Abstract
The Enterococcus faecalis conjugative plasmid pAD1 (60 kb) encodes a mating response to the recipient-produced peptide sex pheromone cAD1. The response involves two key plasmid-encoded regulatory proteins: TraE1, which positively regulates all or most structural genes relating to conjugation, and TraA, which binds DNA and negatively regulates expression of traE1. In vitro studies that included development of a DNA-associated protein-tag affinity chromatography technique showed that TraA (37.9 kDa) binds directly to cAD1 near its carboxyl-terminal end and, as a consequence, loses its affinity for DNA. Analyses of genetically modified TraA proteins indicated that truncations within the carboxyl-terminal 9 residues significantly affected the specificity of peptide-directed association/dissociation of DNA. The data support earlier observations that transposon insertions near the 3′ end of traA eliminated the ability of cells to respond to cAD1.
Enterococcus faecalis and related species (e.g., Enterococcus faecium) are Gram-positive bacteria that inhabit the human intestine and are commonly associated with urinary tract infections, bacteremia, and endocarditis. Enterococci are notorious for their resistance to multiple antibiotics, and the recent emergence of strains resistant to the “last-resort” antibiotic, vancomycin, has generated major concern among clinicians. Conjugation systems involving plasmids and transposons are abundant in these organisms and contribute to the dissemination of both antibiotic resistance and virulence factors (1–3).
A significant percentage of clinical isolates of E. faecalis are hemolytic, a trait that often is associated with an extrachromosomal hemolysin/bacteriocin (cytolysin) element similar to the 60-kb conjugative pAD1 plasmid (4) originally identified in E. faecalis DS16 (5, 6) and found to contribute to virulence in animal models (7, 8). pAD1 encodes a mating response to a peptide sex pheromone (cAD1) produced by plasmid-free strains (9). Donor strains exposed to the pheromone are induced to synthesize a protein adhesin designated “aggregation substance,” which facilitates the initiation of mating-pair formation with recipient cells (for reviews see refs. 10–12); control of this physiological response, however, can be overridden by a reversible phase variation event that can independently switch on all conjugation functions (13, 14). Interestingly, aggregation substance has two RGD motifs (15) that may be responsible for the ability of plasmid-carrying cells to attach to porcine renal kidney epithelial cells (16).
The structures of several different sex pheromones have been determined and correspond to hydrophobic octa- or heptapeptides (see ref. 17); all can be secreted by a single E. faecalis strain, although the presence of a given plasmid in the organism results in “shut-down” of the related pheromone (18). The individual pheromones are quite specific for donors carrying the appropriate plasmid and are active at concentrations as low as 5 × 10−11 M. pAD1 encodes an octapeptide (iAD1) that is secreted and acts as a competitive inhibitor of cAD1 (19, 20); the two peptides share identity at 4 of their 8 aa residues. Whereas a plasmid determinant traB is involved in shutdown of endogenous pheromone (21), iAD1 (determined by iad) serves to prevent self-induction by small amounts of cAD1 still secreted by donor cells (18).
TraC is a plasmid-encoded protein located on the donor cell surface; it increases sensitivity to the exogenous pheromone and is believed to pass the peptide to a less specific, host-encoded peptide transport system (22, 23). TraC has significant similarity with known oligopeptide-binding proteins associated with ABC transport systems in Bacillus subtilis, Escherichia coli, and Salmonella typhimurium. It is also homologous with prgZ carried by the enterococcal pCF10 plasmid, which encodes a response to the pheromone cCF10 (24); PrgZ has been reported to act as a component of an E. faecalis host oligopeptide uptake system (23).
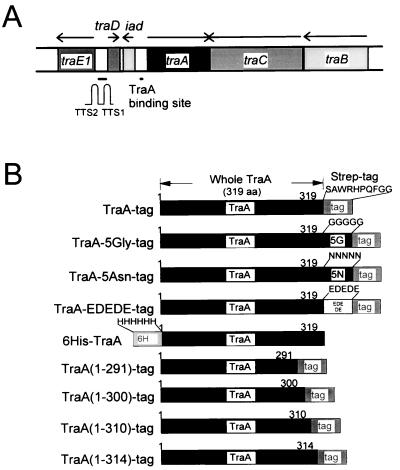
A map showing the locations of important pAD1 regulatory determinants is shown in Fig. 1A. traA, whose 5′ end is adjacent to the 5′ end of iad, and which is divergently transcribed, encodes a key negative regulator (25). Mutations in traA can give rise to constitutive expression of conjugation functions; however, certain mutants are not derepressed, or are only partially derepressed, and in addition exhibit a loss in sensitivity to pheromone (26–28). TraA controls the expression of traE1, whose product is a positive regulator involved in activation of structural genes necessary for conjugation (29, 30). TraA (37.9 kDa) has been shown to bind at the iad promoter (31), where it affects transcription (already occurring at a relatively high basal level) such that there is an elevation of transcripts whose lengths are extended to some extent through the downstream terminators TTS1 and TTS2. TraE1 synthesis is believed to be induced initially via transcriptional read-through of these terminators resulting in production of a minimal amount of TraE1, which can, in turn, activate its own expression from its own promoter, part of which appears to be located within TTS2 (29, 31, 32). traD encodes a putative peptide predicted to consist of 23 aa residues and is transcribed at a high level in the uninduced, but not induced, state (33).
Figure 1.
Map of pAD1 pheromone response regulatory region. (A) Relative locations and transcriptional orientations (arrows) of specific determinants. TTS1 and TTS2 are two transcription terminators located just upstream of traE1. (B) Structure of TraA-related fusion derivatives with modifications at the carboxyl terminus.
In this communication we report that triggering of the induction process involves a direct interaction between the TraA protein and the cAD1 peptide and that this causes TraA to release its binding to DNA. We also show that the carboxyl-terminal end of TraA plays an important role in pheromone specificity and in differentiation between the effects of cAD1, iAD1, and other peptide pheromones.
MATERIALS AND METHODS
Plasmids, Strains, and Cloning Procedures.
The plasmid pASK60-Strep (34) and E. coli strain JM83 (F−araD[lac-proAB]rpsL[Strr]f80dlacD[lacZ]M15, kindly provided by A. Telesnitsky, University of Michigan), were used in all cloning procedures. pAM7500 represents the pAD1 HindIII C fragment (contains traE1, traD, iad, traA, traC, and a portion of traB) cloned in pBluescript SK (25); this plasmid was used as the template in the generation of the various PCR products by using the specific primers described below. The addition of six histidines on the amino terminus or five amino acids to the carboxyl terminus was done by adding appropriate nucleotides to the primers. The derivative without a Strep-tag (i.e., 6His-TraA) was generated by introducing a stop codon. Primers included NruI or EagI sites at their 5′ ends. Amplified DNA fragments were digested with NruI and EagI and ligated with Eco47III-BsaI-digested pASK60-Strep. E. coli JM83 cells were transformed by electrotransformation (35).
Primers.
PCR amplifications were performed with primers noted below. For TraA-tag, TraA(1–291)-tag, TraA(1–300)-tag, TraA(1–310)-tag, and TraA(1–314)-tag, a “forward” primer, PN02 (5′-TTTAACGGCCGGCATGTTTCTTTACGAACT-3′), and reverse primers PR01 (5′-ATTTTTCGCGACTCTAGTCTTTTGGTTATT-3′), PR-291 (5′-ATTTTTCGCGAACCATTATTTTTTTTTGAG-3′), PR-310 (5′-ATTTTTCGCGAGTTTAGTTTTATATGATTA-3′), or PR-314 (5′-ATTTTTCGCGATATTTTCTTTTCGTTTAGTTT-3′) were used, respectively. For 6His-TraA, a forward primer, PN-6His (5′-AAAAACGGCCGGCCACCATCACCATCACCATATGTTTCTTTACGAACT-3′), and a reverse primer, PR-Stop (5′-ATTTTTCGCGAATTATTACTCTAGTCTTTTGGTTA-3′), were used. For TraA-5Gly, TraA-5Asn, and TraA-EDEDE, the forward primer PN02 and reverse primers PR-5Gly (5′-ATTTTTCGCGACCCCCCACCTCCCCCCTCTAGTCTTTTGGTTATTT-3′), PR-5Asn (5′-ATTTTTCGCGAGTTGTTATTGTTGTTCTCTAGTCTTTTGGTTATTT-3′), or PR-EDEDE (5′-ATTTTTCGCGACTCGTCTTCATCTTCCTCTAGTCTTTTGGTTATTT-3′) were used.
Expression and Purification of Proteins.
Protein extracts from bacterial strains carrying specific recombinant plasmids were prepared as follows. Overnight cultures grown at 30°C in Luria–Bertani broth (36) containing ampicillin (100 μg/ml) were diluted 1:50 into 100 ml of the same medium. Cells were grown at 30°C to an A600 of 0.5 and then exposed to 1 mM IPTG for an additional 4 hr. After harvesting by centrifugation and resuspending in 500 μl of buffer (150 mM NaCl/100 mM Tris, pH 8.0/10 mM MgCl2) containing glycerol (10%), benzamidine (1 mM), and/or DTT (1 mM), the cells were disrupted by sonication (approximately 15 W, 1 sec × 12 times with 29-sec intervals). After centrifugation at 14,000 rpm (Brinkmann–Eppendorf Micro Centrifuge model 5415-C) for 30 min, the supernatant was recovered. DNase I (10 μl; 10 mg/ml) and RNase (10 μl; 10 mg/ml) were added, and the sample was incubated on ice for 30 min; EDTA (25 μl; 100 mM) then was added. A streptavidin affinity matrix (Sigma) was utilized as a column by placing 1 ml of the matrix (settled matrix purchased as a 50% vol/vol slurry) into a Bio-Rad Econo-Pac column. The matrix was washed with 10 ml of washing buffer (100 mM Tris, pH 8.0/1 mM EDTA/0.005% Triton X-100/1 mM benzamidine/1 mM DTT/150 mM NaCl). One milliliter of protein extract was applied to the column followed by 20 ml of washing buffer. Proteins were eluted with 5 mM diaminobiotin in the washing buffer as five 1-ml fractions.
cAD1 and iAD1 Affinity Chromatography.
Amino terminus-fixed cAD1 or iAD1 matrix preparations (10-atom spacer arm) were generated by using activated affinity support Affigel-10 (Bio-Rad) as follows. Approximately 3 ml of Affigel-10 slurry was washed four times with 10 ml dry DMSO. Two milligrams of synthetic cAD1 or iAD1 were dissolved in 4 ml dry DMSO and 20 μl triethylamine and incubated at room temperature overnight while being slowly rotated. Ethanolamine (100 μl) then was added and the mixture was incubated 2 hr to fill up unbound activated residues. The matrix then was washed four times with 10 ml DMSO, four times with 10 ml 1 N acetic acid, and finally three times with 10 ml distilled water. The matrix then was resuspended in TSAGE buffer (100 mM Tris, pH 8.0/150 mM NaCl/0.02% sodium azide/10% glycerol/1 mM EDTA).
Carboxyl terminus-fixed cAD1 matrix (6-atom spacer arm) was generated by using Affigel-102 (Bio-Rad) as follows. One milliliter of Affigel-102 (purchased as 50% slurry) was washed and resuspended in 1 ml distilled water. cAD1 (50 μg in 50 μl DMSO) was added to the matrix. The pH was adjusted to 4.7 with 1 M HCl. Ten milligrams of 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide HCl (EDAC; Bio-Rad) was added and the pH was again adjusted to 4.7. The solution then was incubated at room temperature for 8 hr while being slowly rotated. The matrix was washed three times with 10 ml of 1 N acetic acid, four times with 10 ml DMSO, three times with 10 ml distilled water, and finally three times with 10 ml TSAGE buffer. A control matrix was prepared in an identical manner but without cAD1.
Binding of cAD1 to the matrix was confirmed by removing it from the matrix with its fixed spacer by boiling 20 μl of the matrix in 120 μl N2GT for 5 min. After microfuge centrifugation at 14,000 rpm for 1 min, pheromone activity in the supernatant was quantitated by using the microtiter assay (37) and found to have a titer of 256–512 and 64–128 for amino terminus-fixed matrix and carboxyl terminus-fixed matrix, respectively. The pheromone concentration on matrices was estimated to be approximately 2 μg/ml and 1 μg/ml, respectively, on the assumption that the peptide activity is not affected by the covalently attached spacer arm. The control matrix or the sample prepared without boiling did not show any pheromone activity.
cAD1 and iAD1 chromatography were done by using a batch method. A 20-μl portion of the matrix was mixed with 20 μl protein extract or 100 μl purified protein and incubated on ice for 2–5 hr. The matrix was washed three times with 1 ml of 5% DMSO in TSAGE buffer. Elution was with 30 μl of cAD1 or other peptides (50 μg/ml; approximately 60 μM) one to four times. Equivalent volumes of buffer did not release any protein.
DNA-Associated Protein-Tag Affinity Chromatography (DPAC).
In the DPAC procedure, TSAGE with 0.005% Triton X-100, 1 mM benzamidine, and 1 mM DTT was used as the buffer for experiments with the streptavidin matrix. When the Ni-NTA matrix (Qiagen) was used, the buffer was 150 mM NaCl in 50 mM Tris (pH 8.0). Protein extracts were obtained the identical way as described above for protein purification except EDTA was eliminated in the case of the Ni-NTA matrix. One milliliter of buffer was used to wash 20 μl of matrix in a microfuge tube. Then 50 μl of extract was mixed with the matrix and incubated on ice 30 min with occasional mixing. The matrix was washed with 1 ml of the buffer four times. When the effect of pheromones or pheromone-related peptides were tested, the matrix was mixed with 1 ml of 5 μg/ml peptide solution in the buffer and incubated on ice overnight. (The concentration of TraA bound in the matrix was estimated to be about 50 μg/ml or about 1.3 μM, and because the peptide concentration used was 6 μM, the ratio of peptide molecules to protein molecules was about 5:1.) After centrifugation, the supernatant was removed. Restriction enzyme-digested DNA was extracted with phenol/chloroform and precipitated with ethanol. Four microliters corresponding to approximately 2 μg of cleaved DNA in 10 mM Tris (pH 8.0) was added to the matrix and incubated on ice for 2 hr with occasional mixing. The matrix was washed four times with 1 ml of buffer. The protein–DNA complex was eluted with 5 mM diaminobiotin in the buffer in the case of the streptavidin matrix or 100 mM EDTA in the buffer for the Ni-NTA matrix. A part of the eluted solution was subjected to SDS/PAGE in some cases. Size estimates were based on Bio-Rad “Low Range” size markers. The remainder of the sample was extracted with phenol chloroform to avoid the gel-retardation effect and examined by agarose gel electrophoresis. Gels were stained with ethidium bromide.
Batch Method Affinity Chromatography.
This was used for generation of the data in Fig. 3B (streptavidin matrix); it was essentially identical to the method described for DPAC but without mixing with DNA. (In the case of the data relating to Fig. 2A and purified TraA used for Fig. 2B, a scaled-up 1-ml column was used as described above.)
Figure 3.
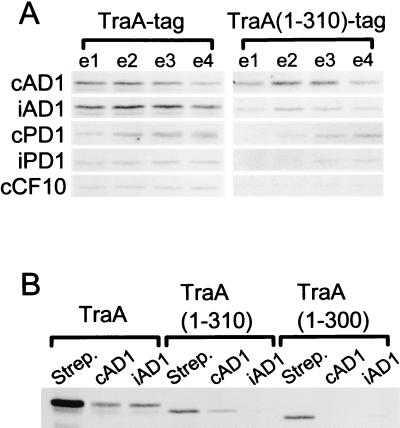
Pheromone specificity of TraA and TraA-related proteins. The proteins were examined by SDS/PAGE and stained with silver stain. (A) Elution of amino terminus-fixed cAD1 affinity matrix-bound TraA-tag or TraA(1–310)-tag with various peptides. Protein extracts were mixed with the cAD1 matrix and, after washing, were eluted with 60 μM cAD1, iAD1, cPD1, iPD1, or cCF10. Each fraction (e1, e2, e3, and e4) represents 30 μl of eluate. (B) Amino terminus-fixed cAD1 and iAD1 affinity chromatography of TraA-tag, TraA(1–310)-tag, and TraA(1–300)-tag. Protein was eluted with 30 μl of 60 μM cAD1 or iAD1 in TSAGE buffer. In the case where protein was bound to the streptavidin matrix, elution was with 5 mM diaminobiotin.
Figure 2.
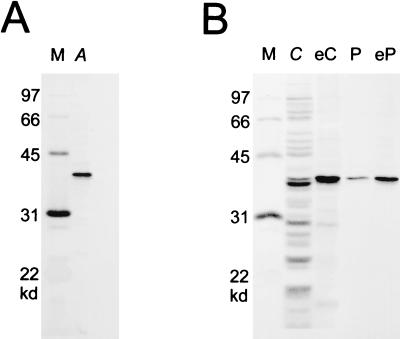
Purification from E. coli of TraA-tag and cAD1 affinity chromatography. The proteins were examined by SDS/PAGE and stained with silver stain. (A) Purified TraA-tag (lane A) with molecular size markers (lane M). (B) Amino terminus-fixed cAD1 affinity chromatography of TraA-tag. Crude extract with overexpressed TraA-Tag (lane C) or purified TraA-tag (lane P) was mixed with amino terminus-fixed cAD1 affinity matrix. After incubation and washing, protein was eluted with 60 μM cAD1 (30 μl). Lane eC represents eluate from the matrix that had been mixed with crude extract. Lane eP is eluate from matrix mixed with purified TraA-Tag. Molecular size markers are in lane M.
RESULTS
Purification of TraA Protein and Binding to cAD1.
TraA fused at its carboxyl terminus to a streptavidin-binding “tag” was purified from a recombinant E. coli derivative by using a streptavidin matrix as described in the Materials and Methods. As shown in Fig. 2A, a protein corresponding closely to the size of TraA (38 kDa; lane A) was eluted from the matrix with diaminobiotin.
Fig. 2B shows that when an affinity matrix, with the amino terminus of cAD1 fixed to the matrix, was prepared and utilized as described in the Materials and Methods, it was able to bind selectively to TraA-tag protein within a crude E. coli protein extract (compare lanes C and eC) as well as to protein previously isolated using the streptavidin matrix (lanes P and eP). When cAD1 was fixed to the matrix at its carboxyl terminus, TraA-tag was not observed to bind to it (not shown).
Fig. 3A shows that TraA-tag protein bound to the cAD1 affinity matrix (where cAD1 is fixed to the matrix at its amino terminus) can be eluted by other peptides, namely iAD1, cPD1, iPD1, and cCF10; however, there appear to be differences in the relative releasing strengths. The ability of the peptides to elute cAD1-bound protein is: cAD1 ≅ iAD1 > cPD1 > iPD1 ≥ cCF10. cPD1 and iPD1 are the pheromone and related inhibitor, respectively, for the pPD1 system; cCF10 is the pheromone responded to by pCF10-bearing bacteria (see ref. 17).
Peptide Modulation of TraA Binding to DNA.
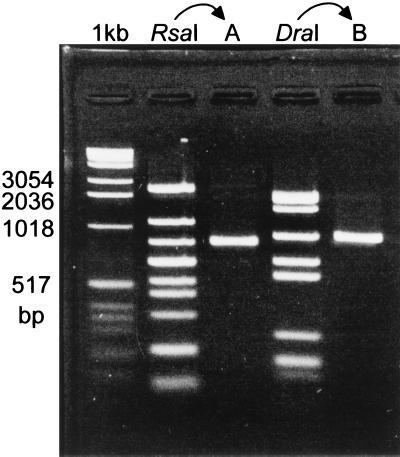
As noted earlier, TraA has been shown to bind to the promoter site of iad (31). To determine whether TraA-tag protein could bind to DNA and the streptavidin matrix simultaneously, the protein first was mixed with the matrix and the matrix–protein complex then was mixed with the chimeric plasmid pAM7500 (contains most of region shown in Fig. 1A; see Materials and Methods) that previously had been treated with the restriction enzyme RsaI or DraI. After washing the matrix and then exposing it to diaminobiotin, the effluent was extracted with phenol-chloroform and analyzed by agarose gel electrophoresis. Fig. 4 shows that in the case of both RsaI- and DraI-treated DNA a single restriction fragment was found to have associated specifically with the matrix complex. The 924-bp DraI fragment and the 839-bp RsaI fragment overlap by 268 bp, identifying a region previously shown to contain a TraA-binding site (31). The method for demonstrating DNA–protein binding subsequently will be referred to as DPAC.
Figure 4.
DNA binding of TraA-tag using the DPAC method. Extracts containing overexpressed TraA-tag and RsaI- or DraI-digested pAM7500 were used in the DPAC procedure as described in the Materials and Methods. Eluted material was extracted with phenol-chloroform, submitted to agarose gel electrophoresis, and stained with ethidium bromide. The restriction fragment “held” to the matrix in the case of RsaI is seen in lane A; the fragment held in the case of DraI is seen in lane B. The lane labeled 1 kb represents marker DNA (1-kb ladder).
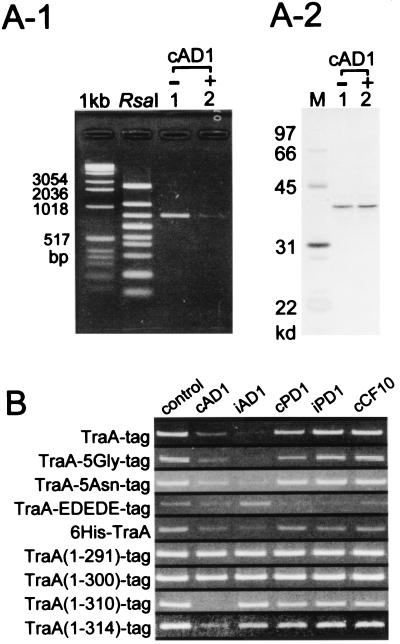
Using the DPAC method we then determined whether cAD1 had any effect on the ability of TraA to bind DNA. As shown in Fig. 5A-1, the exposure of matrix-bound protein to cAD1 considerably reduced the amount of DNA able to bind to the matrix complex. A portion of each sample also was run on an SDS/PAGE gel without phenol-chloroform extraction and then silver-stained; this showed (Fig. 5A-2) that the amount of TraA-tag that had been on the matrix essentially was the same for both the cAD1-exposed and unexposed preparations. The data provide direct evidence that cAD1 can modify the ability of TraA to bind to DNA.
Figure 5.
Modulation of DNA-binding affinity of TraA and TraA-related proteins by pheromone and other peptides. (A) Effect of exposure to cAD1 on the DNA binding of TraA using the DPAC method. After binding the protein to the streptavidin matrix and washing, the bound material was exposed to either 6 μM cAD1 or solvent (0.5% DMSO). RsaI-digested pAM7500 then was added to the matrix complex. After elution with diaminobiotin 25-μl fractions were obtained from which 20 μl was extracted with phenol-chloroform and analyzed by agarose gel electrophoresis (A-1). A 5-μl portion of the eluate was run directly on SDS/PAGE and silver-stained (A-2). (B) Effect of pheromone and other peptides on the DNA binding of TraA and TraA-related proteins. In the case of 6His-TraA, a Ni-NTA matrix was used (see Materials and Methods); all others made use of the streptavidin matrix. Peptide concentrations were all at 6 μM. The bands shown all correspond to the single RsaI band shown above to bind to TraA.
Other peptides similarly were tested for their effects on TraA binding to DNA. As shown in Fig. 5B, iAD1 inhibited DNA binding even stronger than cAD1. iPD1 and cCF10 had no effect, whereas cPD1 appeared to have only a slight effect. Thus, the peptide-directed reduction of TraA-DNA affinity was somewhat specific for cAD1 and iAD1. To see whether perhaps the relative differences between cAD1 and iAD1 were different at lower concentrations, more dilute solutions of each were examined. A comparison of concentrations of 5.0, 2.5, 1.0, 0.5, 0.25, 0.1, 0.05, and 0.025 μg/ml of each showed that the ability to cause release of DNA diluted out similarly. At 0.25 μg/ml, neither showed any activity (data not shown).
Modifications at the Carboxyl Terminus of TraA Affect Peptide Specificity.
A series of derivatives with modifications at the carboxyl terminus was constructed as illustrated in Fig. 1B. The addition of five glycines or five asparagines did not significantly change the peptide specificity (Fig. 5B); however, the insertion of alternating glutamate and aspartate (EDEDE) changed the specificity such that cPD1, iPD1, and cCF10 had some inhibitory effect on DNA binding whereas iAD1 actually may enhance binding. cAD1 activity did not appear to be affected. To determine whether the Strep-tag itself affected peptide specificity, we constructed a chimera that yielded six histidines on the amino terminus with no tag on the carboxyl terminus (6His-TraA). In this case, making use of a nickel-nitrilotriacetic acid (Ni-NTA) matrix, the specificity was observed to be similar to that of TraA-tag (Fig. 5B). Thus, the Strep-tag does not appear to affect the peptide-binding properties of TraA.
TraA contains 319 aa, whereas the tag corresponds to an additional 10 residues. Derivatives that had 5–28 aa residues removed from the carboxyl terminus of TraA were constructed (see Fig. 1B) and examined by DPAC. As shown in Fig. 5B, removal of 19 or 28 residues [TraA(1–300)-tag or TraA(1–291)-tag, respectively] resulted in an inability of any of the peptides to inhibit binding of DNA to the protein. The removal of 9 residues [TraA(1–310)-tag] resulted in strong specificity for cAD1 inhibition of DNA binding without a similar effect by iAD1. Removal of 5 residues [TraA(1–314)-tag] was similar, although cAD1 did not inhibit as strongly.
Examination of the binding of TraA(1–310)-tag and TraA(1–300)-tag to cAD1- or iAD1-fixed matrices showed that the former bound significantly better to cAD1 than iAD1, whereas the latter showed only a slight binding to iAD1 (Fig. 3B). Consistent with this were the data of Fig. 3A, where TraA(1–310)-tag was eluted more readily from the cAD1-matrix by cAD1 than iAD1. In addition, cPD1, iPD1, and cCF10 were progressively less efficient at eluting the cAD1-bound TraA(1–310)-tag protein.
DISCUSSION
It was reported previously that although most transposon insertions in the pAD1 traA determinant resulted in constitutive expression of conjugation functions, a few [e.g., those relating to the derivatives pAM727, pAM728, and pAM2100 (26, 28)] exhibited relatively little derepression. Because the latter mutants also became insensitive to pheromone it was suggested that a site on TraA responsible for interacting with a pheromone “signal” had been removed or altered. Interestingly, all three of these insertions mapped near the 3′ end of traA (25); the one mapping farthest from the 3′ end (pAM728; ref. 25) corresponded to 29 aa residues from the carboxyl terminus of TraA. Although it appeared that the carboxyl-terminal region of TraA “received” the pheromone signal, it was not clear whether this involved a direct interaction with the peptide.
In the present study we have used a combination of biochemical and genetic techniques to show that within its carboxyl-terminal ca. 10%, TraA binds directly to cAD1. In addition, using a technique involving a protein affinity matrix to examine DNA binding (DPAC), we showed that peptide interaction can prevent the adherence of TraA to its specific binding site at the iad promoter on pAD1 DNA. We also found that a region within the carboxyl-terminal 9 residues plays an important role in the specificity of peptide recognition.
Full-length TraA fused to a carboxyl-terminal “strep-tag” could be influenced to release bound DNA by exposure to either cAD1 or iAD1 but not the peptides cPD1, iPD1, or cCF10; however, the addition of charged residues at the carboxyl terminus significantly affected the peptide specificity relating to DNA association. Importantly, the removal of 9 TraA carboxyl-terminal residues resulted in a protein that was highly specific for cAD1-directed prevention of DNA binding; iAD1 had no detectable effect (Fig. 5B). The truncated protein also showed a much stronger affinity for a cAD1-matrix compared with an iAD1-matrix. With the removal of 19 or more residues, TraA became completely insensitive to peptide-facilitated prevention of DNA binding and also lost its ability to bind to the cAD1-affinity matrix. The peptide-binding site would appear to lie within the final 10–20 carboxyl-terminal amino acid residues; whereas the site that adheres to DNA must be located further toward the amino terminus. The results are consistent with the earlier genetic data, which showed that mutants with transposon insertions near the 3′ end of traA maintained a somewhat “repressed” state and were insensitive to pheromone.
It is important to note that in the case of intact cells iAD1 is believed to act extracellularly as a competitive inhibitor of exogenous cAD1. Whereas TraB is believed to be the primary factor in “shutting down” endogenous pheromone production, self-induction by the small amount that is still secreted is prevented by an excess of extracellular iAD1 (18, 21). The plasmid-encoded iAD1 is synthesized as a 22-aa precursor resembling an N-terminal amphipathic signal peptide with the last 8 residues corresponding to iAD1 (19). The particular precursor structure may be designed to ensure that the inhibitor is efficiently secreted and therefore not functional inside the cell. iAD1 is believed to compete with cAD1 via interaction with the pheromone-binding protein TraC on the bacterial surface; there currently is no evidence that it is able to reenter the cell. The ability of iAD1 to bind to TraA and even facilitate DNA release in vitro implies that avoidance of cellular uptake of this peptide is very important. To ensure that TraA may escape interaction with iAD1 and perhaps other peptides that are being produced at maximal levels by pAD1-containing cells (e.g., cPD1 and cCF10), some processing near the carboxyl terminus yielding a truncated protein highly specific for cAD1 would seem highly advantageous. In this regard it is noteworthy that the third and fourth residues from the carboxyl terminus are arginine and lysine, respectively; and the seventh and eighth residues are both lysines (25). It is tempting to suggest that these two potential targets of trypsin-like proteases may play a role in processing TraA to facilitate peptide-binding specificity. Indeed, when E. coli lysates containing a TraA derivative with “6His” fused at the amino terminus and a “tag” at the carboxyl terminus (i.e., a “6His-TraA-tag” construct) were incubated in the absence of protease inhibitor for 5 min at 37°C, degradation products of a size consistent with loss of 5 or 9 carboxyl-terminal residues were observed to bind to a nickel matrix but not the streptavidin matrix (S.F., unpublished data).
In conclusion, we have presented evidence that, after transport into the bacterial cell, the primary target of pheromone is the pAD1-encoded TraA protein and that a conformational shift leads to induction of conjugation functions via an alteration of DNA-binding activity at the iad promoter. Accordingly, a burst of transcription follows that ultimately generates enough read-through into traE1 to allow sufficient TraE1 to be synthesized so that it can activate itself and the overall conjugation system. Although this is a significant aspect of pAD1 conjugation control, there are additional complexities that relate to the region between iad and the transcription terminators just upstream of traE1. There is evidence that a traD product plays an important role as well, and the extensive secondary structure that different transcripts in this region might assume implies that regulation probably involves a number of additional factors (31, 33). Furthermore, a burst in synthesis of the iad-encoded 22-aa precursor of iAD1 may play a feedback role extracellularly in shutting the system down after induction.
pAD1-encoded TraA exhibits strong similarity to the TraA of the E. faecalis pPD1 pheromone system (38, 39), and there is recent evidence that the pPD1-TraA binds to the related pheromone cPD1 (J. Nakayama, personal communication). Whereas the extent of information available on the regulation of the pPD1 system is still limited, there is a significant amount of data published in the case of the pCF10 system; the latter indicate some important differences from pAD1 (11, 40, 41). First, there is no pCF10 determinant that exhibits any similarity with traE1, although a few determinants (e.g., prgQ and prgS) have been tied to positive control of the mating response. traA of pAD1 appears to have some similarity with a portion of the pCF10 determinant prgX (22% identity and 45% similarity in the GenBank database), which is thought to play a negatively controlling role in the mating response (42). However, PrgX does not seem to bind to the cCF10 pheromone (23). pCF10 carries a determinant, prgW, that is significantly similar to repA of pAD1 (43); both are required for plasmid replication. Interestingly, PrgW was reported to bind to cCF10 (23), suggesting that in that system cCF10 may play a role in plasmid replication as well as in the induction of conjugation functions. In the pAD1 system there is currently no evidence for such a connection where pheromone plays a role in replication, although such a possibility thus far cannot be ruled out. An effort to see whether RepA would bind to a matrix with cAD1 fixed at its amino terminus yielded negative results, and using the DPAC technique we showed that although RepA bound specifically to a site within repA DNA, this was not affected by exposure to cAD1 (S.F., unpublished data).
Finally, we note that the DPAC technique presented here may offer advantages with respect to certain types of DNA-binding studies. It is relatively easy to perform and allows for significant flexibility in the manipulation of environmental conditions when studying the binding of specific proteins to DNA. It also provides a means for isolating specific restriction fragments containing sequences that bind to a particular protein.
Acknowledgments
We thank H. Tomita for generous assistance. This work was supported by National Institutes of Health Grant GM33956 and the General Clinical Research Center at the University of Michigan (MO1-RR00042).
ABBREVIATIONS
- DPAC
DNA-associated protein-tag affinity chromatography
- TSAGE buffer
100 mM Tris, pH 8.0/150 mM NaCl/0.02% sodium azide/10% glycerol/1 mM EDTA
References
- 1.Clewell D B. Eur J Clin Microbiol Infect Dis. 1990;9:90–102. doi: 10.1007/BF01963632. [DOI] [PubMed] [Google Scholar]
- 2.Murray B E. Clin Microbiol Rev. 1990;3:46–65. doi: 10.1128/cmr.3.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jett B D, Huycke M M, Gilmore M S. Clin Microbiol Rev. 1994;7:462–478. doi: 10.1128/cmr.7.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ike Y, Clewell D B. J Bacteriol. 1992;174:8172–8177. doi: 10.1128/jb.174.24.8172-8177.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tomich P K, An F Y, Damle S P, Clewell D B. Antimicrob Agents Chemother. 1979;15:828–830. doi: 10.1128/aac.15.6.828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clewell D B, Tomich P K, Gawron-Burke M C, Franke A E, Yagi Y, An F Y. J Bacteriol. 1982;152:1220–1230. doi: 10.1128/jb.152.3.1220-1230.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ike Y, Hashimoto H, Clewell D B. Infect Immunol. 1984;45:528–530. doi: 10.1128/iai.45.2.528-530.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stevens S X, Jensen H G, Jett B D, Gilmore M S. Invest Opthalmol Visual Sci. 1992;33:1650–1656. [PubMed] [Google Scholar]
- 9.Mori M, Sakagami Y, Narita M, Isogai A, Fujino M, Kitada C, Craig R, Clewell D, Suzuki A. FEBS Lett. 1984;178:97–100. doi: 10.1016/0014-5793(84)81248-x. [DOI] [PubMed] [Google Scholar]
- 10.Clewell D B. Cell. 1993;73:9–12. doi: 10.1016/0092-8674(93)90153-h. [DOI] [PubMed] [Google Scholar]
- 11.Dunny G M, Leonard B A B, Hedberg P J. J Bacteriol. 1995;177:871–876. doi: 10.1128/jb.177.4.871-876.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wirth R. Eur J Biochem. 1994;222:235–246. doi: 10.1111/j.1432-1033.1994.tb18862.x. [DOI] [PubMed] [Google Scholar]
- 13.Pontius L T, Clewell D B. Plasmid. 1991;26:172–185. doi: 10.1016/0147-619x(91)90041-t. [DOI] [PubMed] [Google Scholar]
- 14.Heath D G, An F Y, Weaver K W, Clewell D B. J Bacteriol. 1995;177:5453–5459. doi: 10.1128/jb.177.19.5453-5459.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galli D, Lottspeich F, Wirth R. Mol Microbiol. 1990;4:895–904. doi: 10.1111/j.1365-2958.1990.tb00662.x. [DOI] [PubMed] [Google Scholar]
- 16.Kreft B, Marre R, Schramm U, Wirth R. Infect Immunol. 1992;60:25–30. doi: 10.1128/iai.60.1.25-30.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clewell D B. In: Bacterial Conjugation. Clewell D B, editor. New York: Plenum; 1993. pp. 349–367. [Google Scholar]
- 18.Nakayama J, Dunny G M, Clewell D B, Suzuki A. In: Genetics of Streptococci, Enterococci and Lactococci. Ferretti J J, Gilmore M S, Klaenhammer T R, Brown F, editors. Vol. 85. Basel: Karger; 1995. pp. 35–38. [Google Scholar]
- 19.Clewell D B, Pontius L T, An F Y, Ike Y, Suzuki A, Nakayama J. Plasmid. 1990;24:156–161. doi: 10.1016/0147-619x(90)90019-9. [DOI] [PubMed] [Google Scholar]
- 20.Mori M, Isogai A, Sakagami Y, Fujino M, Kitada C, Clewell D B, Suzuki A. Agric Biol Chem. 1986;50:539–541. [Google Scholar]
- 21.An F Y, Clewell D B. Plasmid. 1994;31:215–221. doi: 10.1006/plas.1994.1023. [DOI] [PubMed] [Google Scholar]
- 22.Tanimoto K, An F Y, Clewell D B. J Bacteriol. 1993;175:5260–5264. doi: 10.1128/jb.175.16.5260-5264.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leonard B A B, Podbielski A, Hedberg P J, Dunny G M. Proc Natl Acad Sci USA. 1996;93:260–264. doi: 10.1073/pnas.93.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruhfel R E, Manias D A, Dunny G M. J Bacteriol. 1993;175:5253–5259. doi: 10.1128/jb.175.16.5253-5259.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pontius L T, Clewell D B. J Bacteriol. 1992;174:1821–1827. doi: 10.1128/jb.174.6.1821-1827.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weaver K E, Clewell D B. J Bacteriol. 1988;170:4343–4352. doi: 10.1128/jb.170.9.4343-4352.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weaver K E, Clewell D B. Plasmid. 1989;22:106–119. doi: 10.1016/0147-619x(89)90020-6. [DOI] [PubMed] [Google Scholar]
- 28.Ike Y, Clewell D B. J Bacteriol. 1984;158:777–783. doi: 10.1128/jb.158.3.777-783.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pontius L T, Clewell D B. J Bacteriol. 1992;174:3152–3160. doi: 10.1128/jb.174.10.3152-3160.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muscholl A, Galli D, Wanner G, Wirth R. Eur J Biochem. 1993;214:333–338. doi: 10.1111/j.1432-1033.1993.tb17928.x. [DOI] [PubMed] [Google Scholar]
- 31.Tanimoto K, Clewell D B. J Bacteriol. 1993;175:1008–1018. doi: 10.1128/jb.175.4.1008-1018.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujimoto S, Bastos M, Tanimoto K, An F, Wu K, Clewell D B. In: Streptococci and the Host. Horaud T, Sicard M, Bouve A, de Montelos H, editors. New York: Plenum; 1997. pp. 1037–1040. [Google Scholar]
- 33.Bastos M C F, Tanimoto K, Clewell D B. J Bacteriol. 1997;179:3250–3259. doi: 10.1128/jb.179.10.3250-3259.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmidt T G M, Skerra A. Protein Eng. 1993;6:109–122. doi: 10.1093/protein/6.1.109. [DOI] [PubMed] [Google Scholar]
- 35.Ausubel F M, Brent R, Kingston R E, Moor D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1987. [Google Scholar]
- 36.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1982. [Google Scholar]
- 37.Dunny G M, Craig R A, Carron R L, Clewell D B. Plasmid. 1979;2:454–465. doi: 10.1016/0147-619x(79)90029-5. [DOI] [PubMed] [Google Scholar]
- 38.Tanimoto K, Tomita H, Ike Y. Plasmid. 1996;36:55–61. doi: 10.1006/plas.1996.0032. [DOI] [PubMed] [Google Scholar]
- 39.Nakayama J, Suzuki A. Biosci Biotech Biochem. 1997;61:1796–1799. doi: 10.1271/bbb.61.1796. [DOI] [PubMed] [Google Scholar]
- 40.Chung J W, Dunny G M. Proc Natl Acad Sci USA. 1992;89:9020–9024. doi: 10.1073/pnas.89.19.9020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chung J W, Dunny G M. J Bacteriol. 1995;177:2118–2124. doi: 10.1128/jb.177.8.2118-2124.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hedberg P J, Leonard B A B, Ruhfel R E, Dunny G M. Plasmid. 1996;35:46–57. doi: 10.1006/plas.1996.0005. [DOI] [PubMed] [Google Scholar]
- 43.Weaver K E, Clewell D B, An F. J Bacteriol. 1993;175:1900–1909. doi: 10.1128/jb.175.7.1900-1909.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]