Abstract
The Fox-1 family of RNA-binding proteins are evolutionarily conserved regulators of tissue-specific alternative splicing in metazoans. The Fox-1 family specifically recognizes the (U)GCAUG stretch in regulated exons or in flanking introns, and either promotes or represses target exons. Recent unbiased bioinformatics analyses of alternatively spliced exons and comparison of various vertebrate genomes identified the (U)GCAUG stretch as a highly conserved and widely distributed element enriched in intronic regions surrounding exons with altered inclusion in muscle, heart, and brain, consistent with specific expression of Fox-1 and Fox-2 in these tissues. Global identification of Fox-2 target RNAs in living cells revealed that many of the Fox-2 target genes themselves encode splicing regulators. Further systematic elucidation of target genes of the Fox-1 family and other splicing regulators in various tissues will lead to a comprehensive understanding of splicing regulatory networks.
Keywords: Alternative splicing, Fox-1, UGCAUG, Tissue-specificity, RNA recognition motif, CLIP, Splicing regulatory network
Introduction
Alternative splicing of pre-mRNAs is considered to be a major source of proteomic complexity in metazoans, and many alternative splicing events are controlled in tissue- and cell-type-dependent manners [1–4]. A variety of auxiliary trans-acting factors and cis-acting elements involved in the regulation of alternative splicing have been identified through biochemical, genetic, and bioinformatic analyses. The UGCAUG stretch was identified as a common cis-element critical for the cell-type-dependent regulation of well-studied model exons from alternatively spliced genes [5–11]. Jin et al. [12] first demonstrated that a zebrafish homologue of Caenorhabditis elegans putative RNA-binding protein FOX-1 specifically binds to the GCAUG stretch in vitro and provided evidence that vertebrate Fox-1 family proteins regulate splicing of alternatively spliced exons via (U)GCAUG element(s). Since this discovery, the Fox-1 family proteins have been proved to be responsible for alternative splicing regulation of a variety of alternatively spliced genes with conserved (U)GCAUG element(s). Current literature on the Fox-1 family will be summarized in this review.
Overview of the Fox-1 family proteins
Table 1 summarizes Fox-1 family members in human, mouse, zebrafish, fruitfly, and nematode worm. The Fox-1 family is featured by its highly conserved RNA recognition motif (RRM)-type RNA binding domain (Fig. 1a). Mammals have three family members, Fox-1, Fox-2, and Fox-3 (Table 1). Fox-1 (also known as A2BP1, ataxin-2 binding protein 1) and Fox-2 (also known as RBM9, RNA-binding motif protein 9) have an identical RRM domain (Fig. 1a). The third member, Fox-3, has not yet been well characterized in the literature. The zebrafish has four Fox-1 family members, two of which may not be expressed. A2BP1-like (a2bp1l, also known as zFox-1) was characterized as the first vertebrate homologue of C. elegans FOX-1 by its specific binding to the GCAUG stretch [12]. Drosophila has one homologous gene, CG32062, encoding a nearly identical RRM domain (Fig. 1a). C. elegans has three family members, FOX-1, ASD-1, and SPN-4 (Fig. 1a), as described in detail in this review.
Table 1.
The Fox-1 family genes from human, mouse, zebrafish, Drosophila, and C. elegans
| Protein | Species | Chr | Gene symbol | Aliases |
|---|---|---|---|---|
| Fox-1 | Human | 16 | A2BP1 | A2BP1, FOX1, HRNBP1 |
| Fox-1 | Mouse | 16 | A2bp1 | A2bp1, Hrnbp1, A2bp |
| Fox-2 | Human | 22 | RBM9 | RTA, fxh, HRNBP2, HNRBP2, RBM9, FOX2 |
| Fox-2 | Mouse | 15 | Rbm9 | Rbm9, Fbm2, Fxh, Hrnbp2 |
| a2bp1 | Zebrafish | 3 | a2bp1 | zgc:103635 |
| Fox-3 | Human | 17 | hCG_1776007 | HRNBP3, LOC146713, FOX3 |
| Fox-3 | Mouse | 11 | D11Bwg0517e | Hrnbp3, D11Bwg0517e |
| LOC797814 | Zebrafish | 18 | LOC797814 | |
| a2bp1 l | Zebrafish | 16 | a2bp1l | hm:zeh0082, zFox-1 |
| zgc:85694 | Zebrafish | 6 | zgc:85694 | |
| CG32062 | D. melanogaster | 3L | CG32062 | cg18441, CG14155 |
| FOX-1 | C. elegans | X | fox-1 | T07D1.4 |
| ASD-1 | C. elegans | III | asd-1 | R74.5 |
| SPN-4 | C. elegans | V | spn-4 | GEI-20, PIP-1, ZC404.8 |
Fig. 1.
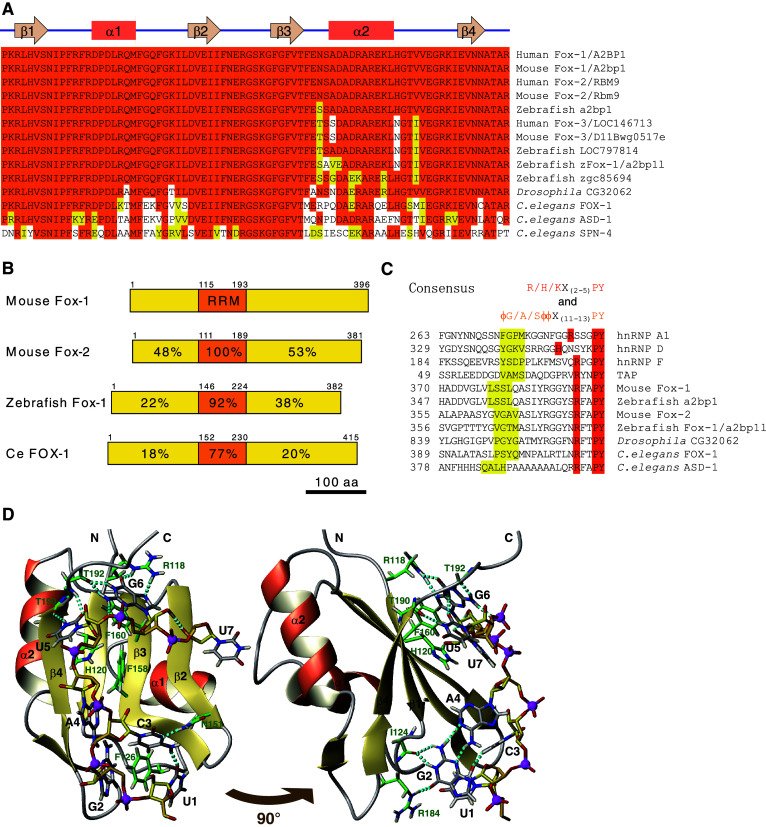
The Fox-1 family RNA-binding proteins are phylogenetically conserved. a Amino acid sequence alignment of the RRM domain of the Fox-1 family proteins in human and mouse (Fox-1, Fox-2, and Fox-3), zebrafish (a2bp1, LOC797814, zFox-1/2bp1l, and zgc85694), fruitfly (Drosophila CG32062), and nematode (C. elegans FOX-1, ASD-1, and SPN-4). Identical amino acid residues are shaded in orange and residues with similar properties are in yellow. The secondary structure of the human Fox-1 RRM domain is indicated above the alignment [23]. b Schematic illustration of the domain structure of mouse Fox-1 and Fox-2, zebrafish Fox-1/A2bp1l, and C. elegans FOX-1. RRM domain is in orange. Identities of amino acid sequences of each domain compared to that of mouse Fox-1 are indicated. Amino acid positions are indicated. c Amino acid sequence alignment of putative hPY-NLS of the Fox-1 family proteins. The C-terminal-most regions of mouse Fox-1, Fox-2, zebrafish a2bp1, zFox-1/2bp1l, Drosophila CG32062, and C. elegans FOX-1 and ASD-1 are aligned with confirmed hPY-NLS of human hnRNP A1, hnRNP D, hnRNP F, and TAP [18]. Residues that match the consensus of hPY-NLS [18] are shaded in orange and yellow. ϕ, hydrophobic side chain. Positions of the N-terminal-most residues are indicated. d Solution structure of the RRM domain of Fox-1/Fox-2 in complex with 5′-UGCAUGU-3′ in ribbon (protein backbone) and stick (RNA) representation. Right panel Image of left panel rotated by 90° as indicated. Important protein side chains and peptide bonds involved in hydrophobic or static interactions with the RNA are represented as sticks. Oxygen, nitrogen, hydrogen, and phosphorus atoms are in red, blue, light yellow, and magenta, respectively. Carbon atoms in the protein, the RNA backbone, and bases are in green, yellow, and gray, respectively. Hydrogen bonds are indicated as cyan dotted sticks. Hydrogen atoms in the RNA backbone are omitted except for one involved in a hydrogen bond. Illustrations were generated from a dataset deposited as ‘2err’ in PDBj database with MOLMOL [78]
Mammalian Fox-1 mRNA was exclusively detected in brain, skeletal muscle, and heart [12–14]. High levels of Fox-2 mRNA were detected in whole embryo and in adult brain, heart, and ovary [14]. Fox-1 and Fox-2 proteins were co-localized in nuclei of neurons but excluded from glial cells in mouse brain [14]. Fox-1 protein was specifically detected in differentiated neuroblastoma and myoblast cell lines, while Fox-2 protein was detected in a variety of cell lines as well as in neuroblastoma and myoblast cell lines [14]. A robust up-regulation of Fox-1 and gradual decrease of Fox-2 were demonstrated along with postnatal mouse heart development [15], although the impact of this switching on alternative splicing patterns needs to be elucidated. Zebrafish Fox-1/a2bp1l is specifically expressed during muscle and heart development as detected by whole mount in situ hybridization [12]. These expression patterns of the vertebrate Fox-1 family proteins are consistent with their functions as tissue-specific alternative splicing regulators mainly in brain, muscle, and heart, and with functions of Fox-2 in some other cell types as described in this review.
The Fox-1 family shares overall domain structure (Fig. 1b), although mammalian Fox-1 and Fox-2 have many splicing isoforms (see below). The single highly conserved RRM domain resides in the middle portion of the protein (Fig. 1b), and the function of the Fox-1 family as a splicing regulator, absolutely relies on the intact RRM domain that can specifically bind to target RNAs [16, 17]. N-terminal and C-terminal portions are less conserved (Fig. 1b) but unique to the Fox-1 family. RF(A/T)PY sequence of the C-terminal end is especially conserved among the family members and matches the consensus of hydrophobic PY nuclear localization signal (hPY-NLS) [18] (Fig. 1c), suggesting that the C-terminal-most region is a conserved nuclear localization signal (NLS). Consistent with this idea, most of the Fox-1 and Fox-2 isoforms were localized to the nucleus in transfected cells [12, 17, 19], and Fox-1 splice variants with a frame-shift in the C-terminal portion were not preferentially localized to the nucleus [17]. C. elegans FOX-1 and ASD-1 also have conserved C-termini (Fig. 1c) and are localized to the nucleus when expressed in HeLa cells, and deletion of C-terminal 7 and 16 amino acid residues, respectively, completely abolished preferred localization to the nucleus (H.K., unpublished observation), suggesting that the hPY-NLS is evolutionarily conserved. Deleting most of the C-terminal portion but not N-terminal portion eliminates activity as a splicing regulator even though the protein was supplemented with an exogenous NLS [12, 20, 21], indicating that the C-terminal portion of the Fox-1 family is indispensable for the splicing regulation.
Fox-1 family proteins specifically bind to (U)GCAUG
A striking feature of the Fox-1 family among other tissue-specific splicing regulators is its exceptionally specific binding to the (U)GCAUG stretch. The strict binding specificity of the Fox-1 family was demonstrated by systematic evolution of ligands by exponential enrichment) (SELEX) analysis. Jin et al. [12] utilized this selection method in vitro to identify target RNA molecules for zebrafish Fox-1/a2bp1l, and reported that 14 out 18 sequenced clones contained a GCAUG pentamer. Ponthier et al. [22] independently performed SELEX experiments with human Fox-1, and reported that 45 out of 47 winner sequences contained a UGCAUG hexamer with an additional preference for U at the seventh position. It is not yet clear what makes the minor difference in the binding specificity between fish and mammalian homologues.
The solution structure of the RRM domain of mammalian Fox-1/Fox-2 in complex with 5′-UGCAUGU-3′ was solved by utilizing nuclear magnetic resonance (NMR) spectroscopy. The Fox-1 family RRM adopts the typical β 1 α 1 β 2 β 3 α 2 β 4 fold of an RRM (Fig. 1a) with the two α-helices packed against a four-stranded antiparallel β-sheet [23, 24] (Fig. 1d). The last three nucleotides, UGU, are recognized by a canonical interface of the RRM, the four-stranded β-sheet, although the base of U7 is not recognized by any specific hydrogen bond [23] (Fig. 1d). The first four nucleotides, UGCA, are bound by two loops independently from the β-sheet binding interface; nucleotides U1, G2, and C3 are wrapped around a single crucial phenylalanine, while G2 and A4 form a mismatch base-pair [23] (Fig. 1d). This unusual molecular mechanism confers the exceptionally high affinity and specificity in target RNA recognition to the Fox-1 family [23].
Fox-1 family proteins either enhance or repress inclusion of alternative exons
To date, several model genes have been extensively studied by utilizing reporter mini-genes. Common methodologies utilized in studies described here are to analyze effects of overexpression and/or knockdown of trans-acting factors including the Fox-1 family on splicing patterns of mRNAs derived from the modified reporter mini-genes or from the endogenous genes by detecting and quantifying alternative isoforms with RT-PCR. Two opposing roles for the Fox-1 family and (U)GCAUG element(s) in alternative splicing regulation are summarized here.
Figure 2a schematically illustrates exon repression by the Fox-1 family via (U)GCAUG element(s) in the upstream intronic flanking (UIF) region of a regulated exon. Exon 9 of the human mitochondrial ATP synthase γ-subunit (F1γ) gene is specifically excluded in skeletal muscle and heart [25]. Expression of Fox-1 induced exon 9 skipping of an F1γ mini-gene via specific binding to several copies of GCAUG stretches in the UIF region in various heterologous cells [12]. Fox-1 and Fox-2 mediated skipping of calcitonin-specific exon 4 in the human calcitonin/calcitonin gene-related peptide (CGRP) gene through intronic and exonic UGCAUG elements in a neuronal context [9, 26], as described in detail later. Expression of Fox-1 and Fox-2 repressed exon 9* of the CaV1.2 L-type calcium channel gene via UGCAUG elements in the UIF and exonic regions [27].
Fig. 2.
Schematic illustration of alternative splicing regulation by the Fox-1 family. a The Fox-1 family represses exon inclusion by binding to (U)GCAUG element(s) in the upstream intronic flanking (UIF) region. b The Fox-1 family enhances exon inclusion by binding to the (U)GCAUG element(s) in the downstream intronic flanking (DIF) region. Boxes indicate exons and horizontal lines indicate introns. Blue horizontal lines indicate the UIF and DIF regions. Orange boxes indicate (U)GCAUG elements. (U)GCAUG elements are frequently found in multiple copies, and increasing the number of UGCAUG elements exerted a stronger effect on the splicing regulation
Figure 2b schematically illustrates Fox-1 family-mediated inclusion of a cassette exon via UGCAUG element(s) in the downstream intronic flanking (DIF) region. Expression of Fox-1 promoted inclusion of EIIIB exon in the rat fibronectin gene via highly repeated and evolutionarily conserved UGCAUG motifs in the DIF region [12]. Expression of Fox-1 and Fox-2 mediated inclusion of a short neuron-specific cassette exon, N30, of the human non-muscle myosin II heavy chain-B (NMHC-B) gene through UGCAUG repeats located approximately 1.5 kb downstream of the N30 exon in Y79 retinoblastoma cells [8, 17]. Expression of Fox-1 or Fox-2 enhanced inclusion of a short neuron-specific exon, N1, of the c-src gene through UGCAUG element in the DIF region in non-neuronal HeLa cells and RNAi-mediated knockdown of Fox-2 inhibited splicing of the endogenous N1 exon in N2A neuroblastoma cells [14]. Fox-2 positively regulated inclusion of exon 16 of protein 4.1R pre-mRNA via conserved three copies of UGCAUG motifs in the DIF region in late erythroleukemia cell differentiation as well as in heterologous HeLa cells [19, 22]. Expression of Fox-2 enhanced splicing of exon 33 of the CaV1.2 L-type calcium channel gene via UGCAUG in the DIF region [27].
Fox-1/UGCAUG-mediated alternative splicing is often involved as a part of regulation mechanisms for mutually exclusive alternative exons. For example, the α-actinin gene undergoes mutually exclusive splicing of an upstream non-muscle-specific (NM) exon and a downstream smooth muscle-specific (SM) exon [28, 29]. Fox-1 not only repressed NM exon via multiple (U)GCAUG elements in the UIF region, but also promoted splicing of the SM exon presumably by antagonizing the effect of PTB [12]. Regulation of mutually exclusive alternative exons of the FGFR2 gene and its C. elegans ortholog, egl-15, by the the Fox-1 family proteins will be discussed later in this review.
Genomic structure of the mammalian the Fox-1 family genes
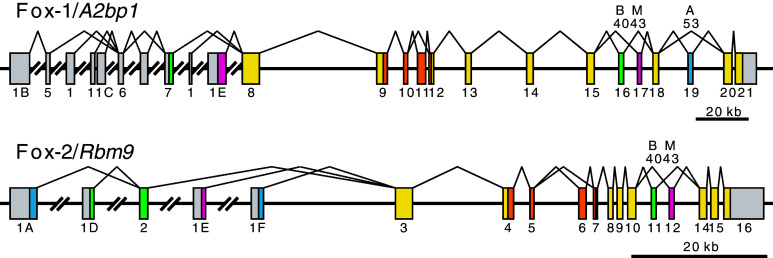
Figure 3 shows the schematic structure of the mouse Fox-1/A2bp1 and Fox-2/Rbm9 genes. Expressed sequence tag (EST) data and RT-PCR analysis revealed that there are multiple first exons in both the Fox-1 and Fox-2 genes [14] (Fig. 3). Mouse brain utilizes exon 1D and skeletal muscle utilizes exon 1E of the Fox-2 gene [19]. Pre-differentiated murine erythroleukemia (MEL) cells predominantly express Fox-2F that utilizes exon 1F, while differentiation-induced MEL cells predominantly express Fox-2A that starts from exon 1A [19]. Fox-2A exerts a much stronger effect than Fox-2F on inclusion of exon 16 of protein 4.1R mRNA, and this isoform switching is consistent with increasing level of exon 16 inclusion during erythroid differentiation [19]. These data suggest that each of the multiple promoters is regulated in tissue- and/or development-specific manners.
Fig. 3.
Schematic structure and major splicing patterns of the mouse Fox-1/A2bp1 and Fox-2/Rbm9 genes. Boxes indicate exons and horizontal lines indicate introns. Use of these exons is supported by cDNA and/or EST sequences in GenBank/EMBL/DDBJ databases, and the exons are designated after [19] for Fox-2/Rbm9 and human orthologs in [14]. Coding regions are coloured; RRM domains in orange, brain-specific region in green, muscle-specific region in magenta, other isoform-specific regions in blue, and common regions in yellow. The size of the exons is not proportional to that of the introns
Fox-1 and Fox-2 genes can produce many mRNA and protein isoforms by variable use of multiple alternatively spliced internal exons and alternative splice sites [14, 17, 19]. Both the Fox-1 and Fox-2 genes have a pair of mutually exclusive internal exons, designated as B40 and M43 (Fig. 3), which are selectively involved in brain and skeletal muscle, respectively [17]. B40 isoforms more potently induced inclusion of N30 exon of the NMHC-B gene than M43 isoforms [17], pointing out specific functions of the isoform-specific regions. The Fox-1 gene has a conserved cassette exon designated as A53 (Fig. 3). Inclusion of the A53 exon causes a frame-shift in the C-terminal portion and, therefore, A53 isoforms lack a conserved RFAPY-end, and are mostly cytoplasmic and less potent in splicing regulation [17]. As the C-terminal portions of the Fox-1 family proteins have a crucial role in splicing regulation besides serving NLS [20, 21], the A53 exon may be utilized for regulating the activity and/or sub-cellular localization of Fox-1 proteins. These and other alternative splicing regulations enable the Fox-1 and Fox-2 genes to produce various isoforms [14, 17, 19]. Expression profiles and functional properties of these isoforms as well as regulation mechanisms of each alternative splicing event are to be elucidated in the future.
Substantial amounts of inactive Fox-1 and Fox-2 isoforms are produced by skipping a cassette exon corresponding to a part of the RRM domain [17] (Fig. 3). Baraniak et al. [21] demonstrated that nucleotide sequences around exon 6, including three UGCAUG elements in the UIF region and one in the DIF region, are highly conserved among human, mouse, and rat Fox-2 genes, and that overexpression of exogenous Fox-2 led to exon 6 skipping of the endogenous Fox-2 gene. Cross-linking experiments (see below) revealed direct binding of Fox-2 protein to its own pre-mRNA in living cells [30]. These data illustrated direct negative auto-regulation of the Fox-2 gene. Similar auto-regulation is observed for C. elegans asd-1 (H.K., unpublished observation), suggesting that negative auto-regulation is a conserved and physiologically critical feature of the the Fox-1 family genes.
(U)GCAUG(U) motif is a conserved intronic element enriched in the proximity of alternatively spliced exons
Early computational analysis of 25 brain-specific alternative cassette exons and adjacent introns demonstrated a highly statistically significant over-representation of UGCAUG hexanucleotide and GCAUG pentanucleotide in the proximal DIF region [31]. UGCAUG was also found at a high frequency in the DIF region of 12 muscle-specific internal exons [31]. Comparative analysis of sequences from various vertebrate genomes revealed that the UGCAUG stretch is phylogenetically and spatially conserved in the proximal DIF region of alternative exons enriched in brain, but not of non-tissue-specific alternative exons or constitutive exons, among orthologous genes [32]. Yeo et al. [33] undertook a genome-wide comparative genomics approach using available mammalian genomes, from human, dog, rat, and mouse, to identify 314 conserved intronic elements proximal to all the internal exons. Many of these elements, including UGCAUG, were enriched near known alternatively spliced exons and actually functioned as splicing regulatory elements in heterologous contexts in human cells [33].
Recently, global analysis of mRNAs from multiple tissues and cell lines by utilizing oligonucleotide microarrays with splicing junction and exon probes facilitated global analysis of tissue specificity profiles of alternative splicing, leading to unbiased identification of candidate regulatory elements in flanking introns for alternative splicing. Sugnet et al. identified 171 cassette exons that are differentially regulated in brain relative to other tissues and found that GCAUG stretch is significantly enriched in the 150-nucleotide (nt) DIF region of brain-included exons [34]. Das et al. [35] identified 56 cassette exons that exhibited higher expression in muscle than in other normal adult tissues by using data from a human exon microarray. They demonstrated that UGCAUG was the most over-represented hexamer in the 200-nt DIF region of the identified exons in human as well as in frog, mouse, and chicken datasets [35]. Kalsotra et al. [15] identified 63 alternative splicing events that are coordinated during mouse heart development. Computational analysis of the 250-nt UIF and the DIF regions identified enriched and conserved pentamer motifs including GCAUG [15].
Most recently, Castle et al. [36] generated the first genome-scale expression compendium of human alternative splicing events using custom whole-transcript microarrays, monitoring 203,672 exons and 178,351 exon–exon junctions in 17,939 human genes, monitoring expression of 24,426 alternative splicing events. They found that 9,516 splicing events were differentially expressed in at least 1 tissue out of 48 diverse human samples. A subsequent, unbiased, systematic screen of 21,760 4-mer to 7-mer words for cis-regulatory motifs identified 143 RNA ‘words’ enriched near regulated cassette exons, including UGCAUG and GCAUGU in the 200-nt DIF region of cassette exons [36]. The compendium of alternative splicing events also illustrated that UGCAUG motif was enriched in the DIF region following upregulated cassette exons in skeletal muscle and heart, with limited enrichment in brain, adipose, and colon [36], consistent with high expression of Fox-1 and Fox-2 in skeletal muscle, brain, and heart.
Whole transcriptome analyses of diverse human tissues and cell lines by deep sequencing of complementary DNA fragments discovered many new exons and exon junctions, and revealed that up to 95% of human multi-exon genes undergo alternative splicing, and many of them are regulated in tissue-specific manners [37, 38]. Analysis of enrichment of hexanucleotides in regions adjacent to tissue-specific exons identified 362 motif/tissue enrichment patterns, and UGCAUG in the DIF region of exons with high inclusion in skeletal muscle was the third most significant motif/tissue pair [38]. The UGCAUG stretch was also substantially enriched in the DIF region of exons with increased inclusion in heart and brain, and in the UIF region of exons that had reduced inclusion in skeletal muscle [38]. The transcriptome analysis by deep sequencing also revealed that patterns of alternative polyadenylation were strongly correlated with those of alternative splicing across tissues, and all eight heptanucleotides NUGCAUG and UGCAUGN were highly enriched in the extension region of tandem 3’ untranslated regions (UTRs) [38].
Venables et al. [39] utilized a sensitive and high-throughput RT-PCR platform to screen 2,186 alternative splicing events found in a human RefSeq database for those associated with ovarian cancer and breast cancer. They identified 115 cancer-associated alternative splicing events common to both ovary and breast tissues, and found an enrichment of (U/A)GCAUG sequences in the DIF region of exons significantly downregulated in cancer [39].
Global analyses of the splicing regulatory networks of the Fox-1 family
Three approaches were applied to comprehensively search for targets of splicing regulation by the Fox-1 family proteins. Two of them relied on the highly specific recognition of the UGCAUG stretch by the Fox-1 family; one study analyzed phylogenetic conservation of UGCAUG stretch(es) in various vertebrate species; the other analyzed effects of Fox-2 knockdown on alternative exons with nearby A/UGCAUG stretch(es) in human cell lines. The third utilized UV-cross-linking and immunoprecipitation (CLIP) of the protein-RNA complex for comprehensive identification of RNA molecules that directly bind to Fox-2 protein in living cells. These studies increased the number of experimentally validated Fox-dependent alternative splicing events to over 120 [39].
Zhang et al. [40] systematically searched all human internal exons and 200-nt UIF and DIF regions for conserved UGCAUG elements by phylogenetic analysis of 28 sequenced vertebrate genomes, and comprehensively predicted 1,457 target exons from 1,103 genes at a false discovery rate of approximately 24%. Microarray analysis of mRNAs from 47 tissues and cell lines on 234 of predicted target cassette exons revealed tissue-dependent inclusion level of these exons, and unbiased hierarchical clustering of tissues grouped brain, muscle, and heart in one cluster [40], confirming the prominent roles of the Fox-1 family in these tissues.
Yeo et al. [30] recently constructed an RNA map of Fox-2-regulated alternative splicing via CLIP coupled with high-throughput sequencing (CLIP-seq) in human embryonic stem cells (hESCs). They used hESCs, which unexpectedly and abundantly express Fox-2, for identifying Fox-2 binding targets because they had previously identified 1,737 internal exons being predicted to undergo alternative splicing in hESCs compared to neural progenitor cells, and GCAUG as a conserved intronic element proximal to the candidate alternatively spliced exons [41]. Using a CLIP-seq technology and computational analyses, they uncovered thousands of Fox-2 binding clusters in 1,876 protein-coding genes, suggesting that ~7% of human genes are subjected to Fox-2 regulation in hESCs [30]. Consistent with former computational sequence analyses of UIF and DIF regions, Fox-2 preferentially bound to exonic, UIF, and DIF regions especially of alternatively spliced exons [30]. The two most significantly enriched hexamers within the clusters were UGCAUG and GCAUGU [30], confirming that Fox-2 recognizes (U)GCAUG(U) in living cells. The Fox-2 CLIP tags were often clustered to regions that lack GCAUG motif [30], implying that Fox-2 may also bind to other elements in certain conditions.
Venables et al. [39] analyzed the impact of Fox-2 knockdown on the splicing of 810 alternative exons that have at least one nearby A/UGCAUG site by utilizing a high-throughput RT-PCR platform, and identified 87 exons responsive to Fox-2 knockdown in ovarian and breast cancer cell lines. The splicing of most of these exons shifted by Fox-2 knockdown in the same direction as they were in ovarian and breast cancer tissues, and expression of Fox-2 itself was downregulated or its splicing was altered in cancer tissues [39], suggesting that cancer-associated modulation of alternative splicing is correlated with the decreased expression of Fox-2 in cancer tissues.
Global identification of many predicted target genes in these studies allowed gene ontology (GO) analysis of the putative target genes to reveal splicing regulatory networks of the Fox-1 family. Zhang et al. [40] reported that predicted targets of the Fox-1 family are enriched in genes with neuromuscular functions, as reflected in top GO terms related to cytoskeleton organization, ion channels, protein phosphorylation, muscle contraction, etc. They also mentioned that the predicted Fox-1 family targets are more likely to be disease genes in the OMIM (Online Mendelian Inheritance in Man) database [40]. As Fox-1 was originally identified as an interacter with ataxin-2 [13], and both Fox-1 and Fox-2 interact with ataxin-1 [42], which are mutated in spinal cerebellar ataxia, characterization of the splicing regulatory network of the Fox-1 family will be important for understanding these neurological, and other diseases. GO analysis of the Fox-2 CLIP tag clusters in hESCs revealed an enrichment of RNA-binding proteins, nuclear mRNA splicing factors, and serine/threonine kinases [30]. Among these Fox-2 target genes were heterogeneous ribonucleoproteins (hnRNPs) such as A2/B1, H1, H2, PTB, and R, alternative splicing regulators including Fox-1, PTB, nPTB, QKI, SRp20, SRp40, SRp55, SFRS11, and Tra2α, and other RNA-binding proteins important for stem-cell biology [30]. The finding that many Fox-2 targets in hESCs are themselves splicing regulators, and that Fox-2 is important for the survival of hESCs but not of other types of cells, implied that Fox-2 may function as an upstream regulator of the splicing network critical for maintaining hESCs [30].
Experimental validation of many predicted target genes for the Fox-1 family confirmed a previously demonstrated trend with respect to exon inclusion or skipping, depending on the location of the UGCAUG element. The alternative exon is included when the Fox-1 family binds to the UGCAUG element in the DIF region, while the exon usage is repressed when the Fox-1 family binds to the UGCAUG element in the UIF region [30, 40] (Fig. 2). The RNA map for the Fox-1/UGCAUG-mediated alternative splicing is reminiscent of an RNA map for a brain-specific splicing regulator family NOVA [43, 44]. These global studies, however, also demonstrated complexity of alternative splicing regulation by the Fox-1 family. The UGCAUG element is often conserved in the UIF and DIF regions of apparent constitutive exons, whose splicing was not necessarily affected by expression or depletion of the Fox-1 family proteins [30, 40]. Neural progenitors differentiated from hESCs and fetal neural stem cells, which also express Fox-2, show different splicing patterns of the validated Fox-2 target genes in hESCs [30]. These facts underscore the importance of experimentally identifying in vivo targets of the Fox-1 family, and presumably of other splicing regulators, in each cell and tissue context.
Coordinated alternative splicing regulation by the Fox-1 family and other splicing regulators
Several studies focusing on molecular mechanisms with which the Fox-1 family together with other splicing regulators ‘represses’ inclusion of regulated exons will be summarized in this section. Little is known, on the other hand, about how the Fox-1 family ‘promotes’ inclusion of cassette exons via binding to UGCAUG element(s) in the DIF region, as is the case with other tissue-specific alternative splicing regulators. A clue to elucidating the promotion mechanism may be interaction between Fox-1/Fox-2 and a U1 small nuclear ribonucleoprotein (snRNP)-specific protein U1-C in a yeast two-hybrid system [45], which is reminiscent of the case of TIA-1 recruiting U1 snRNP to the 5′ splice site [46].
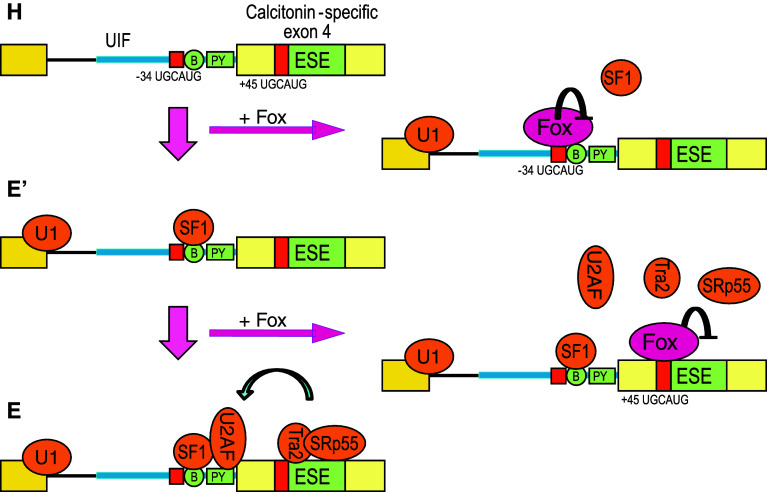
Zhou et al. [26, 49] focused on the alternative processing of calcitonin/CGRP pre-mRNA. Calcitonin-specific exon 4 is regulated by a balance between competing effects of the Fox-1 family proteins binding to UGCAUG elements at positions −34 in the UIF region and +45 in exon 4, and of Tra2β and SRp55 binding to exonic splicing enhancers (ESEs) [26, 47, 48]. Zhou et al. [49] demonstrated that the Fox-1 family proteins bound to the −34 UGCAUG silencer element to prevent SF1 from binding to the branch point without affecting U1 snRNP binding to the pre-mRNA, and that the −34 UGCAUG element repressed formation of pre-spliceosome E′ complex (Fig. 4), a pre-spliceosome complex formed in U2AF-depleted HeLa nuclear extracts prior to early (E) complex formation [50]. They also demonstrated that the Fox-1 family proteins interfered with binding of Tra2β and SRp55 to the ESEs via the +45 UGCAUG element and that the +45 UGCAUG element blocked recruitment of U2AF65 and formation of the pre-spliceosome E complex [49] (Fig. 4). These results raised a fail-safe two-step model of repression of calcitonin-specific exon 4 by the Fox-1 family and two UGCAUG elements [49].
Fig. 4.
A model for repression of prespliceosome complex formation by the Fox-1 family. The repression of calcitonin-specific exon 4 of calcitonin/CGRP pre-mRNA in neuronal cells by the Fox-1 family involves two distinct regulatory events. First, the −34 element in the UIF region prevents E′ complex formation through repressing SF1 binding to the branch point (B). Second, the +45 exonic element blocks transition to E complex via inhibiting U2AF65 binding to polypyrimidine tract (PY). This figure is modified from [49]
Fukumura et al. [20, 51] utilized exon 9 of the human F1γ gene as a model system of Fox-1-mediated exon skipping. Fox-1 repressed F1γ exon 9 by inhibiting splicing of the downstream intron 9 via GCAUG elements in the UIF region without affecting efficiency of splicing of intron 8 where the GCAUG elements reside. Unexpectedly, U1 snRNP components were specifically absent from pre-spliceosomal E complex on the intron 9 formed [51], and Fox-1 prevented formation of the E complex in the in vitro splicing reaction with HeLa nuclear extract [20]. F1γ intron 9 was efficiently spliced in U1-disrupted Xenopus oocytes and in nuclear extracts from U1-disrupted HeLa cells, confirming that F1γ intron 9 was the natural substrate of U1-independent and U2-dependent splicing [51]. Mutations in the 5′ splice site of intron 9 conferred U1 dependency and concomitantly impaired Fox-1-mediated regulation, indicating that U1-independent splicing and presumably suboptimal 5′ splice site sequence is indispensable for the Fox-1-mediated repression of F1γ exon 9 [51]. It is worth investigating whether Fox-1-mediated repression of alternative exons in other target genes is generally accompanied by U1-independent splicing.
The fibroblast growth factor receptor 2 (FGFR2) transcripts occur in a cell-type-specific manner leading to the mutually exclusive use of exon IIIb in epithelia or exon IIIc in mesenchyme, and the alternative exons determine the ligand-specificity of the receptor. Baraniak et al. [21, 52] demonstrated that expression of Fox-2 can modestly activate exon IIIb and strongly repress exon IIIc via UGCAUG elements in the UIF and exonic regions of exon IIIc in epithelial cells. Mauger et al. [53] demonstrated that hnRNP H and hnRNP F proteins act as silencers of exon IIIc via binding to exonic GGG motifs overlapping with a critical ESE, and that hnRNP H1 forms a complex with Fox-2 to better antagonize ASF/SF2 binding to the ESE. Fox-2 is a critical but insufficient regulator for mesenchymal-epithelial transitions, as mesenchyme-like cells expressing Fox-2 take FGFR2(IIIc) form [21]. Recent cell-based cDNA expression screening identified epithelial cell-type-specific regulators epithelial splicing regulatory proteins 1 and 2 (ESRP1 and ESRP2) that specifically bind to the ISE/ISS-3 element in the UIF region of exon IIIc as critical regulators for FGFR2(IIIc)-to-(IIIb) switching [54]. The mutually exclusive alternative splicing of the FGFR2 gene has been utilized as a model system for visualization of the splicing patterns with fluorescent proteins [55–57], which will be reviewed elsewhere (H.K., in press).
Fox-1 family/UGCAUG-mediated alternative splicing is evolutionarily conserved
Caenorhabditis elegans is an excellent model organism for studying the regulation mechanisms of alternative splicing in vivo. At least 5% of total genes in C. elegans are alternatively spliced in manners similar to vertebrates [58, 59]. Kabat et al. [60] compared the sequence of C. elegans genome with that of a related nematode species C. briggsae and identified 147 alternatively spliced cassette exons that exhibit high nucleotide conservation in the UIF and DIF regions. They analyzed frequency of pentamers and hexamers in the conserved intronic elements and found that high-scoring nematode motifs corresponded to known mammalian splicing regulatory elements, including (U)GCAUG [60]. In vivo experiments with transgenic reporter mini-genes confirmed involvement of these conserved elements in alternative splicing regulation [60], suggesting that mechanisms of alternative splicing regulation are well conserved in metazoans.
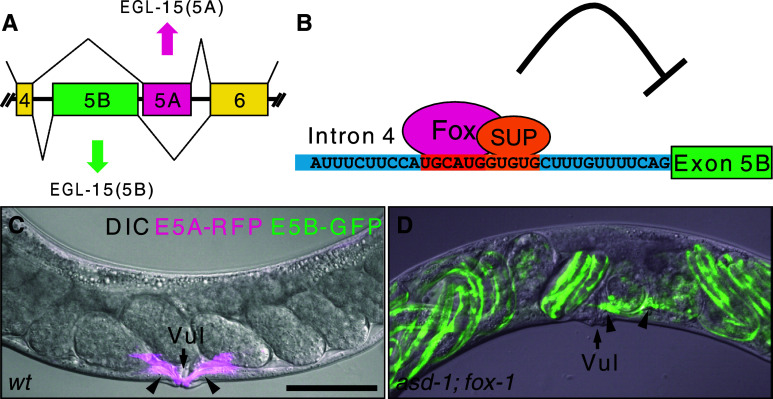
Recent genetic analysis demonstrated that the Fox-1 family regulates tissue-specific alternative splicing in C. elegans. The target gene egl-15 encodes the sole homolog of fibroblast growth factor receptors (FGFRs) in C. elegans [61], and selection of its mutually exclusive alternative exons, 5A and 5B (Fig. 5a), confers ligand-specificity to the receptor [62–65]. We visualized tissue-specific expression profiles of the egl-15 alternative exons in vivo by utilizing green fluorescent protein (GFP) and red fluorescent protein (RFP) [16]. Screening for mutants defective in expression of the fluorescent splicing reporters in muscles led to identification of the Fox-1 family proteins ASD-1 (alternative-splicing-defective-1) and FOX-1 and muscle-specific RNA-binding protein SUP-12 as regulators of muscle-specific repression of egl-15 exon 5B [16, 66]. In muscles, the Fox-1 family and SUP-12 cooperatively bind to UGCAUG and GUGUG stretches, respectively, in the UIF region to repress exon 5B presumably by interfering with recognition of a branch point, leading to inclusion of exon 5A [16, 66] (Fig. 5a, b). A remarkable phenotype of the asd-1; fox-1 double mutant is the egg-laying defective (Egl) due to aberrant migration of sex myoblasts in hermaphrodites [16] (Fig. 5c, d). The same phenotype has been reported for mutants that specifically lack EGL-15 (5A) isoform [65] or lack EGL-15 (5A)-specific ligand EGL-17 [63, 64], indicating that the major role of the Fox-1 family in C. elegans is to repress exon 5B and promote exon 5A of the egl-15 gene.
Fig. 5.
Fox-1 family proteins ASD-1 and FOX-1 regulate tissue-specific mutually exclusive alternative splicing of the FGFR gene, egl-15, in C. elegans. a Tissue-specific mutually exclusive selection exons of the egl-15 gene of C. elegans. Note that muscle-specific exon 5A resides downstream of exon 5B. b Schematic illustration of muscle-specific repression of exon 5B. Nucleotide sequence of the UIF region is presented. The cis-elements for tissue-specific regulation are in orange. Fox, ASD-1 or FOX-1; SUP, SUP-12. (c,d) Left lateral view of adult hermaphrodite worms expressing egl-15 alternative splicing reporter in body wall muscles and vulval muscles in the wild-type (wt) background (c) and the asd-1; fox-1 double mutant background (d). Merged images of DIC and confocal images of GFP (green) and RFP (magenta). The egl-15 alternative splicing reporter worms express either RFP, representing exon 5A selection, or GFP, representing exon 5B selection, in body wall muscles and vulval muscles (see [16] for detail). Note that in the asd-1; fox-1 double mutant, vulval muscles (arrowheads) express GFP and are mislocalized off the vulva (Vul, arrows), and late embryos with reporter expression in body wall muscles retained in the uterus. Anterior is to the left. Scale bar in (c) 50 μm. (c,d) are modified from [16]
RRM domain of C. elegans FOX-1 is 77% identical to that of human Fox-1 (Fig. 1b), and not only the amino acid residues whose side chains directly contact with the UGCAUGU heptamer RNA but also the complete side of the RRM domain facing the UGCAUGU are conserved [23] (Fig. 1a, d). It is therefore natural that C. elegans FOX-1 and ASD-1 also specifically recognize UGCAUG [16]. The studies on alternative splicing regulation of the egl-15 gene provided the first genetic evidence that two families of trans-factors cooperatively regulate tissue-specific alternative splicing of a specific target gene in vivo through cooperative binding to their juxtaposed cis-elements in a subset of tissues where the trans-factors are co-expressed [16, 66]. The coordination of multiple trans-factors via multiple cis-elements would be a common mechanism for specific and robust regulation of tissue-specific alternative splicing in higher organisms. Furthermore, determination of the ligand specificity of FGFRs by Fox-1 family-mediated alternative splicing shows marked analogy between vertebrates and nematodes, suggesting selective pressure of conservation. The fully-sequenced genomes of C. elegans and other nematodes combined with its elegant genetics thus offers unique advantages for exploring alternative splicing regulation in metazoans [67, 68].
C. elegans SPN-4 is an outlying member of the Fox-1 family (Fig. 1a). Genetic evidence indicated that SPN-4 positively regulates translation of maternal GLP-1 mRNA in early blastomeres by counteracting a CCCH zinc-finger protein POS-1 via direct binding to 3’UTR of the GLP-1 mRNA, which does not have UGCAUG stretch [69]. Consistent with the function as a translational regulator and the fact that it lacks PY-NLS, SPN-4 was exclusively localized in the cytoplasm in immunohistochemical staining [69]. Although SPN-4 is most closely related to the Fox-1 family among RNA-binding protein families, its apparent counterpart is missing in higher eukaryotes and, therefore, its evolutionary origin is unclear.
Role of the fox-1 gene in sex determination of C. elegans
The founder member of the Fox-1 family, FOX-1 of C. elegans, was originally identified as a numerator element of its sex chromosome. Sex in C. elegans is determined by the ratio of X chromosome number to autosomal set number; the chromosome-counting mechanism reliably distinguishes the twofold difference in X-chromosome dose between males (XO, one X chromosome) and hermaphrodites (XX, two X chromosomes) [70]. Genetic studies with X chromosome duplications and transgenic overexpression suggested that FOX-1 is one of only a small number of major numerator sites on the X chromosome; when overexpressed, XX animals were viable, but XO animals were lethal and feminized [71].
The target gene of FOX-1 in sex determination is believed to be xol-1 (XO Lethal), the master sex-determination switch gene that specifies the male fate when active and the hermaphrodite fate when inactive [70]. The dose-sensitive signal elements on chromosome X control xol-1 through two different molecular mechanisms; a nuclear hormone receptor-like protein SEX-1 represses xol-1 transcription [72] and FOX-1 post-transcriptionally downregulates xol-1 level [73]. The small quantitative difference in X chromosome dosage is thus translated into the ‘on/off’ response of the xol-1 gene [70, 74]. xol-1 pre-mRNA is alternatively spliced into three mRNA isoforms and only one of them is necessary and sufficient for xol-1 activity in XO animals [75], and disruption and overexpression of fox-1 affected the functional xol-1 mRNA level in XO animals [76]. These genetic studies and the presence of multiple (U)GCAUG elements in the xol-1 pre-mRNA strongly suggest that FOX-1 regulates processing of xol-1 pre-mRNA in a sex-dependent manner. However, it has not yet been experimentally demonstrated which processing event is enhanced or repressed by FOX-1 expression. This is at least in part due to low expression level and rapid degradation of xol-1 mRNA in hermaphrodites in which SEX-1 and FOX-1 are active. fox-1 single mutant shows no apparent phenotype and even asd-1; fox-1 double mutant is normal in hermaphrodite fate specification [16], suggesting that the sex-determination of C. elegans does not strictly rely on the post-transcriptional regulation of xol-1 by the Fox-1 family.
Perspectives
As described in this review, the (U)GCAUG element is one of the most widely distributed, and evolutionarily conserved splicing regulatory elements in metazoans, and the Fox-1 family is the only subset of RNA-binding proteins that are known to specifically recognize the (U)GCAUG element and regulate splicing of nearby exons. One of the remaining questions on the Fox-1 family is a functional difference between Fox-1 and Fox-2, which has not yet been described in the literature. Conditional knockout models will reveal specific and redundant functions of each of the Fox-1 family proteins in vivo.
Deep sequencing analysis of transcriptome revealed strong correlation in alternative splicing and alternative cleavage and polyadenylation, and enrichment of UGCAUG stretch in 3′ UTRs [38], raising an interesting hypothesis that the Fox-1 family is also involved in 3′ UTR-related functions such as regulation of alternative cleavage and polyadenylation, mRNA stability, localization, or translation. Indeed, recent purification of human mRNA 3′ processing complexes revealed that Fox-2 is one of ~85 protein components of the complex [77]. These stories are reminiscent of the CLIP-seq analysis of NOVA that led to the discovery that NOVA interacts with 3′ UTRs of target mRNAs and regulates alternative polyadenylation in the brain [43]. Further biochemical analysis will reveal how these tissue-specific trans-acting factors coordinate with core machineries for splicing and 3′-end processing at molecular levels.
Another challenging problem is refinement of the ‘RNA map’ or location-dependent activity of the Fox-1 family via (U)GCAUG element(s). Recent global analysis of target genes indicated that the Fox-1 family is not necessarily the major regulator for all target genes. Indeed, coordinating or antagonizing activities of other RNA-binding proteins are described in this review. Future global analyses should be directed at identifying direct target genes for multiple trans-factors and elucidating ‘RNA maps’ for cooperating and/or antagonizing cis-elements in various tissues. Such systematic analyses will lead to a comprehensive understanding of splicing regulatory networks of the Fox-1 family and other tissue-specific regulators in metazoans.
Acknowledgment
I thank Akihide Takeuchi of TMDU and Kunio Inoue of Kobe University for critically reading and making comments on the manuscript.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- ASD-1
Alternative-splicing-defective-1
- CGRP
Calcitonin gene-related peptide
- CLIP
Cross-linking and immunoprecipitation
- DIC
Differential interference contrast
- DIF
Downstream intronic flanking
- Egl
Egg-laying defective
- ESE
Exonic splicing enhancer
- EST
Expressed sequence tag
- nt
Nucleotide
- FGFR
Fibroblast growth factor receptor
- GFP
Green fluorescent protein
- GO
Gene ontology
- hESC
Human embryonic stem cell
- hnRNP
Heterogeneous ribonucleoprotein
- MEL
Murine erythroleukemia
- NLS
Nuclear localization signal
- RFP
Red fluorescent protein
- RRM
RNA recognition motif
- SELEX
Systematic evolution of ligands by exponential enrichment
- snRNP
Small nuclear ribonucleoprotein
- UIF
Upstream intronic flanking
- UTR
Untranslated region
References
- 1.Black DL. Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology. Cell. 2000;103:367–370. doi: 10.1016/S0092-8674(00)00128-8. [DOI] [PubMed] [Google Scholar]
- 2.Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. doi: 10.1016/S0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- 3.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 4.Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 5.Huh GS, Hynes RO. Regulation of alternative pre-mRNA splicing by a novel repeated hexanucleotide element. Genes Dev. 1994;8:1561–1574. doi: 10.1101/gad.8.13.1561. [DOI] [PubMed] [Google Scholar]
- 6.Hu Y, Zhou Z, Huang X, Xu M, Lu L, Xu Z, Li J, Sha J. Expression of a novel DnaJA1 alternative splicing in human testis and sperm. Int J Androl. 2004;27:343–349. doi: 10.1111/j.1365-2605.2004.00492.x. [DOI] [PubMed] [Google Scholar]
- 7.Lim LP, Sharp PA. Alternative splicing of the fibronectin EIIIB exon depends on specific TGCATG repeats. Mol Cell Biol. 1998;18:3900–3906. doi: 10.1128/mcb.18.7.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawamoto S. Neuron-specific alternative splicing of nonmuscle myosin II heavy chain-B pre-mRNA requires a cis-acting intron sequence. J Biol Chem. 1996;271:17613–17616. [PubMed] [Google Scholar]
- 9.Hedjran F, Yeakley JM, Huh GS, Hynes RO, Rosenfeld MG. Control of alternative pre-mRNA splicing by distributed pentameric repeats. Proc Natl Acad Sci USA. 1997;94:12343–12347. doi: 10.1073/pnas.94.23.12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Modafferi EF, Black DL. A complex intronic splicing enhancer from the c-src pre-mRNA activates inclusion of a heterologous exon. Mol Cell Biol. 1997;17:6537–6545. doi: 10.1128/mcb.17.11.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deguillien M, Huang SC, Moriniere M, Dreumont N, Benz EJ, Jr, Baklouti F. Multiple cis elements regulate an alternative splicing event at 4.1R pre-mRNA during erythroid differentiation. Blood. 2001;98:3809–3816. doi: 10.1182/blood.V98.13.3809. [DOI] [PubMed] [Google Scholar]
- 12.Jin Y, Suzuki H, Maegawa S, Endo H, Sugano S, Hashimoto K, Yasuda K, Inoue K. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 2003;22:905–912. doi: 10.1093/emboj/cdg089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shibata H, Huynh DP, Pulst SM. A novel protein with RNA-binding motifs interacts with ataxin-2. Hum Mol Genet. 2000;9:1303–1313. doi: 10.1093/hmg/9.9.1303. [DOI] [PubMed] [Google Scholar]
- 14.Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol Cell Biol. 2005;25:10005–10016. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalsotra A, Xiao X, Ward AJ, Castle JC, Johnson JM, Burge CB, Cooper TA. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc Natl Acad Sci USA. 2008;105:20333–20338. doi: 10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuroyanagi H, Kobayashi T, Mitani S, Hagiwara M. Transgenic alternative-splicing reporters reveal tissue-specific expression profiles and regulation mechanisms in vivo. Nat Methods. 2006;3:909–915. doi: 10.1038/nmeth944. [DOI] [PubMed] [Google Scholar]
- 17.Nakahata S, Kawamoto S. Tissue-dependent isoforms of mammalian Fox-1 homologs are associated with tissue-specific splicing activities. Nucleic Acids Res. 2005;33:2078–2089. doi: 10.1093/nar/gki338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee BJ, Cansizoglu AE, Suel KE, Louis TH, Zhang Z, Chook YM. Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell. 2006;126:543–558. doi: 10.1016/j.cell.2006.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang G, Huang SC, Wu JY, Benz EJ., Jr Regulated Fox-2 isoform expression mediates protein 4.1R splicing during erythroid differentiation. Blood. 2008;111:392–401. doi: 10.1182/blood-2007-01-068940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fukumura K, Kato A, Jin Y, Ideue T, Hirose T, Kataoka N, Fujiwara T, Sakamoto H, Inoue K. Tissue-specific splicing regulator Fox-1 induces exon skipping by interfering E complex formation on the downstream intron of human F1gamma gene. Nucleic Acids Res. 2007;35:5303–5311. doi: 10.1093/nar/gkm569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baraniak AP, Chen JR, Garcia-Blanco MA. Fox-2 mediates epithelial cell-specific fibroblast growth factor receptor 2 exon choice. Mol Cell Biol. 2006;26:1209–1222. doi: 10.1128/MCB.26.4.1209-1222.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ponthier JL, Schluepen C, Chen W, Lersch RA, Gee SL, Hou VC, Lo AJ, Short SA, Chasis JA, Winkelmann JC, Conboy JG. Fox-2 splicing factor binds to a conserved intron motif to promote inclusion of protein 4.1R alternative exon 16. J Biol Chem. 2006;281:12468–12474. doi: 10.1074/jbc.M511556200. [DOI] [PubMed] [Google Scholar]
- 23.Auweter SD, Fasan R, Reymond L, Underwood JG, Black DL, Pitsch S, Allain FH. Molecular basis of RNA recognition by the human alternative splicing factor Fox-1. EMBO J. 2006;25:163–173. doi: 10.1038/sj.emboj.7600918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 25.Hayakawa M, Sakashita E, Ueno E, Tominaga S, Hamamoto T, Kagawa Y, Endo H. Muscle-specific exonic splicing silencer for exon exclusion in human ATP synthase gamma-subunit pre-mRNA. J Biol Chem. 2002;277:6974–6984. doi: 10.1074/jbc.M110138200. [DOI] [PubMed] [Google Scholar]
- 26.Zhou HL, Baraniak AP, Lou H. Role for Fox-1/Fox-2 in mediating the neuronal pathway of calcitonin/calcitonin gene-related peptide alternative RNA processing. Mol Cell Biol. 2007;27:830–841. doi: 10.1128/MCB.01015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang ZZ, Zheng S, Nikolic J, Black DL. Developmental control of CaV1.2 L-Type calcium channel splicing by fox proteins. Mol Cell Biol. 2009;29:4757–4765. doi: 10.1128/MCB.00608-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki H, Jin Y, Otani H, Yasuda K, Inoue K. Regulation of alternative splicing of alpha-actinin transcript by Bruno-like proteins. Genes Cells. 2002;7:133–141. doi: 10.1046/j.1356-9597.2001.00506.x. [DOI] [PubMed] [Google Scholar]
- 29.Southby J, Gooding C, Smith CW. Polypyrimidine tract binding protein functions as a repressor to regulate alternative splicing of alpha-actinin mutually exclusive exons. Mol Cell Biol. 1999;19:2699–2711. doi: 10.1128/mcb.19.4.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brudno M, Gelfand MS, Spengler S, Zorn M, Dubchak I, Conboy JG. Computational analysis of candidate intron regulatory elements for tissue-specific alternative pre-mRNA splicing. Nucleic Acids Res. 2001;29:2338–2348. doi: 10.1093/nar/29.11.2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Minovitsky S, Gee SL, Schokrpur S, Dubchak I, Conboy JG. The splicing regulatory element, UGCAUG, is phylogenetically and spatially conserved in introns that flank tissue-specific alternative exons. Nucleic Acids Res. 2005;33:714–724. doi: 10.1093/nar/gki210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yeo GW, Nostrand EL, Liang TY. Discovery and analysis of evolutionarily conserved intronic splicing regulatory elements. PLoS Genet. 2007;3:e85. doi: 10.1371/journal.pgen.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sugnet CW, Srinivasan K, Clark TA, O’Brien G, Cline MS, Wang H, Williams A, Kulp D, Blume JE, Haussler D, Ares M., Jr Unusual intron conservation near tissue-regulated exons found by splicing microarrays. PLoS Comput Biol. 2006;2:e4. doi: 10.1371/journal.pcbi.0020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Das D, Clark TA, Schweitzer A, Yamamoto M, Marr H, Arribere J, Minovitsky S, Poliakov A, Dubchak I, Blume JE, Conboy JG. A correlation with exon expression approach to identify cis-regulatory elements for tissue-specific alternative splicing. Nucleic Acids Res. 2007;35:4845–4857. doi: 10.1093/nar/gkm485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Castle JC, Zhang C, Shah JK, Kulkarni AV, Kalsotra A, Cooper TA, Johnson JM. Expression of 24,426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nat Genet. 2008;40:1416–1425. doi: 10.1038/ng.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 38.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Venables JP, Klinck R, Koh C, Gervais-Bird J, Bramard A, Inkel L, Durand M, Couture S, Froehlich U, Lapointe E, Lucier JF, Thibault P, Rancourt C, Tremblay K, Prinos P, Chabot B, Elela SA. Cancer-associated regulation of alternative splicing. Nat Struct Mol Biol. 2009;16:670–676. doi: 10.1038/nsmb.1608. [DOI] [PubMed] [Google Scholar]
- 40.Zhang C, Zhang Z, Castle J, Sun S, Johnson J, Krainer AR, Zhang MQ. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev. 2008;22:2550–2563. doi: 10.1101/gad.1703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yeo GW, Xu X, Liang TY, Muotri AR, Carson CT, Coufal NG, Gage FH. Alternative splicing events identified in human embryonic stem cells and neural progenitors. PLoS Comput Biol. 2007;3:1951–1967. doi: 10.1371/journal.pcbi.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lim J, Hao T, Shaw C, Patel AJ, Szabo G, Rual JF, Fisk CJ, Li N, Smolyar A, Hill DE, Barabasi AL, Vidal M, Zoghbi HY. A protein–protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125:801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 43.Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, Darnell JC, Darnell RB. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe BJ, Darnell RB. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- 45.Ohkura N, Takahashi M, Yaguchi H, Nagamura Y, Tsukada T. Coactivator-associated arginine methyltransferase 1, CARM1, affects pre-mRNA splicing in an isoform-specific manner. J Biol Chem. 2005;280:28927–28935. doi: 10.1074/jbc.M502173200. [DOI] [PubMed] [Google Scholar]
- 46.Forch P, Puig O, Martinez C, Seraphin B, Valcarcel J. The splicing regulator TIA-1 interacts with U1-C to promote U1 snRNP recruitment to 5′ splice sites. EMBO J. 2002;21:6882–6892. doi: 10.1093/emboj/cdf668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tran Q, Coleman TP, Roesser JR. Human transformer 2beta and SRp55 interact with a calcitonin-specific splice enhancer. Biochim Biophys Acta. 2003;1625:141–152. doi: 10.1016/s0167-4781(02)00600-0. [DOI] [PubMed] [Google Scholar]
- 48.Tran Q, Roesser JR. SRp55 is a regulator of calcitonin/CGRP alternative RNA splicing. Biochemistry. 2003;42:951–957. doi: 10.1021/bi026753a. [DOI] [PubMed] [Google Scholar]
- 49.Zhou HL, Lou H. Repression of prespliceosome complex formation at two distinct steps by Fox-1/Fox-2 proteins. Mol Cell Biol. 2008;28:5507–5516. doi: 10.1128/MCB.00530-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kent OA, Ritchie DB, Macmillan AM. Characterization of a U2AF-independent commitment complex (E′) in the mammalian spliceosome assembly pathway. Mol Cell Biol. 2005;25:233–240. doi: 10.1128/MCB.25.1.233-240.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fukumura K, Taniguchi I, Sakamoto H, Ohno M, Inoue K. U1-independent pre-mRNA splicing contributes to the regulation of alternative splicing. Nucleic Acids Res. 2009;37:1907–1914. doi: 10.1093/nar/gkp050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baraniak AP, Lasda EL, Wagner EJ, Garcia-Blanco MA. A stem structure in fibroblast growth factor receptor 2 transcripts mediates cell-type-specific splicing by approximating intronic control elements. Mol Cell Biol. 2003;23:9327–9337. doi: 10.1128/MCB.23.24.9327-9337.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mauger DM, Lin C, Garcia-Blanco MA. hnRNP H and hnRNP F complex with Fox2 to silence fibroblast growth factor receptor 2 exon IIIc. Mol Cell Biol. 2008;28:5403–5419. doi: 10.1128/MCB.00739-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell. 2009;33:591–601. doi: 10.1016/j.molcel.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Newman EA, Muh SJ, Hovhannisyan RH, Warzecha CC, Jones RB, McKeehan WL, Carstens RP. Identification of RNA-binding proteins that regulate FGFR2 splicing through the use of sensitive and specific dual color fluorescence minigene assays. RNA. 2006;12:1129–1141. doi: 10.1261/rna.34906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oltean S, Sorg BS, Albrecht T, Bonano VI, Brazas RM, Dewhirst MW, Garcia-Blanco MA. Alternative inclusion of fibroblast growth factor receptor 2 exon IIIc in Dunning prostate tumors reveals unexpected epithelial mesenchymal plasticity. Proc Natl Acad Sci USA. 2006;103:14116–14121. doi: 10.1073/pnas.0603090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bonano VI, Oltean S, Brazas RM, Garcia-Blanco MA. Imaging the alternative silencing of FGFR2 exon IIIb in vivo. RNA. 2006;12:2073–2079. doi: 10.1261/rna.248506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nagasaki H, Arita M, Nishizawa T, Suwa M, Gotoh O. Species-specific variation of alternative splicing and transcriptional initiation in six eukaryotes. Gene. 2005;364:53–62. doi: 10.1016/j.gene.2005.07.027. [DOI] [PubMed] [Google Scholar]
- 59.Kim E, Magen A, Ast G. Different levels of alternative splicing among eukaryotes. Nucleic Acids Res. 2007;35:125–131. doi: 10.1093/nar/gkl924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kabat JL, Barberan-Soler S, McKenna P, Clawson H, Farrer T, Zahler AM. Intronic alternative splicing regulators identified by comparative genomics in nematodes. PLoS Comput Biol. 2006;2:e86. doi: 10.1371/journal.pcbi.0020086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.DeVore DL, Horvitz HR, Stern MJ. An FGF receptor signaling pathway is required for the normal cell migrations of the sex myoblasts in C. elegans hermaphrodites. Cell. 1995;83:611–620. doi: 10.1016/0092-8674(95)90101-9. [DOI] [PubMed] [Google Scholar]
- 62.Birnbaum D, Popovici C, Roubin R. A pair as a minimum: the two fibroblast growth factors of the nematode Caenorhabditis elegans. Dev Dyn. 2005;232:247–255. doi: 10.1002/dvdy.20219. [DOI] [PubMed] [Google Scholar]
- 63.Burdine RD, Chen EB, Kwok SF, Stern MJ. egl-17 encodes an invertebrate fibroblast growth factor family member required specifically for sex myoblast migration in Caenorhabditis elegans. Proc Natl Acad Sci USA. 1997;94:2433–2437. doi: 10.1073/pnas.94.6.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Burdine RD, Branda CS, Stern MJ. EGL-17(FGF) expression coordinates the attraction of the migrating sex myoblasts with vulval induction in C. elegans . Development. 1998;125:1083–1093. doi: 10.1242/dev.125.6.1083. [DOI] [PubMed] [Google Scholar]
- 65.Goodman SJ, Branda CS, Robinson MK, Burdine RD, Stern MJ. Alternative splicing affecting a novel domain in the C. elegans EGL-15 FGF receptor confers functional specificity. Development. 2003;130:3757–3766. doi: 10.1242/dev.00604. [DOI] [PubMed] [Google Scholar]
- 66.Kuroyanagi H, Ohno G, Mitani S, Hagiwara M. The Fox-1 family and SUP-12 coordinately regulate tissue-specific alternative splicing in vivo. Mol Cell Biol. 2007;27:8612–8621. doi: 10.1128/MCB.01508-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zahler AM (2005) Alternative splicing in C. elegans. WormBook 1–13 [DOI] [PMC free article] [PubMed]
- 68.David CJ, Manley JL. The search for alternative splicing regulators: new approaches offer a path to a splicing code. Genes Dev. 2008;22:279–285. doi: 10.1101/gad.1643108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ogura K, Kishimoto N, Mitani S, Gengyo-Ando K, Kohara Y. Translational control of maternal glp-1 mRNA by POS-1 and its interacting protein SPN-4 in Caenorhabditis elegans . Development. 2003;130:2495–2503. doi: 10.1242/dev.00469. [DOI] [PubMed] [Google Scholar]
- 70.Meyer BJ. Sex in the worm: counting and compensating X-chromosome dose. Trends Genet. 2000;16:247–253. doi: 10.1016/S0168-9525(00)02004-7. [DOI] [PubMed] [Google Scholar]
- 71.Hodgkin J, Zellan JD, Albertson DG. Identification of a candidate primary sex determination locus, fox-1, on the X chromosome of Caenorhabditis elegans . Development. 1994;120:3681–3689. doi: 10.1242/dev.120.12.3681. [DOI] [PubMed] [Google Scholar]
- 72.Carmi I, Kopczynski JB, Meyer BJ. The nuclear hormone receptor SEX-1 is an X-chromosome signal that determines nematode sex. Nature. 1998;396:168–173. doi: 10.1038/24164. [DOI] [PubMed] [Google Scholar]
- 73.Nicoll M, Akerib CC, Meyer BJ. X-chromosome-counting mechanisms that determine nematode sex. Nature. 1997;388:200–204. doi: 10.1038/40669. [DOI] [PubMed] [Google Scholar]
- 74.Gladden JM, Farboud B, Meyer BJ. Revisiting the X:A signal that specifies Caenorhabditis elegans sexual fate. Genetics. 2007;177:1639–1654. doi: 10.1534/genetics.107.078071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rhind NR, Miller LM, Kopczynski JB, Meyer BJ. xol-1 acts as an early switch in the C. elegans male/hermaphrodite decision. Cell. 1995;80:71–82. doi: 10.1016/0092-8674(95)90452-2. [DOI] [PubMed] [Google Scholar]
- 76.Skipper M, Milne CA, Hodgkin J. Genetic and molecular analysis of fox-1, a numerator element involved in Caenorhabditis elegans primary sex determination. Genetics. 1999;151:617–631. doi: 10.1093/genetics/151.2.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR, 3rd, Frank J, Manley JL. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell. 2009;33:365–376. doi: 10.1016/j.molcel.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14(51–55):29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]