Abstract
Objective
Genetic polymorphisms that modify the detoxifying activity of glutathione S-transferases (GSTs) can affect the level of carcinogenic metabolites created by endogenous steroid hormones and exogenous chemical substances. Although the GSTM1 null genotype has been shown to increase prostate cancer mortality in Caucasians, potential associations between GST polymorphisms and prostate cancer biochemical recurrence (BCR) have not been well studied, particularly in African-Americans.
Methods
We examined potential associations between the GSTM1 null, GSTT1 null and GSTP1 Ile105Val polymorphisms and BCR, after prostatectomy, in 168 African-American and 226 Caucasian patients treated at Henry Ford Hospital in Detroit, Michigan using Cox proportional hazards modeling.
Results
We found that African-Americans with the GSTT1 null genotype had increased BCR risk compared to those having GSTT1 present (hazard ratio (HR) = 2.30; 95% CI = 1.01–5.18; p = 0.04); and African-Americans with the GSTT1 null genotype and high grade tumors had an even greater risk (HR = 7.82; 95% CI = 2.49–24.50; p < 0.001). In Caucasians, an increased risk was observed in those patients with high grade tumors and the GSTM1 null genotype (HR = 2.88; 95% CI = 1.16–7.14; p = 0.02). Similar associations were observed for advanced stage and more aggressive (high grade or advanced stage) disease.
Conclusion
Our results suggest GSTs may hold promise as therapeutic targets in more advanced prostate cancers, particularly, in African-Americans.
Keywords: GSTT1, GSTM1, GSTP1, Prostate cancer, Biochemical recurrence
Introduction
Prostate cancer is the most commonly diagnosed non-skin cancer and the third leading cause of cancer death among men in the United States [1], with higher mortality rates among African-Americans compared to Caucasians [2]. Clinical diagnostic characteristics also affect prostate cancer mortality and biochemical recurrence rates. Those men present with a biopsy Gleason score of eight or greater, clinical tumor stage of T2c or greater, or serum prostate specific antigen (PSA) level of greater than 20 ng/ml are at high risk of disease recurrence [3] and mortality [4] even after radical prostatectomy, while men with a Gleason score of less than seven, tumor stage of T2a or less, or PSA of less than 10 ng/ml are unlikely to die from prostate cancer [5]. Furthermore, compared to Caucasians with similar clinical presentation, African-American men continue to show higher rates of biochemical recurrence after prostatectomy [6], suggesting other genetic and/or environmental factors contribute to prostate cancer recurrence and mortality.
Glutathione S-transferases (GSTs) are an important family of enzymes involved in the biosynthesis and metabolism of many substances [7] including the detoxification of exogenous carcinogenic chemicals such as polycyclic aromatic hydrocarbons (PAH), which are found in many common exposures such as cigarette smoke, diesel fuel and grilled meats. GSTs may also detoxify reactive endogenous steroid hormone metabolites such as estradiol (E2)-quinones and reactive oxygen species (ROS), which may arise from multiple sources including inflammation and the futile redox cycling of E2- and PAH-quinones [8, 9]. Specific GST isoforms in the μ (M1), θ (T1) and π (P1) classes are highly expressed in the prostate [10, 11]; therefore, genetic polymorphisms that modify activity of these GSTs can affect the level of carcinogenic metabolites in the prostate, which, in turn, may alter the risk of prostate cancer recurrence and mortality as well as its incidence.
Functional genetic variants in GSTs, predominantly the GSTM1 null, GSTT1 null and GSTP1 Ile105Val polymorphisms, have been associated inconsistently with prostate cancer incidence; and authors of a recent meta-analysis concluded that the GSTM1 null, GSTT1 null and GSTP1 Ile105Val polymorphisms were unlikely to be major determinants of prostate cancer susceptibility on a wide population basis [12]. However, this meta-analysis was conducted using Caucasian and Asian populations only and did not include any studies with subjects of African descent. This is a critical point because we have previously shown that allele frequencies and associations between GST polymorphisms and PAH-DNA adduct levels are significantly different among African-American compared to Caucasian men [13]. Furthermore, the GSTM1-null genotype has been shown to increase risk of prostate cancer mortality in Caucasian men [14]; however, no prior studies have examined potential effects of these GST polymorphisms on prostate cancer biochemical recurrence risk in African-American men.
Therefore, we examined the potential associations between the GSTM1-null, GSTT1-null and GSTP1 Ile105Val polymorphisms and prostate cancer biochemical recurrence within 5 years after prostatectomy surgery in 168 African-American and 226 Caucasian patients treated at the Henry Ford Hospital in Detroit, Michigan. We also evaluated potential differential associations between these GST polymorphisms and prostate cancer BCR by ethnicity and clinical measures of disease severity at diagnosis.
Materials and methods
Study population
The study population from which the subjects used in this analysis were derived has been previously described [15]. Briefly, 637 cases with a histologically confirmed prostate cancer diagnosis within the last 2 years that were treated at the Henry Ford Hospital in Detroit, Michigan were enrolled within 2 years after diagnosis between 1 July 2001 and 31 December 2004 for an observational prostate cancer case–control study. Of the 637 cases, 429 (67%) underwent radical prostatectomy and were followed from the date of surgery forward using electronic medical records to retrieve all prostate specific antigen (PSA) test results. We excluded the 20 cases that had no follow-up or only one PSA test after surgery and 14 men who also underwent hormone treatment. One case was excluded because of missing tumor grade information. The remaining 394 men comprised the analytic study sample, which had data on 4,459 follow-up PSA test results, ranging from 2 to 54 tests and a median of ten PSA tests per subject. All protocols used in this study were reviewed and approved by the Henry Ford Hospital Institutional Review Board and all study participants provided informed consent.
Genotyping
Standard venipuncture was used to collect blood samples from all study participants in tubes with EDTA as an anticoagulant. Genomic DNA was extracted from buffy coats using QIAmp DNA Blood kit (Qiagen Inc., Valencia, CA). All purified DNA samples were diluted to a constant DNA concentration in 10 mmol/L Tris, 1 mmol/L EDTA buffer (pH 8).
The GSTP1 Ile105Val (rs947894) polymorphism was detected using the Invader assay with reagents developed by Third Wave Technologies, Inc. (Madison, WI), which uses primers 5′-ACC CCA GGG CTC TAT GGG AA-3′ and 5′-TGA GGG CAC AAG AAG CCC CT-3′ followed by digestion with Alw261. Separation of PCR products, either a 176 bp (GSTP1 Ile) or 91 and 85 bp (GSTP Val), was done on a 3.5% agarose gel.
The full deletion of GSTM1 and GSTT1 was detected by PCR with β-globin as an internal control by the method of Arand et al. with modifications [16, 17]. Primers used were 5′-GAACTCCCTGAAAAGCTAAAGC-3′ and 5′-GTTGGGCTCAAATATACGGTGG-3′ for GSTM1 or 5′-TTCCTTACTGGTCCTCACATCT-3′ and 5′-TCACCGGATCATGGCCAGCA-3′ for GSTT1 and 5′-GAAGAGCCAAGGACAGGTAC-3′ and 5′-CAACTTCATCCACGTTCACC-3′ for β-globin. DNA (50 ng) was amplified in a reaction volume of 25 μl with 10 pmol of each of the primers, 1.5 mM MgCl2 and 1.5 U AmpliTaq Gold DNA polymerase. Cycling conditions were 10 min at 94°C, followed by 35 cycles (GSTM1) or 30 cycles (GSTT1) of 94°C for 1 min, 62°C for 1 min, 72°C for 1 min and a final extension at 72°C for 5 min. PCR products were separated on a 2% agarose gel and GSTM1 identified by a 215 bp band, GSTT1 by a 473 bp band and β-globin by a 268 bp band.
Statistical analysis
Hazard ratios (HRs) for prostate cancer biochemical recurrence (BCR) by genotype status for the GSTM1-null, GSTT1-null and GSTP1 Ile105Val polymorphisms were estimated with a Cox proportional hazard model using PROC PHREG in SAS v9.1 (SAS Institute, Inc., Cary, NC). Following Freedland et al. [18] and Kupelian et al. [19], we defined a BCR event as having two consecutive detectable (PSA > 0.2 ng/mL) increasing PSA levels for four or more weeks after surgery. Time to event was defined as the duration between the date of surgery and the second PSA test that defined the recurrence event. Patients did not recur were censored at the last post-operative PSA test. Differences in survival curves were tested using the Wilcoxon rank test using PROC LIFETEST in SAS v9.1 (SAS Institute, Inc., Cary, NC). All models were adjusted for age, race, smoking, tumor stage, tumor (Gleason score) grade and PSA level at diagnosis. We evaluated potential confounding by race (population stratification) in a subset of African-American cases (n = 146) with African ancestry scores, which we estimated using ADMIXMAP software (http://homepages.ed.ac.uk/pmckeigu/admixmap) [20] and a standard panel of ancestry informative markers (http://www.illumina.com/pages.ilmn?ID=235). We also examined potential confounding by PAH-DNA adduct levels in a subset of patients (N = 368) with PAH-DNA adduct data available. We investigated whether GST polymorphisms were associated with BCR of prostate cancer in subgroups defined by race (Caucasian, African-American) and clinical risk factors (clinical tumor stage, tumor grade (from biopsy Gleason score) and PSA level at diagnosis). All reported p-values are from two-sided tests.
Results
Characteristics of the study population are shown in Table 1. Biochemical recurrence (BCR) of prostate cancer was experienced by 76 men (19.3%). Men with a BCR event had an average follow-up time of 66.9 months, which was not significantly different from the mean follow-up time of men not having a BCR event (61.0 months); however, for purposes of the analyses and presentation of survival data, follow-up time was censored at 60 months (5 years). Those men that experienced BCR were more likely to have tumors with an advanced stage (T3 or higher), high tumor grade (total Gleason score ≥8 or primary Gleason ≥4), and higher PSA level at diagnosis. The mean age at diagnosis, smoking status and race were not significantly different between men with a BCR event and those not experiencing BCR. Genotype frequencies for the GSTM1 null, GSTT1 null and GSTP1 Ile105Val polymorphisms were not significantly different between men with and without recurrence. In addition, the number of GST variant (risk) alleles was also not significantly different between the two groups.
Table 1.
Characteristics of 394 prostate cancer cases by biochemical recurrence (BCR) status following prostatectomy surgery
| Characteristic | BCR (n = 76) | No BCR (n = 318) | p-valuea |
|---|---|---|---|
| Age (mean ± SE) | 60.8 ± 6.0 | 61.0 ± 6.8 | 0.73 |
| African-Americans (%) | 30 (39.5%) | 138 (43.4%) | 0.53 |
| Average time of follow-up/observation (months)b | 66.9 ± 24.7 | 61.0 ± 22.7 | 0.09 |
| PSA at diagnosis (ng/ml) | 10.6 ± 9.6 | 6.0 ± 4.3 | <0.001 |
| High tumor gradec | 35 (46.1%) | 77 (24.2%) | <0.001 |
| Advanced tumor staged | 30 (39.7%) | 43 (13.5%) | <0.001 |
| Cigarette smoking status | |||
| Never | 23 (33.3%) | 119 (38.1%) | 0.42 |
| Former | 46 (60.5%) | 156 (51.7%) | |
| Current | 7 (9.2%) | 33 (10.3%) | |
| GSTM1 null | 32 (42.1%) | 123 (38.7%) | 0.58 |
| GSTT1 null | 15 (19.7%) | 67 (21.1%) | 0.80 |
| GSTP1 Ile105Val | |||
| Ile/Ile | 26 (34.2%) | 117 (36.8%) | 0.22 |
| Ile/Val | 34 (44.7%) | 159 (50.0%) | |
| Val/Val | 16 (21.1%) | 42 (13.2%) | |
| Total number of high risk genotypes (combined)e | |||
| 0 | 11 (14.5%) | 53 (16.7%) | 0.96 |
| 1 | 36 (47.4%) | 149 (46.9%) | |
| 2 | 26 (32.4%) | 106 (33.3%) | |
| 3 | 3 (4.0%) | 10 (3.1%) | |
a p-value from t-test or χ2 test, as applicable
bTime from study entry to date of last PSA test for the entire cohort
cHigh-grade defined as a total Gleason score of eight or higher or a primary Gleason score of four or higher
dAdvanced tumor stage defined as Stage 3a or higher
eNumber of GSTM1 null, GSTT1 null and GSTP1 105Val alleles
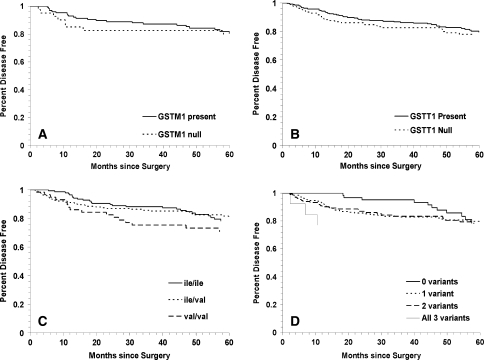
Kaplan-Meier curves for prostate cancer BCR in the GSTM1, GSTT1 and GSTP1 Ile105Val polymorphisms were not significantly different (Fig. 1a–d). For the GSTM1 null (Fig. 1, Panel a) and GSTP1 105 Val/Val (Fig. 1, Panel c) genotypes, BCR was higher at 2 to 3 years of follow-up, but the survival curves tended to move toward each other at 5 years. In models adjusted for age, race, smoking, tumor stage, tumor grade and PSA at baseline (Table 2), a non-statistically significant increased risk of BCR was observed in men with the GSTM1 null (HR = 1.41; 95% CI 0.88–2.26; p = 0.16) and GSTT1 null (HR = 1.11; 95% CI 0.63–1.96; p = 0.72) genotypes compared to those with GSTM1 and GSTT1 present, respectively. Similarly, we observed no statistically significant association between the GSTP1 Ile105Val polymorphism and prostate cancer BCR, with the largest effect size observed with a recessive genetic model (i.e., GSTP1 105 Val/Val versus Ile/Ile or Ile/Val: HR = 1.62; 95% CI 0.87–2.68; p = 0.14). We also examined the total number of putative high risk (variant) GST genotypes and under an additive model (i.e., having 1, 2, or all 3 GST variants (GSTM1 null, GSTT1 null and/or GSTP1 105 Ile/Val or Val/Val) versus having no GST variants (GSTM1 present, GSTT1 present and GSTP1 105 Ile/Ile) we observed no significant increased BCR risk (HR = 1.15; 95% CI 0.85–1.56; p = 0.37).
Fig. 1.
Kaplan–Meier survival curves for biochemical recurrence of prostate cancer for GST polymorphisms: a GSTM1 null versus GSTM1 present (log rank p = 0.4); b GSTT1 null versus GSTT1 present (log rank p = 0.9); c GSTP1 codon 105 Ile/Ile versus Ile/Val versus Val/Val (log rank p = 0.2) and d total number of GST high risk (variant) genotypes (log rank p = 0.9)
Table 2.
Risk of prostate cancer biochemical recurrence after prostatectomy associated with GST polymorphisms by race
| Polymorphism | Total sample (n = 394) | Caucasians (n = 226) | African-Americans (n = 168) | P-valuea |
|---|---|---|---|---|
| GSTM1 null |
1.41 (0.88–2.26); p = 0.16 (N H1 = 32; N R = 44)b |
1.61 (0.89–2.96); p = 0.11 (N H1 = 24; N R = 22) |
1.11 (0.44–2.40); p = 0.95 (N H1 = 8; NR = 22) |
0.62 |
| GSTT1 null |
1.11 (0.63–1.96); p = 0.72 (N H1 = 15; N R = 61) |
0.55 (0.21–1.40); p = 0.20 (N H1 = 5; N R = 41) |
2.30 (1.01–5.18); p = 0.04 (N H1 = 10; N R = 20) |
0.02 |
| GSTP1 Ile105Val | ||||
| Co-dominant | ||||
| Ile/Val vs. Ile/Ile |
0.81 (0.48–1.39); p = 0.44 (N H1 = 16; N R = 34) |
0.54 (0.27–1.08); p = 0.08 (N H1 = 9; N R = 17) |
1.71 (0.64–4.55); p = 0.28 (N H1 = 7; N R = 17) |
0.22 |
| Val/Val vs. Ile/Ile |
1.35 (0.71–2.57); p = 0.37 (N H1 = 16; N R = 26) |
0.96 (0.40–2.28); p = 0.93 (N H1 = 9; N R = 20) |
2.10 (0.66–6.67); p = 0.21 (N H1 = 7; N R = 6) |
0.50 |
| Recessive | ||||
| Val/Val vs. Ile/Val or Ile/Ile |
1.62 (0.87–2.68); p = 0.14 (N H1 = 16; N R = 60) |
1.39 (0.64–3.00); p = 0.42 (N H1 = 9; NR = 37) |
1.45 (0.59–3.55); p = 0.42 (N H1 = 7; N R = 23) |
0.96 |
| Dominant | ||||
| Ile/Val or Val/Val vs. Ile/Ile |
0.93 (0.57–1.53); p = 0.78 (N H1 = 50; N R = 26) |
0.62 (0.33–1.18); p = 0.15 (N H1 = 26; N R = 20) |
1.81 (0.71–4.63); p = 0.22 (N H1 = 24; N R = 6) |
0.26 |
| Additive | ||||
| Ile/Val vs. Val/Val vs. Ile/Ile |
1.11 (0.79–1.57); p = 0.54 (N H1 = 16; N H2 = 34; N R = 26) |
0.87 (0.55–1.39); p = 0.56 (N H1 = 9; N H2 = 17; N R = 20) |
1.44 (0.83–2.52); p = 0.20 (N H1 = 7; N H2 = 17; N R = 6) |
0.35 |
| Total number of high risk GST genotypesc |
1.15 (0.85–1.56); p = 0.37 (N H1 = 3; N H2 = 26; N H3 = 36; N R = 11) |
0.92 (0.64–1.32); p = 0.65 (N H1 = 0; N H2 = 17; N H3 = 21; N R = 8) |
1.89 (1.06–3.40); p = 0.03 (N H1 = 3; N H2 = 9; N H3 = 15; N R = 3) |
0.04 |
Hazard ratio (HR) and the 95% confidence interval (CI) adjusted for age, race, smoking, tumor stage, tumor grade and PSA at diagnosis
a p-value for race × genotype interaction
bNumber of events in putative high risk groups (N h1, N h2, N h3) versus low risk referent group (N R)
cAdditive risk of having 1, 2 or 3 of the GSTM1 null, GSTT1 null and GSTP1 105 Ile/Val or Val/Val genotypes versus having no variant genotypes
When we stratified by race (Table 2), however, we found that the GSTT1 null genotype increased risk in African-Americans (HR = 2.30; 95% CI = 1.01–5.18; p = 0.04) but not in Caucasians (p-value for the race × genotype interaction term (p int) = 0.02). The GSTM1 null genotype conferred a modest, non-statistically significant increased risk of BCR in Caucasians (HR = 1.61; 95% CI = 0.89–2.96; p = 0.11), but the effect size in African-Americans was not much different than 1.0 (HR = 1.11; 95% CI = 0.44–2.40; p = 0.95). No significant association was observed for either Caucasian or African-American men with the GSTP1 105Val variant allele under any genetic model (recessive, dominant, or additive). Interestingly, in African-Americans, the total number of high risk GST genotypes under an additive genetic model was associated with an increased BCR risk (HR = 1.89; 95% CI 1.06–3.40; p = 0.03), but this association was not observed in Caucasians (p int = 0.04).
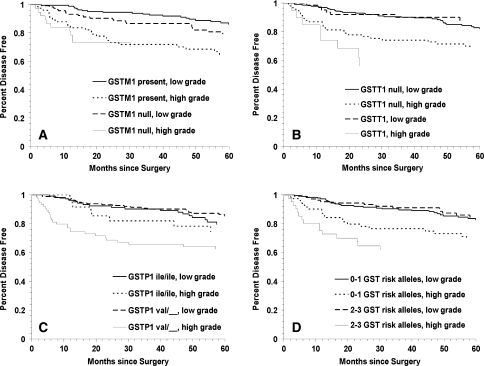
We next examined potential associations between GST polymorphisms and prostate cancer BCR by measures of disease severity at diagnosis. Kaplan–Meier curves for BCR for each of the three GST polymorphisms and the combined high risk genotypes by tumor grade are shown in Fig. 2a–d. When stratifying by genotype, patients with high grade tumors had increased prostate cancer BCR if they had the GSTT1 null (Fig. 2, Panel b: Log Rank p = 0.003) or the GSTP1 105 Ile/Val or Val/Val (Panel c: Log Rank p = 0.003) genotypes. However, further stratification by race revealed that the risk associated with the GSTT1 null and GSTP1 105 Ile/Val or Val/Val genotypes was even greater in African-Americans and essentially absent in Caucasians (Table 3). More specifically, African-Americans with high grade tumors and the GSTT1 null genotype had nearly an eightfold increased risk of BCR compared to African-Americans with low grade tumors and GSTT1 present (HR = 7.82; 95% CI = 2.49–24.50; p < 0.001). In addition, African-American patients with high grade tumors and the GSTP1 105 Ile/Val or Val/Val genotypes had over a threefold increased risk of BCR compared to African-Americans with low grade tumors and the GSTP1 Ile/Ile genotype (HR = 3.68; 95% CI = 1.13–12.04; p = 0.03). Caucasian men with high grade tumors and the GSTM1 null genotype had nearly a threefold increased risk of BCR compared to Caucasians with low grade tumors and GSTM1 present (HR = 2.88; 95% CI = 1.16–7.14; p = 0.02). Moreover, having two or three compared to one or no high risk (variant) GST genotypes only conferred increased BCR risk in African-Americans with high grade tumors (HR = 4.78; 95% CI = 1.58–14.43; p = 0.006).
Fig. 2.
Kaplan–Meier survival curves for biochemical recurrence of prostate cancer for GST polymorphisms in high versus low tumor grade: a GSTM1 null versus GSTM1 present (log rank p = 0.0002); b GSTT1 null versus GSTT1 present (log rank p = 0.0001); c GSTP1 codon 105 Ile/Ile and Ile/Val versus Val/Val (log rank p < 0.00001) and d total number of GST high risk (variant) genotypes (log rank p = 0.0001)
Table 3.
Risk of prostate cancer BCR after prostatectomy associated with GST polymorphisms by race and tumor grade
| Genotype | Total sample | Caucasians | African-Americans | |||
|---|---|---|---|---|---|---|
| Low grade | High grade | Low grade | High grade | Low grade | High grade | |
| GSTM1 |
1.00 (referent) (N = 20)b |
2.45 (1.32–4.53) p = 0.004 (N = 21) |
1.00 (referent) (N = 11) |
2.08 (0.87–4.96) p = 0.10 (N = 15) |
1.00 (referent) (N = 9) |
4.00 (1.60–9.99) p = 0.003 (N = 6) |
| GSTM1 null |
1.83 (0.97–3.44) p = 0.06 (N = 24) |
2.48 (1.16–5.30) p = 0.02 (N = 11) |
1.84 (0.81–4.18) p = 0.14 (N = 11) |
2.88 (1.16–7.14) p = 0.02 (N = 9) |
2.04 (0.71–5.83) p = 0.19 (N = 13) |
1.49 (0.31–7.24) p = 0.62 (N = 2) |
| GSTT1 |
1.00 (referent) (N = 34) |
1.58 (0.93–2.70) p = 0.09 (N = 7) |
1.00 (referent) (N = 24) |
1.49 (0.78–2.87) p = 0.23 (N = 2) |
1.00 (referent) (N = 10) |
2.21 (0.86–5.64) p = 0.10 (N = 5) |
| GSTT1 null |
0.72 (0.32–1.64) p = 0.44 (N = 27) |
3.05 (1.38–6.72) p = 0.006 (N = 8) |
0.32 (0.08–1.35) p = 0.12 (N = 17) |
1.59 (0.45–5.67) p = 0.48 (N = 3) |
1.60 (0.53–4.80) p = 0.40 (N = 10) |
7.82 (2.49–24.50) p < 0.001 (N = 5) |
| GSTP1 Ile/Ile |
1.00 (referent) (N = 18) |
1.12 (0.48–2.63) p = 0.79 (N = 23) |
1.00 (referent) (N = 14) |
1.30 (0.50–3.43) p = 0.59 (N = 12) |
1.00 (referent) (N = 4) |
0.80 (0.14–4.52) p = 0.80 (N = 11) |
| GSTP1 Ile/Val or Val/Val |
0.68 (0.36–1.27) p = 0.22 (N = 8) |
1.68 (0.87–3.24) p = 0.12 (N = 27) |
0.50 (0.22–1.13) p = 0.09 (N = 6) |
1.13 (0.50–2.57) p = 0.77 (N = 14) |
0.84 (0.26–2.67) p = 0.76 (N = 2) |
3.68 (1.13–12.04) p = 0.03 (N = 13) |
| 0–1 High risk GST allelesa |
1.00 (referent) (N = 27) |
1.61 (0.88–2.96) p = 0.12 (N = 14) |
1.00 (referent) (N = 18) |
1.53 (0.70–3.35) p = 0.28 (N = 8) |
1.00 (referent) (N = 9) |
2.34 (0.88–6.22) p = 0.09 (N = 6) |
| 2–3 High risk GST alleles |
0.95 (0.49–1.82) p = 0.87 (N = 20) |
2.25 (1.22–4.53) p = 0.01 (N = 15) |
0.74 (0.31–1.74) p = 0.49 (N = 11) |
1.67 (0.73–3.85) p = 0.23 (N = 9) |
1.43 (0.50–4.05) p = 0.51 (N = 9) |
4.78 (1.58–14.43) p = 0.006 (N = 6) |
Hazard ratio (HR) and the 95% confidence interval (CI) adjusted for age, race, smoking, tumor stage and PSA at diagnosis
aThe number of high risk alleles refers to the total number of GST (GSTM1 null, GSTT1 null and GSTP1 105 Ile/Val or Val/Val) variants and individual carries (0, 1, 2 or 3)
bNumber of biochemical failure events (N) in each stratum
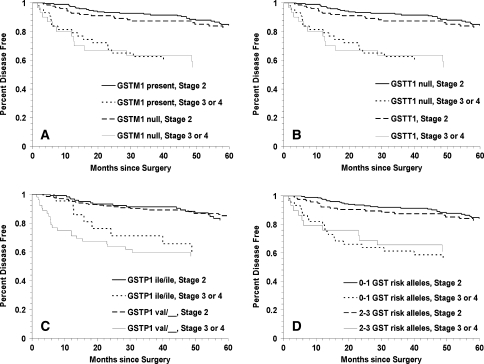
We observed similar findings when examining associations for the various GST polymorphisms by clinical tumor stage subgroups. Specifically, we found that patients with high stage tumors had increased prostate cancer BCR if they had the GSTT1 null (Fig. 3, Panel b: Log Rank p < 0.0001) or the GSTP1 105 Ile/Val or Val/Val (Panel c: Log Rank p = <0.0001) genotypes. Furthermore, as shown in Table 4, men with high stage tumors and the GSTM1 null genotype had an increased BCR risk compared to those with low stage tumors and GSTM1 present (HR = 3.58; 95% CI = 1.82–7.05; p < 0.001); however, stratifying by race revealed that this association only remained significant among Caucasians (HR = 4.89; 95% CI = 2.10–11.43; p < 0.001). African-Americans with high stage tumors, however, had increased BCR when they had the GSTT1 null (HR = 6.20; 95% CI = 1.63–23.58; p = 0.008) or the GSTP1 105 Ile/Val or Val/Val (HR = 3.94; 95% CI = 1.25–12.43) genotype compared to African-Americans with low stage tumors and GSTT1 present or GSTP1 105 Ile/Ile, respectively. Similar to the results with high tumor grade, only African-American patients with high stage tumors and two or three compared to one or no high risk (variant) GST genotypes had increased risk of BCR (HR = 3.60; 95% CI = 1.11–11.65; p = 0.03).
Fig. 3.
Kaplan–Meier survival curves for biochemical recurrence of prostate cancer for GST polymorphisms in high versus low tumor stage: a GSTM1 null versus GSTM1 present (log rank p < 0.0001); b GSTT1 null versus GSTT1 present (log rank p < 0.0001); c GSTP1 codon 105 Ile/Ile and Ile/Val versus Val/Val (log rank p < 0.0001) and d total number of GST high risk (variant) genotypes (log rank p < 0.0001)
Table 4.
Risk of prostate cancer BCR after prostatectomy associated with GST polymorphisms by race and tumor stage
| Genotype | Total sample | Caucasians | African-Americans | |||
|---|---|---|---|---|---|---|
| Low stage | High stage | Low stage | High stage | Low stage | High stage | |
| GSTM1 |
1.00 (referent) (N = 27)b |
2.48 (1.31–4.69) p = 0.005 (N = 19) |
1.00 (referent) (N = 13) |
2.98 (1.23–7.21) p = 0.02 (N = 14) |
1.00 (referent) (N = 14) |
2.97 (1.16–7.62) p = 0.02 (N = 5) |
| GSTM1 null |
1.38 (0.75–2.53) p = 0.30 (N = 17) |
3.58 (1.82–7.05) p < 0.001 (N = 13) |
1.61 (0.73–3.53) p = 0.24 (N = 9) |
4.89 (2.10–11.43) p < 0.001 (N = 10) |
1.36 (0.48–3.86) p = 0.57 (N = 8) |
2.01 (0.55–7.30) p = 0.29 (N = 3) |
| GSTT1 |
1.00 (referent) (N = 35) |
2.88 (1.68–4.95) p < 0.001 (N = 11) |
1.00 (referent) (N = 23) |
3.46 (1.82–6.58) p < 0.001 (N = 4) |
1.00 (referent) (N = 12) |
2.42 (0.91–6.45) p = 0.08 (N = 7) |
| GSTT1 null |
1.34 (0.68–2.65) p = 0.40 (N = 26) |
2.14 (0.73–6.21) p = 0.16 (N = 4) |
0.82 (0.28–2.40) p = 0.72 (N = 18) |
0.76 (0.10–5.97) p = 0.80 (N = 1) |
2.21 (0.86–5.68) p = 0.10 (N = 8) |
6.20 (1.63–23.58) p = 0.008 (N = 3) |
| GSTP1 Ile/Ile |
1.00 (referent) (N = 18) |
2.39 (1.03–5.54) p = 0.04 (N = 28) |
1.00 (referent) (N = 13) |
3.53 (1.39–8.97) p = 0.008 (N = 14) |
1.00 (referent) (N = 5) |
0.81 (0.09–7.08) p = 0.85 (N = 14) |
| GSTP1 Ile/Val or Val/Val |
0.89 (0.49–1.63) p = 0.70 (N = 8) |
2.42 (1.23–4.76) p = 0.01 (N = 22) |
0.66 (0.30–1.46) p = 0.30 (N = 7) |
2.01 (0.86–4.71) p = 0.11 (N = 12) |
1.24 (0.43–3.56) p = 0.69 (N = 1) |
3.94 (1.25–12.43) p = 0.02 (N = 10) |
| 0–1 High risk GST allelesa |
1.00 (referent) (N = 28) |
2.85 (1.56–5.23) p < 0.001 (N = 18) |
1.00 (referent) (N = 17) |
3.66 (1.72–7.78) p < 0.001 (N = 10) |
1.00 (referent) (N = 11) |
2.69 (0.95–7.65) p = 0.06 (N = 8) |
| 2–3 High risk GST allelesa |
1.29 (0.71–2.35) p = 0.40 (N = 19) |
2.78 (1.35–5.75) p = 0.006 (N = 11) |
1.08 (0.48–2.40) p = 0.85 (N = 12) |
2.47 (0.98–6.24) p = 0.06 (N = 7) |
1.91 (0.75–4.84) p = 0.17 (N = 7) |
3.60 (1.11–11.65) p = 0.03 (N = 4) |
Hazard ratio (HR) and the 95% confidence interval (CI) adjusted for age, race, smoking, tumor grade and PSA at diagnosis
aThe number of high risk alleles refers to the number of GST (GSTM1 null, GSTT1 null and GSTP1 105 Ile/Val or Val/Val) variants carried
bNumber of biochemical failure events (N) in each stratum
Men with more aggressive (high grade or high stage) tumors carrying the GSTM1 null genotype had an increased risk of BCR compared to those with less aggressive (low grade or low stage) disease and GSTM1 present (HR = 3.75; 95% CI = 1.85–7.61; p < 0.001); however, after stratifying by race, this association only remained statistically significant in Caucasians (HR = 4.26; 95% CI = 1.73–10.48; p = 0.002) (Table 5). African-Americans with more aggressive disease carrying the GSTT1 null genotype had increased BCR compared to those with less aggressive disease and GSTT1 present (HR = 5.61; 95% CI = 1.72–18.36; p = 0.004). Furthermore, men with more aggressive disease carrying 2–3 GST variants compared to those with less aggressive disease carrying none or one GST variant had an increased risk of BCR (HR = 3.00; 95% CI = 1.55–5.78; p = 0.001), but this association was stronger in African-Americans (HR = 5.14; 95% CI = 1.65–16.07; p = 0.005) than in Caucasians (HR = 2.29; 95% CI = 1.02–5.13; p = 0.05).
Table 5.
Risk of prostate cancer BCR after prostatectomy associated with GST polymorphisms by race and tumor aggressiveness status
| Genotype | Total sample | Caucasians | African-Americans | |||
|---|---|---|---|---|---|---|
| Low aggressive | High aggressive | Low aggressive | High aggressive | Low aggressive | High aggressive | |
| GSTM1 present |
1.00 (referent) (N = 15)b |
2.85 (1.52–5.34) p = 0.001 (N = 29) |
1.00 (referent) (N = 8) |
2.73 (1.13–6.56) p = 0.03 (N = 14) |
1.00 (referent) (N = 7) |
3.44 (1.37–8.65) p = 0.009 (N = 15) |
| GSTM1 null |
1.95 (0.96–4.00) p = 0.07 (N = 16) |
3.75 (1.85–7.61) p < 0.001 (N = 16) |
1.98 (0.76–5.14) p = 0.16 (N = 11) |
4.26 (1.73–10.48) p = 0.002 (N = 13) |
2.06 (0.64–6.62) p = 0.22 (N = 5) |
2.72 (0.70–10.61) p = 0.15 (N = 3) |
| GSTT1 present |
1.00 (referent) (N = 24) |
2.38 (1.42–4.01) p = 0.001 (N = 37) |
1.00 (referent) (N = 17) |
2.34 (1.25–4.38) p = 0.008 (N = 24) |
1.00 (referent) (N = 7) |
2.85 (1.10–7.43) p = 0.03 (N = 13) |
| GSTT1 null |
1.10 (0.48–2.56) p = 0.82 (N = 7) |
2.53 (1.13–5.64) p = 0.02 (N = 8) |
0.52 (0.12–2.26) p = 0.38 (N = 2) |
1.34 (0.39–4.62) p = 0.65 (N = 3) |
2.20 (0.69–6.97) p = 0.18 (N = 5) |
5.61 (1.72–18.36) p = 0.004 (N = 5) |
| GSTP1 Ile/Ile |
1.00 (referent) (N = 14) |
1.44 (0.66–3.15) p = 0.35 (N = 12) |
1.00 (referent) (N = 11) |
1.83 (0.75–4.48) p = 0.18 (N = 9) |
1.00 (referent) (N = 3) |
1.07 (0.21–3.04) p = 0.94 (N = 3) |
| GSTP1 Ile/Val or Val/Val |
0.62 (0.30–1.27) p = 0.19 (N = 17) |
1.92 (1.00–3.68) p = 0.05 (N = 33) |
0.49 (0.19–1.28) p = 0.14 (N = 8) |
1.56 (0.71–3.44) p = 0.27 (N = 18) |
0.82 (0.22–3.04) p = 0.77 (N = 9) |
3.01 (0.85–10.71) p = 0.09 (N = 15) |
| 0–1 High risk GST allelesa |
1.00 (referent) (N = 19) |
2.29 (1.27–4.12) p = 0.006 (N = 28) |
1.00 (referent) (N = 13) |
2.30 (1.10–4.80) p = 0.03 (N = 16) |
1.00 (referent) (N = 6) |
2.81 (1.02–7.76) p = 0.05 (N = 12) |
| 2–3 High risk GST allelesa |
1.20 (0.58–2.49) p = 0.62 (N = 12) |
3.00 (1.55–5.78) p = 0.001 (N = 17) |
0.90 (0.34–2.40) p = 0.83 (N = 6) |
2.29 (1.02–5.13) p = 0.05 (N = 11) |
1.87 (0.59–5.87) p = 0.29 (N = 6) |
5.14 (1.65–16.07) p = 0.005 (N = 6) |
High aggressive = High stage or high grade; Low aggressive = Low stage or low grade; Hazard ratio and 95% confidence interval adjusted for age, race, smoking and PSA at diagnosis
aThe number of high risk alleles refers to the number of GST (GSTM1 null, GSTT1 null and GSTP1 105 Ile/Val or Val/Val) variants carried
bNumber of biochemical failure events (N) in each stratum
In a subset of African-American cases for which we also had ancestry informative markers (N = 146), we additionally adjusted all models for African ancestry scores (see “Methods: Statistical Analysis”) and observed no material differences (data not shown). Furthermore, because we previously observed that PAH-DNA adducts in prostate cells were associated with an increased risk of prostate cancer BCR [21], we also performed all of the aforementioned analyses with additional adjustment for PAH-DNA adduct levels in a subset of patients with PAH-DNA adduct data also available (N = 368); and we observed no material differences in any of the results presented (data not shown).
Discussion
We found that the GSTT1 null and GSTP1 105 Ile/Val and Val/Val variant genotypes increased risk of prostate cancer BCR in African-Americans but not Caucasians with high grade and high stage tumors; and the GSTM1 null variant genotype increased risk of BCR in Caucasians but not African-Americans with high grade and high stage tumors. Effect sizes (HRs) in African-Americans were markedly smaller for the GSTP1 polymorphism (3.68 and 3.94 for high grade and high stage tumors, respectively) compared to those observed for the GSTT1 null polymorphism (7.82 and 6.20 for high grade and high stage tumors, respectively).
In the only prior report examining the potential association between GST polymorphisms and prostate cancer biochemical recurrence, Agalliu et al. [14] found that in Caucasian men the GSTM1 null genotype increased the risk prostate cancer recurrence by 56%, but this was not statistically significant. Our findings are consistent with these prior results in that we observed a modest (61%) but non-statistically significant increased risk of BCR with the GSTM1 null genotype in our Caucasian patient population. However, we did observe a significant increased risk with the GSTM1 null genotype in Caucasian patients with high grade and advanced (high) stage disease. Agalliu et al. [14] also reported that neither the GSTT1 null or GSTP1 Ile105Val polymorphisms were associated with recurrence or mortality, which is consistent with our findings among Caucasians but not in African-Americans, where we observed a significant association between the GSTT1 null genotype and BCR, which was markedly increased among African-Americans with high grade and advanced stage disease. Importantly, their study population comprised only a small percentage of patients with high grade tumors (total Gleason score of eight or greater), which may help explain the differences in our results. Furthermore, the Agalliu et al. population was comprised predominantly of Caucasians (95%), which limited their ability to potentially detect the differences we observed when stratifying by race and tumor grade, particularly since African-American men generally present with more advanced disease [2] and have higher rates of recurrence than Caucasian men [6]. Moreover, our study population was restricted to those who treated with radical prostatectomy alone, whereas the Agalliu et al. [14] study population included men who underwent androgen deprivation therapy (ADT) (8.4% with BCR, 2.5% without BCR), ADT and radiation (21.0% with BCR, 16.7% without BCR), and a few men who did not undergo any treatment (3.5% with BCR, 6.2% without BCR) for prostate cancer. Differences in treatment regimes could potentially affect the association between GST polymorphisms and biochemical recurrence due to the numerous functions of GSTs including metabolism of steroid hormones and detoxification of carcinogenic metabolites [7]. When Agalliu et al. [14] restricted their analysis to only those treated with radical prostatectomy, they found an even higher risk of recurrence with the GSTM1 null polymorphism, but this result was not statistically significant.
Although a recent meta-analysis suggests polymorphisms in GSTM1 null, GSTT1 null and GSTP1 Ile105Val are unlikely to be major determinants of prostate cancer incidence [12], our results suggest that GST polymorphisms may play a greater role in recurrence of advanced prostate cancer and effects may differ by race. Specifically, we found that the GSTT1 null and GSTP1 Ile105Val polymorphisms were associated with increased BCR in African-American men but not Caucasian men with high grade and high stage tumors and that the GSTM1 null polymorphism was associated with BCR in Caucasian men but not African-American men with high grade and high stage tumors. Although the total number of high risk (variant) GST genotypes increased BCR risk almost twofold in African-Americans, we did not observe a significant effect with having increasing numbers of GST variants in Caucasians. We hypothesize that the potential role of the GSTP1 polymorphism is complicated by the consistent finding that the GSTP1 promoter region CpG islands are hypermethylated, leading to inactivation of GSTP1 in prostate tumor cells [22], which may serve as an indicator of biochemical recurrence after radical prostatectomy [23]. Perhaps, the silencing of GSTP1 in prostate cancer speaks to an even greater role for the GSTM1 null and GSTT1 null polymorphisms in prostate cancer progression, since the partial or complete lack of GSTP1 function would require that GSTM1 or GSTT1 serve as substitutes where there is overlap in their substrate specificities [7]. Although, why GSTM1 might be more biologically important in Caucasians and GSTT1 in African-Americans is unclear. Differences in patterns of exposure may play a role. For example, we [13] and others [24] have observed interactions between the GSTM1 null polymorphism and smoking in Caucasians, but larger studies are needed to better understanding of these and other possible gene–environment interactions.
Strengths of our study include the use of patients who only underwent surgery (radical prostatectomy) for treatment, which prevented confounding by treatment and enabled a uniform definition of disease recurrence. Our study population also had a fairly large representation of African-American patients (42.6%). However, our sample size diminished considerably after further stratifying by genotype and clinical measures. Furthermore, due to the exploratory nature of our analyses, we did not correct for multiple statistical tests. Therefore, our results should be interpreted with caution and require validation in a larger independent study population.
In summary, we found that the GSTT1 null genotype increased risk of BCR in African-American men with prostate cancer, particularly those patients with high grade and high stage tumors. In Caucasians with high grade and high stage tumors, the GSTM1 null genotype increased risk of BCR. Overall, the GST polymorphisms were more strongly associated with BCR risk in African-Americans, suggesting GST-targeted therapeutics may have their greatest impact on treatment of advanced prostate cancers in African-Americans.
Acknowledgments
This work was supported, in part, by the following grants: NIEHS R01-ES011126, NCI K07-CA129162 and DOD W81XWH-06-1-0181.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Contributor Information
Nora L. Nock, Email: nln@case.edu
Benjamin A. Rybicki, Phone: +1-313-8746399, FAX: +1-313-8746730, Email: brybick1@hfhs.org
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Thompson I, Tangen C, Tolcher A, Crawford E, Eisenberger M, Moinpour C. Association of African-American ethnic background with survival in men with metastatic prostate cancer. J Natl Cancer Inst. 2001;93:219–225. doi: 10.1093/jnci/93.3.219. [DOI] [PubMed] [Google Scholar]
- 3.Grossfeld GD, Latini DM, Lubeck DP, Mehta SS, Carroll PR. Predicting recurrence after radical prostatectomy for patients with high risk prostate cancer. J Urol. 2003;169:157–163. doi: 10.1016/S0022-5347(05)64058-X. [DOI] [PubMed] [Google Scholar]
- 4.D’Amico AV, Cote K, Loffredo M, Renshaw AA, Chen MH. Pretreatment predictors of time to cancer specific death after prostate specific antigen failure. J Urol. 2003;169:1320–1324. doi: 10.1097/01.ju.0000049200.30192.d1. [DOI] [PubMed] [Google Scholar]
- 5.Ercole B, Marietti SR, Fine J, Albertsen PC. Outcomes following active surveillance of men with localized prostate cancer diagnosed in the prostate specific antigen era. J Urol. 2008;180:1336–1339. doi: 10.1016/j.juro.2008.06.027. [DOI] [PubMed] [Google Scholar]
- 6.Wood HM, Reuther AM, Gilligan TD, Kupelian PA, Modlin CS, Jr, Klein EA. Rates of biochemical remission remain higher in black men compared to white men after radical prostatectomy despite similar trends in prostate specific antigen induced stage migration. J Urol. 2007;178:1271–1276. doi: 10.1016/j.juro.2007.05.127. [DOI] [PubMed] [Google Scholar]
- 7.Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 8.Flowers L, Bleczinski WF, Burczynski ME, Harvey RG, Penning TM. Disposition and biological activity of benzo[a]pyrene-7, 8-dione. A genotoxic metabolite generated by dihydrodiol dehydrogenase. Biochemistry. 1996;35:13664–13672. doi: 10.1021/bi961077w. [DOI] [PubMed] [Google Scholar]
- 9.Yager JD, Liehr JG. Molecular mechanisms of estrogen carcinogenesis. Annu Rev Pharmacol Toxicol. 1996;36:203–232. doi: 10.1146/annurev.pa.36.040196.001223. [DOI] [PubMed] [Google Scholar]
- 10.Di Paolo OA, Teitel CH, Nowell S, Coles BF, Kadlubar FF. Expression of cytochromes P450 and glutathione S-transferases in human prostate, and the potential for activation of heterocyclic amine carcinogens via acetyl-coA-, PAPS- and ATP-dependent pathways. Int J Cancer. 2005;117:8–13. doi: 10.1002/ijc.21152. [DOI] [PubMed] [Google Scholar]
- 11.Elek J, Park KH, Narayanan R. Microarray-based expression profiling in prostate tumors. In Vivo. 2000;14:173–182. [PubMed] [Google Scholar]
- 12.Ntais C, Polycarpou A, Ioannidis JP. Association of GSTM1, GSTT1, and GSTP1 gene polymorphisms with the risk of prostate cancer: a meta-analysis. Cancer Epidemiol Biomarkers Prev. 2005;14:176–181. [PubMed] [Google Scholar]
- 13.Nock NL, Tang D, Rundle A, et al. Associations between smoking, polymorphisms in polycyclic aromatic hydrocarbon (PAH) metabolism and conjugation genes and PAH-DNA adducts in prostate tumors differ by race. Cancer Epidemiol Biomarkers Prev. 2007;16:1236–1245. doi: 10.1158/1055-9965.EPI-06-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agalliu I, Lin DW, Salinas CA, Feng Z, Stanford JL. Polymorphisms in the glutathione S-transferase M1, T1, and P1 genes and prostate cancer prognosis. Prostate. 2006;66:1535–1541. doi: 10.1002/pros.20491. [DOI] [PubMed] [Google Scholar]
- 15.Rybicki BA, Nesland-Dudas C, Nock NL, et al. Prostate cancer risk from occupational exposure to polycyclic aromatic hydrocarbons interacting with the GSTP1 Ile105Val polymorphism. Cancer Detect Prev. 2006;30:412–422. doi: 10.1016/j.cdp.2006.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arand M, Muhlbauer R, Hengstler J, Jager E, Fuchs J, Winkler L. A multiplex polymerase chain reaction protocol for the simultaneous analysis of the glutathione S-transferase GSTM1 and GSTT1 polymorphisms. Anal Biochem. 1996;236:184–186. doi: 10.1006/abio.1996.0153. [DOI] [PubMed] [Google Scholar]
- 17.Kim JW, Lee CG, Park YG, et al. Combined analysis of germline polymorphisms of p53, GSTM1, GSTT1, CYP1A1, and CYP2E1: relation to the incidence rate of cervical carcinoma. Cancer. 2000;88:2082–2091. doi: 10.1002/(SICI)1097-0142(20000501)88:9<2082::AID-CNCR14>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 18.Freedland SJ, Sutter ME, Dorey F, Aronson WJ. Defining the ideal cutpoint for determining PSA recurrence after radical prostatectomy. Prostate-specific antigen. Urology. 2003;61:365–369. doi: 10.1016/S0090-4295(02)02268-9. [DOI] [PubMed] [Google Scholar]
- 19.Kupelian PA, Elshaikh M, Reddy CA, Zippe C, Klein EA. Factors affecting recurrence rates after prostatectomy or radiotherapy in localized prostate carcinoma patients with biopsy Gleason score 8 or above. Cancer. 2002;95:2302–2307. doi: 10.1002/cncr.10977. [DOI] [PubMed] [Google Scholar]
- 20.Hoggart CJ, Parra EJ, Shriver MD, et al. Control of confounding of genetic associations in stratified populations. Am J Hum Genet. 2003;72:1492–1504. doi: 10.1086/375613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rybicki BA, Neslund-Dudas C, Bock CH, et al. Polycyclic aromatic hydrocarbon–DNA adducts in prostate and biochemical recurrence after prostatectomy. Clin Cancer Res. 2008;14:750–757. doi: 10.1158/1078-0432.CCR-07-0986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meiers I, Shanks JH, Bostwick DG. Glutathione S-transferase pi (GSTP1) hypermethylation in prostate cancer: review 2007. Pathology. 2007;39:299–304. doi: 10.1080/00313020701329906. [DOI] [PubMed] [Google Scholar]
- 23.Ellinger J, Bastian PJ, Jurgan T, et al. CpG island hypermethylation at multiple gene sites in diagnosis and prognosis of prostate cancer. Urology. 2008;71:161–167. doi: 10.1016/j.urology.2007.09.056. [DOI] [PubMed] [Google Scholar]
- 24.Agalliu I, Langeberg WJ, Lampe JW, Salinas CA, Stanford JL. Glutathione S-transferase M1, T1, and P1 polymorphisms and prostate cancer risk in middle-aged men. Prostate. 2006;66:146–166. doi: 10.1002/pros.20305. [DOI] [PubMed] [Google Scholar]