ribozyme inhibitors using fluorescence-based technology. Nat Biotechnol. 2001;19:56–61. doi: 10.1038/83513. [DOI] [PubMed] [Google Scholar]
- 82.Feig AL, Scott WG, Uhlenbeck OC. Inhibition of the hammerhead ribozyme cleavage reaction by site-specific binding of Tb(III) Science. 1998;279:81–84. doi: 10.1126/science.279.5347.81. [DOI] [PubMed] [Google Scholar]
- 83.Horton TE, DeRose VJ. Cobalt hexammine inhibition of the hammerhead ribozyme. Biochemistry. 2000;39:11408–11416. doi: 10.1021/bi001141g. [DOI] [PubMed] [Google Scholar]
- 84.Hearschlag D, Khosla M, Tsuchihashi Z, Karpel RL. An RNA chaperone activity of non-specific RNA binding proteins in hammerhead ribozyme catalysis. EMBO J. 1994;13:2913–2924. doi: 10.1002/j.1460-2075.1994.tb06586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bertrand EL, Rossi JJ. Facilitation of hammerhead ribozyme catalysis by the nucleocapsid protein of HIV-1 and the heterogeneous nuclear ribonucleoprotein A1. EMBO J. 1994;13:2904–2912. doi: 10.1002/j.1460-2075.1994.tb06585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sioud M, Jespersen L. Enhancement of hammerhead ribozyme catalysis by glyceraldehyde-3-phosphate dehydrogenase. J Mol Biol. 1996;257:775–789. doi: 10.1006/jmbi.1996.0201. [DOI] [PubMed] [Google Scholar]
- 87.Tsuchihashi Z, Khosla M, Herschlag D. Protein enhancement of hammerhead ribozyme catalysis. Science. 1993;262:99–102. doi: 10.1126/science.7692597. [DOI] [PubMed] [Google Scholar]
- 88.Kuciak M, Gabus C, Ivanyi-Nagy R, Semrad K, Storchak R, Chaloin O, Muller S, Mély Y, Darlix JL. The HIV-1 transcriptional activator Tat has potent nucleic acid chaperoning activities in vitro. Nucleic Acids Res. 2008;36:3389–3400. doi: 10.1093/nar/gkn177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Huang ZS, Wu HN. Identification and characterization of the RNA chaperone activity of hepatitis delta antigen peptides. J Biol Chem. 1998;273:26455–26461. doi: 10.1074/jbc.273.41.26455. [DOI] [PubMed] [Google Scholar]
- 90.Jankowsky E, Schwenger B. Oligonucleotide facilitators may inhibit or activate a hammerhead ribozyme. Nucleic Acids Res. 1996;24:423–429. doi: 10.1093/nar/24.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang DY, Lai BH, Feldman AR, Sen D. A general approach for the use of oligonucleotide effectors to regulate the catalysis of RNA-cleaving ribozymes and DNAzymes. Nucleic Acids Res. 2002;30:1735–1742. doi: 10.1093/nar/30.8.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Porta H, Lizardi PM. An allosteric hammerhead ribozyme. Biotechnology. 1995;13:161–164. doi: 10.1038/nbt0295-161. [DOI] [PubMed] [Google Scholar]
- 93.Burke DH, Ozerova ND, Nilsen-Hamilton M. Allosteric hammerhead ribozyme TRAPs. Biochemistry. 2002;41:6588–6594. doi: 10.1021/bi0201522. [DOI] [PubMed] [Google Scholar]
- 94.Komatsu Y, Yamashita S, Kazama N, Nobuoka K, Ohtsuka E. Construction of new ribozymes requiring short regulator oligonucleotides as a cofactor. J Mol Biol. 2000;229:1231–1243. doi: 10.1006/jmbi.2000.3825. [DOI] [PubMed] [Google Scholar]
- 95.Tang J, Breaker RR. Rational design of allosteric ribozymes. Chem Biol. 1997;4:453–459. doi: 10.1016/s1074-5521(97)90197-6. [DOI] [PubMed] [Google Scholar]
- 96.Tang J, Breaker RR. Mechanism for allosteric inhibition of an ATP-sensitive ribozyme. Nucleic Acids Res. 1998;26:4214–4221. doi: 10.1093/nar/26.18.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Araki M, Okuno Y, Hara Y, Sugiura Y. Allosteric regulation of a ribozyme activity through ligand-induced conformational change. Nucleic Acids Res. 1998;26:3379–3384. doi: 10.1093/nar/26.14.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Soukup GA, Breaker RR. Design of allosteric hammerhead ribozymes activated by ligand-induced structure stabilization. Structure. 1999;15:783–791. doi: 10.1016/s0969-2126(99)80102-6. [DOI] [PubMed] [Google Scholar]
- 99.Wang DY, Lai BH, Sen D. A general strategy for effector-mediated control of RNA-cleaving ribozymes and DNA enzymes. J Mol Biol. 2002;318:33–43. doi: 10.1016/S0022-2836(02)00046-3. [DOI] [PubMed] [Google Scholar]
- 100.Jose AM, Soukup GA, Breaker RR. Cooperative binding of effectors by an allosteric ribozyme. Nucleic Acids Res. 2001;29:1631–1637. doi: 10.1093/nar/29.7.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wieland M, Hartig JS. Turning inhibitors into activators: a hammerhead ribozyme controlled by a guanine quadruplex. Angew Chem Int Ed Engl. 2006;45:5875–5878. doi: 10.1002/anie.200600909. [DOI] [PubMed] [Google Scholar]
- 102.Vaish NK, Dong F, Andrews L, Schweppe RE, Ahn NG, Blatt L, Seiwert SD. Monitoring post-translational modification of proteins with allosteric ribozymes. Nat Biotechnol. 2002;20:810–815. doi: 10.1038/nbt719. [DOI] [PubMed] [Google Scholar]
- 103.Cho S, Kim JE, Lee BR, Kim JH, Kim BG. Bis-aptazyme sensors for hepatitis C virus replicase and helicase without blank signal. Nucleic Acids Res. 2005;33:e177. doi: 10.1093/nar/gni174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang DY, Sen D. Rationally designed allosteric variants of hammerhead ribozymes responsive to the HIV-1 Tat protein. Comb Chem High Throughput Screen. 2002;5:301–312. doi: 10.2174/1386207023330273. [DOI] [PubMed] [Google Scholar]
- 105.Young DD, Deiters A. Photochemical hammerhead ribozyme activation. Bioorg Med Chem Lett. 2006;16:2658–2661. doi: 10.1016/j.bmcl.2006.02.034. [DOI] [PubMed] [Google Scholar]
- 106.Win MN, Smolke CD. A modular and extensible RNA-based gene-regulatory platform for engineering cellular function. Proc Natl Acad Sci USA. 2007;104:14283–14288. doi: 10.1073/pnas.0703961104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Koizumi M, Soukup GA, Kerr JN, Breaker RR. Allosteric selection of ribozymes that respond to the second messengers cGMP and cAMP. Nat Struct Biol. 1999;6:1062–1071. doi: 10.1038/14947. [DOI] [PubMed] [Google Scholar]
- 108.Marshall KA, Ellington AD. Training ribozymes to switch. Nat Struct Biol. 1999;6:992–994. doi: 10.1038/14872. [DOI] [PubMed] [Google Scholar]
- 109.Ferguson A, Boomer RM, Kurz M, Keene SC, Diener JL, Keefe AD, Wilson C, Cload ST. A novel strategy for selection of allosteric ribozymes yields RiboReporterTM sensors for caffeine and aspartame. Nucleic Acids Res. 2004;32:1756–1766. doi: 10.1093/nar/gkh336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Soukup GA, Emilsson GA, Breaker RR. Altering molecular recognition of RNA aptamers by allosteric selection. J Mol Biol. 2000;298:623–632. doi: 10.1006/jmbi.2000.3704. [DOI] [PubMed] [Google Scholar]
- 111.Lee HW, Robinson SG, Bandyopadhyay S, Mitchell RH, Sen D. Reversible photo-regulation of a hammerhead ribozyme using a diffusible effector. J Mol Biol. 2007;371:1163–1173. doi: 10.1016/j.jmb.2007.06.042. [DOI] [PubMed] [Google Scholar]
- 112.Zivarts M, Liu Y, Breaker RR. Engineered allosteric ribozymes that respond to specific divalent metal ions. Nucleic Acids Res. 2005;33:622–631. doi: 10.1093/nar/gki182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Soukup GA, Breaker RR. Engineering precision RNA molecular switches. Proc Natl Acad Sci USA. 1999;96:3584–3589. doi: 10.1073/pnas.96.7.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Komatsu Y, Nobuoka K, Karino-Abe N, Matsuda A, Ohtsuka E. In vitro selection of hairpin ribozymes activated with short oligonucleotides. Biochemistry. 2002;41:9090–9098. doi: 10.1021/bi020012s. [DOI] [PubMed] [Google Scholar]
- 115.Hampel A, Tritz R. RNA catalytic properties of the minimum (-)sTRSV sequence. Biochemistry. 1989;28:4929–4933. doi: 10.1021/bi00438a002. [DOI] [PubMed] [Google Scholar]
- 116.Rupert PB, Ferré-D’Amaré AR. Crystal structure of a hairpin ribozyme–inhibitor complex with implications for catalysis. Nature. 2001;410:780–786. doi: 10.1038/35071009. [DOI] [PubMed] [Google Scholar]
- 117.Fedor MJ. Structure and function of the hairpin ribozyme. J Mol Biol. 2000;297:269–291. doi: 10.1006/jmbi.2000.3560. [DOI] [PubMed] [Google Scholar]
- 118.Ferré-D’Amaré AR. The hairpin ribozyme. Biopolymers. 2004;73:71–78. doi: 10.1002/bip.10516. [DOI] [PubMed] [Google Scholar]
- 119.Wilson TJ, Nahas M, Araki L, Harusawa S, Ha T, Lilley DM. RNA folding and the origins of catalytic activity in the hairpin ribozyme. Blood Cells Mol Dis. 2007;38:8–14. doi: 10.1016/j.bcmd.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 120.Komatsu Y, Koizumi M, Nakamura H, Ohtsuka E. Loop-size variation to probe a bent structure of a hairpin ribozyme. J Am Chem Soc. 1994;116:3692–3696. [Google Scholar]
- 121.Komatsu Y, Kanzaki I, Ohtsuka E. Enhanced folding of hairpin ribozymes with replaced domains. Biochemistry. 1996;35:9815–9820. doi: 10.1021/bi960627n. [DOI] [PubMed] [Google Scholar]
- 122.Butcher SE, Heckman JE, Burke JM. Reconstitution of hairpin ribozyme activity following separation of functional domains. J Biol Chem. 1995;270:29648–29651. doi: 10.1074/jbc.270.50.29648. [DOI] [PubMed] [Google Scholar]
- 123.Berzal-Herranz A, Joseph S, Burke JM. In vitro selection of active hairpin ribozymes by sequential RNA-catalyzed cleavage and ligation reactions. Genes Dev. 1992;6:129–134. doi: 10.1101/gad.6.1.129. [DOI] [PubMed] [Google Scholar]
- 124.Chowrira BM, Berzal-Herranz A, Burke JM. Novel guanosine requirement for catalysis by the hairpin ribozyme. Nature. 1991;354:320–322. doi: 10.1038/354320a0. [DOI] [PubMed] [Google Scholar]
- 125.Hampel A, Tritz R, Hicks M, Cruz P. “Hairpin” catalytic RNA model: evidence for helices and sequence requirement for substrate RNA. Nucleic Acids Res. 1990;18:299–304. doi: 10.1093/nar/18.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Fedor MJ. Tertiary structure stabilization promotes hairpin ribozymes ligation. Biochemistry. 1999;38:11040–11050. doi: 10.1021/bi991069q. [DOI] [PubMed] [Google Scholar]
- 127.Meli M, Vergne J, Maurel MC. In vitro selection of adenine-dependent hairpin ribozymes. J Biol Chem. 2003;278:9835–9842. doi: 10.1074/jbc.M213058200. [DOI] [PubMed] [Google Scholar]
- 128.Strohbach D, Novak N, Müller S. Redox-active riboswitching: allosteric regulation of ribozyme activity by ligand-shape control. Angew Chem Int Ed Engl. 2006;45:2127–2129. doi: 10.1002/anie.200503820. [DOI] [PubMed] [Google Scholar]
- 129.Najafi-Shoushtari SH, Famulok M. DNA aptamer-mediated regulation of the hairpin ribozyme by human alpha-thrombin. Blood Cells Mol Dis. 2007;38:19–24. doi: 10.1016/j.bcmd.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 130.Vauléon S, Müller S. External regulation of hairpin ribozyme activity by an oligonucleotide effector. Chembiochem. 2003;4:220–224. doi: 10.1002/cbic.200390035. [DOI] [PubMed] [Google Scholar]
- 131.Lipfert J, Ouellet J, Norman DG, Doniach S, Lilley DM. The complete VS ribozyme in solution studied by small-angle X-ray scattering. Structure. 2008;16:1357–1367. doi: 10.1016/j.str.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Guo HC, Collins RA. Efficient trans-cleavage of a stem-loop RNA substrate by a ribozyme derived from neurospora VS RNA. EMBO J. 1995;14:368–376. doi: 10.1002/j.1460-2075.1995.tb07011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lilley DM. The Varkud satellite ribozyme. RNA. 2004;10:151–158. doi: 10.1261/rna.5217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rastogi T, Beattie TL, Olive JE, Collins RA. A long-range pseudoknot is required for activity of the Neurospora VS ribozyme. EMBO J. 2006;15:2820–2825. [PMC free article] [PubMed] [Google Scholar]
- 135.Andersen AA, Collins RA. Rearrangement of a stable RNA secondary structure during VS ribozyme catalysis. Mol Cell. 2000;5:469–478. doi: 10.1016/s1097-2765(00)80441-4. [DOI] [PubMed] [Google Scholar]
- 136.Hiley SL, Collins RA. Rapid formation of a solvent-inaccessible core in the Neurospora Varkud satellite ribozyme. EMBO J. 2001;20:5461–5469. doi: 10.1093/emboj/20.19.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Collins RA, Olive JE. Reaction conditions and kinetics of self-cleavage of a ribozyme derived from Neurospora VS RNA. Biochemistry. 1993;32:2795–2799. doi: 10.1021/bi00062a009. [DOI] [PubMed] [Google Scholar]
- 138.Olive JE, De Abreu DM, Rastogi T, Andersen AA, Mittermaier AK, Beattie TL, Collins RA. Enhancement of Neurospora VS ribozyme cleavage by tuberactinomycin antibiotics. EMBO J. 1995;14:3247–3251. doi: 10.1002/j.1460-2075.1995.tb07327.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jones FD, Ryder SP, Strobel SA. An efficient ligation reaction promoted by a Varkud Satellite ribozyme with extended 5′- and 3′-termini. Nucleic Acids Res. 2001;29:5115–5120. doi: 10.1093/nar/29.24.5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Klein DJ, Ferré-D’Amaré AR. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science. 2006;313:1745–1747. doi: 10.1126/science.1129666. [DOI] [PubMed] [Google Scholar]
- 141.Cochrane JC, Lipchock SV, Strobel SA. Structural investigation of the GlmS ribozyme bound to its catalytic cofactor. Chem Biol. 2007;14:97–105. doi: 10.1016/j.chembiol.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.McCarthy TJ, Plog MA, Floy SA, Jansen JA, Soukup JK, Soukup GA. Ligand requirements for glmS ribozyme self-cleavage. Chem Biol. 2005;12:1221–1226. doi: 10.1016/j.chembiol.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 143.Roth A, Nahvi A, Lee M, Jona I, Breaker RR. Characteristics of the glmS ribozyme suggest only structural roles for divalent metal ions. RNA. 2006;12:607–619. doi: 10.1261/rna.2266506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Link KH, Guo L, Breaker RR. Examination of the structural and functional versatility of glmS ribozymes by using in vitro selection. Nucleic Acids Res. 2006;34:4968–4975. doi: 10.1093/nar/gkl643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Leontis NB, Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]