Abstract
The human opportunistic pathogen Bacillus cereus belongs to the B. cereus group that includes bacteria with a broad host spectrum. The ability of these bacteria to colonize diverse hosts is reliant on the presence of adaptation factors. Previously, an IVET strategy led to the identification of a novel B. cereus protein (IlsA, Iron-regulated leucine rich surface protein), which is specifically expressed in the insect host or under iron restrictive conditions in vitro. Here, we show that IlsA is localized on the surface of B. cereus and hence has the potential to interact with host proteins. We report that B. cereus uses hemoglobin, heme and ferritin, but not transferrin and lactoferrin. In addition, affinity tests revealed that IlsA interacts with both hemoglobin and ferritin. Furthermore, IlsA directly binds heme probably through the NEAT domain. Inactivation of ilsA drastically decreases the ability of B. cereus to grow in the presence of hemoglobin, heme and ferritin, indicating that IlsA is essential for iron acquisition from these iron sources. In addition, the ilsA mutant displays a reduction in growth and virulence in an insect model. Hence, our results indicate that IlsA is a key factor within a new iron acquisition system, playing an important role in the general virulence strategy adapted by B. cereus to colonize susceptible hosts.
Author Summary
Iron is an essential compound for almost all living organisms, taking part in basic cellular homeostasis. Preventing access to iron sources for invading pathogens is one of the defense systems used by hosts to avoid pathogen colonization. To counteract this, pathogens have developed mechanisms to acquire nutrient iron during infection. Bacillus cereus is an opportunistic bacterium able to infect both insects and mammals; thus, it should have systems enabling iron uptake from these hosts. Here we describe, for the first time, a unique surface protein, called IlsA, which is essential for iron uptake from two very different iron binding molecules: ferritin and hemoglobin. IlsA is only produced in iron limited environments. We show that during insect infection, its expression is specific to insect hemocoel (blood), where ferritin is the major iron-binding molecule. Interestingly, the IlsA mutant has reduced survival in in vivo infection and in vitro when heme, hemoglobin and ferritin are the sole iron sources available. Thus, as IlsA is important for iron uptake from the major iron rich molecules in insects and mammals, we suggest that this new iron acquisition system may be a key factor that is evolutionary adapted to infection of such diverse hosts.
Introduction
Iron is an essential element for most organisms, including bacteria, because it is involved in many cellular processes including aerobic respiration, amino acid and nucleotide biosynthesis [1],[2]. Since free iron is highly toxic for the cells, its homeostasis is strictly regulated in living organisms. Protection against iron is achieved by iron sequestration in carrier proteins such as transferrin, lactoferrin, ferritin or as iron-binding to the heme in hemoproteins. Thus, the lack of free iron is an obstacle that bacteria must overcome, when invading a host. In order to scavenge iron from the host iron-binding proteins, bacteria have developed two principal high affinity iron-uptake systems, which are considered to be important virulence factors. One system is based on the secretion of siderophores that capture iron from iron-binding proteins by the virtue of a superior binding strength. The siderophores are then recognized by specific membrane anchored binding proteins and internalized into the cytosol [3],[4]. The second system involves direct binding to host iron rich proteins via specific bacterial surface receptors which subsequently interact with membrane bound ABC transporters and permeases allowing iron transfer into the cytosol. These systems have been more studied in Gram-negative compared to Gram-positive bacteria [5]–[8]. The majority of these iron-uptake systems are under the control of the repressor Fur (Ferric uptake regulator) [3]. In Gram-positive bacteria, the best characterized system relying on bacterial surface proteins is the iron-regulated surface determinants (Isd) of Staphylococcus aureus. The Isd system implements cell wall proteins that act as hemoprotein receptors [9],[10] because of the presence of several copies of the conserved NEAT domains (for NEAr iron Transport), which play a key role in heme and hemoproteins binding and transport [11],[12]. An Isd system has been also studied in Bacillus anthracis. In addition to cell wall proteins, the B. anthracis Isd system uses secreted proteins that contain NEAT domains which act as hemophores, enabling heme acquisition from hemoglobin [13],[14].
The Gram-positive, spore-forming and human opportunistic pathogen, Bacillus cereus, belongs to the Bacillus cereus group, which also includes the entomopathogen, Bacillus thuringiensis, and the etiological agent of anthrax in mammals, B. anthracis. These three closely related species share a large number of chromosomal determinants, whereas their host-specific toxins are carried on plasmids [15]–[17]. B. cereus is generally associated with human food poisoning, resulting from the diarrhea and the emetic toxins [18]. However, B. cereus can also cause serious infections such as endophthalmitis, pneumonia and meningitis [19]–[21]. In addition, a new B. cereus species was found to cause severe respiratory illness resembling anthrax [22]. To date, virulence of B. cereus has been ascribed to different extracellular factors that are under the control of the pleiotropic regulator PlcR, which is part of a quorum sensing system [23],[24]. The PlcR regulon is important for virulence in both mice (intranasal infection) and insects [25]. However, other factors are implicated in the pathogenesis of B. cereus. Indeed, little research has been carried out on virulence genes involved in the adaptation and the persistence of B. cereus in the host and specifically on genes involved in iron acquisition.
Previous studies have shown that B. cereus secretes two siderophores: Bacillibactin and Petrobactin [26]. A recent study has identified a surface binding receptor protein as a potential receptor to these siderophores [27]. The ability of B. cereus, to acquire iron from host iron-rich sources, has received little attention; two independent research groups have reported contradictory results on the capacity of B. cereus to use transferrin [28],[29]. In addition, other studies have reported that B. cereus can use hemoglobin, hemin and other hemoproteins such as hemoglobin-haptoglobin and hemin-albumin for growth [28],[30],[31]. The majority of these studies focused on the identification of iron sources that can be used by B. cereus, whereas mechanisms used by B. cereus to scavenge iron from the host and especially those involving surface proteins have not yet been reported, except for the recently described siderophore binding protein [27].
In our previous study, an IVET strategy (In Vivo Expression Technology) enabled us to identify a B. cereus gene specifically expressed in insect larvae infected via the oral route [32]. It was shown that the transcription of this gene was induced in response to iron restrictive conditions. The presence of a Fur-binding box in the gene promoter region and a conserved NEAT domain in the protein are in agreement with an iron-dependent regulation. The presence of a N-terminal peptide signal and SLH domains (S-layer homology) suggest that the protein might be located on the bacterial surface (Figure 1A). This protein was designated IlsA, for Iron regulated leucine-rich surface protein. Moreover, IlsA was involved in the virulence of B. cereus in the lepidopteran Galleria mellonella larva infected by the oral route [32]. A BLAST search indicates that IlsA-like proteins are restricted to B. cereus group bacteria.
Figure 1. Schematic representation of the protein IlsA and its localization.
(A) The predicted conserved domain of the protein IlsA. The N-terminal putative signal peptide (SP) for secretion is indicated by a bold square. The three conserved domains: NEAT (NEAr iron Transporter), LRR (Leucine-Rich Repeat) and SLH (Surface Layer Homology) are indicated. (B) Localization of IlsA on the surface of B. cereus. The wild-type grown in LB medium (panels i–iii) and in LB+150 µM 2,2′-dipyridyl medium (panels iv–vi), the mutant ΔilsA grown in LB+150 µM 2,2′-dipyridyl medium (panels vii–ix), and the complemented ΔilsA+pHT304ΩilsA strains grown in LB+150 µM 2,2′-dipyridyl medium (panels x–xii), were analyzed by light microscopy with phase contrast (panels i,iv,vii,x) and immunofluorescence (panels ii,v,viii,xi) using the IlsA polyclonal antibody. The merged images (panels iii,vi,ix,xii) show the DAPI stained DNA in red and IlsA in green. IlsA is localized at the bacterial surface of the wild-type and only in iron depleted condition (panels v,vi). Interruption of ilsA abolished the presence of IlsA on the bacterial surface (panels viii,ix) and the complementation of the mutation restored the localisation (panel xi,xii). White bars represent 2 µm.
Here, we demonstrate that IlsA is a surface protein absolutely required for iron-uptake from several host proteins by direct binding to hemoglobin, hemin and ferritin. This is the first time that a NEAT domain protein has been shown to be involved in iron-uptake from ferritin. Furthermore, in vivo studies show that IlsA is specifically expressed in the hemocoel of G. mellonella and that the reduced virulence of an ilsA mutant correlates with a decrease of its in vivo growth capacity. Our results suggest that IlsA is an important adaptation factor required for the development of B. cereus in the host. Finally, these findings provide new insights into the ecology of this bacterial group that is able to colonize hosts as diverse as mammals and insects.
Materials and Methods
Bacterial stains and growth conditions
Bacillus cereus strain ATCC 14579 (laboratory stock) was used throughout this study.
The mutant B. cereus ATCC 14579 ΔilsA was previously constructed by allelic exchange through a double cross-over event by introducing a tetracycline resistant cassette [32]. E. coli K12 strain TG1 was used as a host for cloning experiments. Dam–, Dcm– E. coli strains ET 12567 (laboratory stock) were used to generate unmethylated DNA for electro-transformation in B. cereus. For electro-transformation, B. cereus was grown in BHI (Brain Heart Infusion, Difco) broth. E. coli and B. cereus strains were transformed by electroporation as previously described [33],[34]. E. coli BL-21 [F−, ompT, HsdS (rB −, mB −), gal] strain was used for expression and purification of the Glutathione S-transferase (GST) fusion protein (GST-IlsA). E. coli and B. cereus were cultured in LB (Luria–Bertani) broth, with vigorous shaking (175 rpm) at 37°C. Antibiotics for bacterial selection were used at the following concentrations: ampicillin (100 µg ml−1 for E. coli), erythromycin (10 µg ml−1 for B. cereus) and tetracycline (10 µg ml−1 for B. cereus). The iron chelator, 2,2′-dipyridyl and the iron sources, hemoglobin, hemin, ferritin, transferrin and lactoferrin, were obtained from Sigma-Aldrich, St. Quentin Fallavier, France. The iron sources were used at a concentration (see below) that permits the growth of B. cereus. The ferric chloride (FeCl3) was used at a final concentration of 0.2 mM. All iron solutions were sterilized by passage through 0.22 µm pore size filter.
DNA manipulations
Chromosomal DNA was extracted from B. cereus cells with the Puregene DNA Purification Kit (Gentra, Minneapolis, USA). Plasmid DNA was extracted from E. coli and B. cereus using QIAprep spin columns (QIAgen, France). For B. cereus, 5 mg ml−1 of lysosyme was added and cells were incubated at 37°C for 1 h. Oligonucleotide primers were synthesized by Proligo (Paris, France). PCRs were performed in a thermocycler PTC-100™ (MJ-Research, Inc., USA). Amplified fragments were purified using the QIAquick PCR purification Kit (QIAgen). Restriction enzymes (New England Biolabs, USA) and T4 DNA ligase (Invitrogen, USA) were used as recommended by the manufacturer. Digested DNA fragments were separated by electrophoresis on 1% agarose gels and extracted from gels using the QIAquick gel extraction Kit (QIAgen).
Complementation of the ilsA mutant strain
The genetic complementation of the strain B. cereus ΔilsA was carried out as follows. A DNA fragment corresponding to the gene ilsA was amplified by PCR using the B. cereus ATCC 14579 genomic DNA as a template and the primers ilsA-forward (5′-AACTGCAGGGGCTTTTTTATTTTGTACC-3′) and ilsA-reverse (5′-CGGAATTC GTGAGGGCTACTAATCAGTTG-3′). The PCR product was digested with EcoRI and Pst1 restriction enzymes and inserted into the plasmid pHT304 by ligation using the T4 DNA ligase [35]. The resulting plasmid (pHT304ΩilsA) was amplified in E. coli and then introduced into the mutant strain B. cereus ΔilsA by electroporation.
IlsA purification and antibody production
Glutathione S-transferase (GST) and GST-IlsA were purified as recombinant proteins from E. coli. The expression plasmid pgst-ilsA was constructed by PCR amplification of the ilsA sequence from the B. cereus genome using the primer pair ilsA-forward-2 (5′-CGGAATTCTTGAAAAAAAATTATATGAAGG-3′) and ilsA-reverse-2 (5′-CCCTCGAGTTATTTCTTTATTGCATTATAC-3′). The DNA fragment was digested with EcoRI/XhoI, cloned into pGEX6P1 (Amersham Biosciences) digested with the same restriction enzymes and transferred into E. coli Bl-21 by electroporation. E. coli Bl-21 strain harboring pGEX6P1-ilsA was grown to log phase in LB medium with ampicillin at 37°C. The expression of the fusion protein GST-IlsA was induced by adding IPTG (isopropyl-β-d-thiogalactopyranoside) to a final concentration of 1 mM and the cultures were incubated for 3 hours at 30°C. The bacteria were collected following centrifugation at 7700 rpm for 15 min in 50 ml tubes and suspended in 1X PBS containing 1% Triton X-100. Bacteria were lysed on ice by sonication using a Branson sonicator 250. Bacterial lysate was centrifuged at 7600 rpm for 15 min at 4°C, and the pellet containing the fusion protein IlsA-GST was solubilized in 50 mM Tris (pH 8.0) containing 8 M urea. After solubilization in 8 M urea overnight at 4°C, the insoluble pellet was removed by centrifugation at 13000 rpm for 20 min at 4°C. The supernatant containing the solubilized protein was dialyzed against buffer containing Tris 50 mM (pH 8.0) and 1 M urea, for about 5 hours with stirring at 4°C to remove the residual detergent. The solubilized and dialyzed fusion protein was loaded onto a glutathione Sepharose 4B column equilibrated with 1X PBS. After binding the fusion protein, the column was washed with PBS 1X and the target protein GST-IlsA was eluted with 10 mM reduced glutathione.
Purified GST-IlsA was then dialyzed against the PreScission protease buffer (50 mM Tris HCl pH 7.0, 150 mM NaCl, 1 mM EDTA, 1 mM DTT) at 4°C overnight. The GST was then cleaved using the PreScission protease at a concentration of 2 Units µl−1 for 4 hours at 5°C. After digestion, the mix was separated on a 10% gel SDS PAGE and the IlsA protein band was cut from the gel and used to raise anti-IlsA antibodies (Proteogenix, Oberhausbergen, France). Purification of IlsA was monitored by SDS PAGE electrophoresis (Figure S1). The GST was prepared using the strain E. coli Bl21 harbouring the plasmid pGEX6P1. After induction using IPTG, the bacteria were lysed by sonication and GST present in the cytosol of the bacteria was purified using the glutathione Sepharose 4B column as described above (Figure S1).
Immunofluorescence analysis
For the localization of IlsA at the bacterial surface, overnight cultures of B. cereus wild-type, B. cereus ΔilsA and the complemented strain B. cereus ΔilsA pHT304ΩilsA were grown in LB medium or in LB medium treated with 150 µM 2,2′-dipyridyl at 37°C and used immediately. Bacteria from stationary phase culture (109) concentrated in 50 µl, were washed twice in phosphate buffered saline (PBS 1X) and fixed with 4% paraformaldhehyde dissolved in PBS 1X on a cover slip. After fixation, bacteria were washed in PBS and then labelled with an anti-IlsA polyclonal antibody diluted at 1∶500 in 1% BSA, followed by an anti-rabbit secondary antibody Alexa 488-conjugated at a dilution of 1∶500 in 1% BSA. Bacterial DNA was also labelled with DAPI at a dilution of 1∶300 in 1% BSA. Labelled bacteria were then sticked with Mowiol to a glass slide and dried at 37°C for 30 min. Preparations were examined under a Zeiss Axiovert 135 microscope. Image acquisition from the Zeiss microscope was carried out with a cooled charge-coupled device camera (Princeton), and the images were processed with Metamorph software (Universal Imaging Corporation).
Growth assay
B. cereus cultures were grown overnight under low iron conditions by inoculating strains in LB medium supplemented with 200 µM 2,2′-dipyridyl. Overnight cultures were centrifuged and washed twice in LB medium supplemented with 500 µM 2,2-dipyridyl. Washed bacteria were then inoculated to a final optical density (OD) of about 0.01 into LB medium containing 2,2′-dipyridyl (450 µM) only or supplemented separately with hemoglobin (2 µM), hemin (16.5 µM), ferritin (0.3 µM), transferrin (1.5 µM) or lactoferrin (1.5 µM). Knowing that both the content of iron per molecule and the biochemical structure largely vary among these sources; the concentration of these iron sources were determined to give the optimal conditions for B. cereus growth (data not shown). Cultures were grown at 37°C with aeration and bacterial growth was monitored at an optical density of 600 nm over 10 hours.
Growth index
B. cereus wild-type strain, the mutant strain B. cereus ΔilsA and the complemented strain B. cereus ΔilsA pHT304ΩilsA were inoculated into LB medium supplemented with 200 µM 2,2′-dipyridyl and grown overnight. Overnight cultures were inoculated into LB medium supplemented with 200 µM 2,2′-dipyridyl at a final OD of 0.1. After incubation for 8 hours at 37°C, the resulting cultures were washed with 500 µM 2,2′-dipyridyl and inoculated at an OD of about 0.01 into fresh iron-restricted LB medium containing the appropriate antibiotics in the presence of 450 µM 2,2′-dipyridyl with or without either hemoglobin (2 µM), or hemin (16.5 µM), or ferritin (0.3 µM) or FeCl3 (200 µM) as a positive control. After 24 h of incubation, bacterial growth was recorded by measuring the absorbance at 600 nm (OD600 nm) and the data were generated as the growth index (growth in iron-restricted LB with or without the iron sources divided by growth in iron-rich LB). The assays were repeated three times.
Enzyme Linked Immuno Sorbent Assay (ELISA)
The binding assay was performed by using increasing amounts of hemoglobin (0–180 nM) and ferritin (0–180 nM) for coating 24-well ELISA plates with buffer (0.1 M phosphate buffer pH 7.4). After incubation for 18 hours at 4°C, wells were blocked with 1% BSA dissolved in the coating buffer for 1 h at 37°C. After blocking and between each further step, wells were washed three times with 0.15 M NaCl dissolved in the coating buffer with 0.1% Tween 20. Purified IlsA was added at a concentration of 30 nM and incubated with hemoglobin and ferritin for 2 hours at 37°C. Interaction between hemoglobin and IlsA or ferritin and IlsA was detected with the polyclonal anti-IlsA antibody at a 1∶500 dilution and the horseradish peroxidase-labelled secondary anti-rabbit immunoglobulin G (IgG) at a dilution of 1∶10 000. Binding of IlsA was quantified by measuring the conversion of the chromogenic substrate, otho-phenylendiamine dihydrochloride (OPD), to the coloured product based on optical density readings at 492 nm. Readings were transformed and used for estimation of the dissociation constant, K d, using the computer program Origin.
Surface Plasmon Resonance (SPR)
Real time binding kinetics experiments were conducted on a BIAcore 3000 (GE Healthcare Europe). GST-IlsA fusion protein was immobilized via its GST-tag in a flow cell of a CM5 sensor chip. For this purpose, a goat anti-GST antibody was first immobilized on the sensor chip using amine-coupling chemistry. The surface was activated for 7 min with a mixture of 0.05 M NHS and 0.2 M EDC. Anti-GST was then covalently linked to the surface giving up to 8000 resonance units (RU). Ethanolamine (1 M, pH 8.5) was injected for 7 min to block the remaining activated groups. Capture of GST-IlsA (1 µM) was performed at an injection flow of 5 µl min−1 in 0.1 M Phosphate buffer (pH 7.4), until stabilization of the immobilization level (6–8 min). For binding kinetics, a concentration of 200 nM of each IlsA ligand (hemoglobin, hemin, ferritin, and transferrin) was injected on the captured IlsA for 8 min and dissociation was registered for 10 min after the end of injections. Regeneration of the anti-GST capture antibody was achieved by a 2 min injection of glycine buffer (pH 2.2). The sensorgram from a cell with immobilized GST-IlsA was corrected by subtracting the response sensorgram from a cell with immobilized only anti-GST (reference surface) and normalized to a baseline of 0 RU. All measurements were performed at 20°C. Sensorgrams were analyzed using BIAevaluation Software. SPR tests were run 5 times on three different chips.
Heme detection by chemiluminescence
Hemin was dissolved immediately before use in a minimal volume of 0.1 M NaOH and diluted with phosphate buffer (Na2HPO4 0.1 M, pH 7.4) to a concentration of 10−4 M. Twenty microliters of purified GST-IlsA (2.5×10−7 M) were incubated with heme or with the buffer at room temperature for 30 min. Purified GST (2.5×10−7 M) was also used as negative control. Mixtures were separated by non-denaturing 8% PAGE in the absence of SDS at 4°C and the proteins were electrotransferred to a nitrocellulose membrane. The presence of heme complexed to IlsA was detected by chemiluminescence due to the heme peroxidase activity [36], using the Pierce ECL Super signal system.
In vivo expression using the gfp reporter gene
In order to follow the expression of ilsA in vivo, we constructed a plasmid carrying the pilsA-gfp transcriptional fusion. The plasmid pHT315-gfp was obtained by cloning the gene gfp-mut1 into the plasmid pHT315 [35] between XbaI and HindIII restriction sites. gfp-mut1 was amplified from the template plasmid pNF8 of Listeria monocytogenes [37] by using the primers F-gfp (5′-GCTCTAGAGAAAGGAGGTTATTAAAATGAGTAAAGGAGAAGAACTT-3′) and R-gfp (5′-CCCAAGCTTTTATTTGTATAGTTCATCCATGCCA-3′). The ilsA promoter region (pilsA) was generated by PCR with oligonucleotide pairs Fw-pilsagfp (5′-CCGGAATTCGGGCTTTTTTATTTTGTACC-3′) and Rv-I1 (5′-CCCTGAACAGTGTTCTCGG-3′) using the pHT304-pivi29′-I from the IVET screen [32] as a template. The PCR fragments were digested by EcoRI and XbaI and were inserted between the EcoRI and XbaI restriction sites of pHT315-gfp plasmid to give the plasmid pHT315-pilsA'-gfp. The resulting plasmid pHT315-pilsA'-gfp was cloned in E. coli and was then used to transform B. cereus. The strain harboring the plasmid pHT315-pilsA'-gfp was used to infect the lepidopteran larvae of G. mellonella orally in order to follow the kinetics of ilsA expression in vivo. The insect larvae were infected with 5×106 mid-log phase bacteria (OD600 ∼1) in association with 3 µg of Cry1C toxin and incubated at 37°C, according to the protocol previously described [38]. Larvae were dissected and bacteria were recovered from the insect gut at 9 hours after infection and from the hemocoel at 24 hours after infection. The bacteria were then subjected to bright field (DIC) and epifluoresence microscope observation (NIKON Eclipse E600 equipped with UV mercury disposal and observed with an FITC filter). For a positive control we used the B. cereus strain harbouring the plasmid pHT315-paphA3'-gfp that contains a transcriptional fusion between the constitutive promoter paphA3 obtained by PCR amplified from the pGDG783 plasmid [39] and the gfp gene (C. Nielsen-LeRoux, unpublished data).
Growth kinetics of B. cereus in the hemocoel of G. mellonella
The growth (number of bacteria) was assessed at 2, 6 and 24 hours after injecting doses of 500 mid-log phase bacteria of the wild type or the ilsA mutant strains into the hemocoel of the larvae G. mellonella. At each time-point, three alive larvae were recovered, surface-sterilized by rinsing briefly in 70% ethanol, crushed, and homogenized separately in 10 ml of phosphate buffered saline (PBS) pH 7.4, using a polytron homogenizer (Kinematica, Switzerland). Serial dilutions were plated on LB medium, in order to estimate the number of bacteria present in the larvae. At each time point, the results are the means of nine independent larvae from three independent tests. The resulting data were analysed using the Student T-test.
Virulence assays
Contribution of IlsA to B. cereus virulence was assayed by comparing the killing effect of the wild-type and the ilsA mutant B. cereus strains, after injecting them separately into the hemocoel of G. mellonella larvae. Last-instar larvae weighing about 200 mg were injected with 10 µl of mid-log phase bacteria suspended in PBS, using the microinjector (Buckard Scientific, UK.) as previously described [40]. Various doses of B. cereus wild-type and the ilsA mutant (103 to 102 bacteria/larvae) were used, and each dose was repeated three times on at least 20 larvae. A control group of larvae was injected with PBS only. Infected larvae were kept at 37°C and mortality was recorded after 24 hours. The LD50s values were estimated using Probit analysis [41],[42]. This program tests for the linearity of dose-mortality curves, provides lethal doses and the slope of each dose-mortality line. It tests the parallelism of two or more dose-mortality lines and determines the virulence ratio between the bacterial strains. The ratio is considered to be significantly different from 1 (P<0.05) when the confidence limits do not include the value 1.
Results
Localization of IlsA on the bacterial surface
The analysis of the IlsA sequence indicated a possible SLH-domain (S-Layer Homology) presumably binding the protein to the peptidoglycan (Figure 1A) and our previous results showed that IlsA was expressed in iron depleted conditions [32], thus we analyzed its localization in such conditions. The localization of IlsA was determined by immunofluorescence using the polyclonal antibody anti-IlsA (Figure 1B). IlsA was detected on the surface of B. cereus wild-type grown in iron depleted condition (LB supplemented with iron chelator) (Figure 1B iv–vi), whereas IlsA was not present on the surface of either the wild-type strain grown in LB medium (standard conditions) (Figure 1B i–iii) or the mutant strain B. cereus ΔilsA grown in iron depleted medium (Figure 1B vii–ix). Transformation of the B. cereus ΔilsA mutant with the plasmid pHT304-ilsA restored the production of IlsA at the surface of the bacteria grown in iron-depleted medium (Figure 1B x–xii). In addition, we observed an accumulation of IlsA on the division site of the bacteria. These results indicate that IlsA is present on the surface of the bacteria and only in iron depleted conditions, which is in agreement with the iron regulated expression of ilsA [32].
B. cereus uses hemoglobin, hemin and ferritin as iron sources
The ability of B. cereus ATCC 14579 to utilize host iron sources has not been previously investigated. To determine which iron sources B. cereus is able to use, we measured its ability to grow on hemoglobin, hemin, ferritin, transferrin and lactoferrin, in iron depleted LB medium (Figure 2). B. cereus growth was very weak in iron depleted condition, but the addition of either hemoglobin, or hemin, or ferritin to the iron depleted medium increased significantly the growth kinetics of B. cereus. In contrast, transferrin and lactoferrin addition to iron depleted medium yielded only growth of B. cereus similar to that in iron depleted medium.
Figure 2. Growth kinetics of B. cereus wild-type using iron-rich host proteins as sole iron sources.
B. cereus was grown in LB medium treated with 2,2′-dipyridyl without addition of iron sources (black square) or supplemented with hemoglobin (white diamond), or hemin (black diamond), or ferritin (black circle) or transferrin (white triangle) or lactoferrin (white square). Bacterial growth was determined by measuring the optical density (OD) at 600 nm. Curves are representative of three independent experiments.
This result indicates that B. cereus is able to use hemoglobin, hemin and ferritin as iron sources for growth, whereas transferrin and lactoferrin cannot be used.
IlsA is required for efficient iron acquisition from hemoglobin, hemin and ferritin
To test the hypothesis that IlsA is important for iron scavenging from the host iron sources used by B. cereus, the growth rates of the wild-type and ilsA mutant strains were compared after inoculation in iron depleted medium where either hemoglobin, hemin or ferritin were provided as the sole iron source. Bacterial growth was measured by spectrophotometric analysis of culture samples at an optical density of 600 nm (OD600) (Figure 3). Iron depleted LB did not support the growth of the wild-type and the ΔilsA mutant strains, confirming that iron is an essential nutrient for B. cereus growth. In contrast, the wild-type strain grew well in iron depleted LB that had been supplemented with hemoglobin, hemin or ferritin, while B. cereus lacking ilsA showed significant growth defects in these media. However, the mutant strain grew as the wild-type when iron was provided in its inorganic form (FeCl3). Therefore, IlsA is not required for the uptake of inorganic iron. To verify that this phenotype was specifically due to ilsA, the mutant strain was complemented with a plasmid harboring ilsA. The growth of the bacteria was restored to the wild-type level when hemoglobin, hemin or ferritin was added to iron depleted LB. These data indicate that IlsA is very important for efficient growth of B. cereus under iron-restricted conditions; they suggest that IlsA is involved in iron acquisition from the host during infection.
Figure 3. Growth index phenotype of B. cereus wild-type strain and the mutant B. cereus ΔilsA using iron-rich host proteins as the sole iron sources.
B. cereus wild-type, the mutant ΔilsA and the complemented ΔilsA+pHT304ΩilsA strains were grown in iron restricted medium treated with 2,2′-dipyridyl (450 µM) or supplemented with host iron sources: hemoglobin (2 µM) or hemin (16.5 µM) or ferritin (0.3 µM) for 24 h. Ferric chloride (200 µM) was used as a control for inorganic iron source. The optical density was recorded at 600 nm and the growth index was quantified (growth in indicated conditions, compared to growth in LB (iron-rich medium). The mutant ΔilsA strain was unable to grow in the presence of any of the tested host iron sources and complementation of ilsA to the mutant strain ΔilsA restores the bacterial growth phenotype. The mutant ΔilsA strain was able to grow with ferric chloride. Results are the means of three independent experiments and error bars indicate the standard deviations of the mean.
IlsA binds hemoglobin and ferritin
To address the question of whether IlsA can directly interact with hemoglobin and ferritin, we performed binding studies using the purified recombinant protein GST-IlsA. In these assays, increasing amounts of hemoglobin and ferritin were immobilized on ELISA plates (hemin can not bind to these plates) and IlsA was added. The amount of bound IlsA was detected using anti-IlsA polyclonal antibody. By comparing the signal intensities generated by the binding of IlsA to the different proteins, we detected a slightly stronger binding of IlsA to hemoglobin (K d = 3±1 nM) relative to ferritin (K d = 10±3 nM) (Figure 4A). However, the Student T-test showed no significant difference (p<0.05) for IlsA to bind hemoglobin and ferritin.
Figure 4. IlsA binds hemoglobin and ferritin.
(A) Binding of purified GST-IlsA to hemoglobin and ferritin was performed in an ELISA based assay. Hemoglobin (white square) and ferritin (black square) were coated on ELISA plates at increasing amounts, and IlsA was added at 3 pmole. Binding was quantified by measuring the absorbance after incubation with anti-IlsA and the secondary antibody peroxidase labelled. Data shown are the mean of three independent experiments. (B) Real-time kinetics of interaction between heme, ferritin, hemoglobin and transferrin with GST-IlsA fusion protein. Overlay plot of sensorgrams corresponding to interaction of these various proteins with GST-IlsA fusion protein. GST-IlsA fusion protein was first captured to saturation by anti-GST antibody linked covalently to the carboxymethyldextran chip. Then various iron sources (200 nM) were injected for 8 min after which dissociation was followed. Note the absence of transferrin binding. The sensorgrams are representative for five experiments.
To investigate whether binding also occurs in a real time interaction, we used the Surface Plasmon Resonance (SPR) system. The interaction between chip immobilized GST-IlsA and different host iron-rich molecules (hemin, hemoglobin, ferritin and transferrin) under flow conditions shows typical SPR curves (Figure 4B). These SPR curves are from one experiment, but the assays were run 5 times on 3 different chips for which the curves shape were similar to the ones shown. Computational analysis showed that our data don′t fit into a typical association-dissociation model with a 1∶1 molar ratio. This is probably resulting from protein multivalencies and potential steric hindrance of the various sizes of the ligands (ferritin 440 KDa, hemoglobin 68 KDa and heme 651 Da). Then, quantitative information about the relative stability of the complex, was deduced from the dissociation phase only, independent from the analyte concentration. Then, assuming a simplified kinetic scheme, the indicative kinetic dissociation constants (koff, expressed in s−1) were calculated: 1.71±0.83×10−3 for ferritin, 1.48±0.98×10−3 for hemoglobin and 1.34±0.44×10−3 for hemin. Thus, the results showed similar koff values for the three iron sources. These findings are in accordance with the ELISA experiments showing that soluble ferritin and hemoglobin bound to immobilized IlsA, but transferrin did not (result not shown). The lack of direct interaction between IlsA and transferrin could be expected, because the growth experiment (Figure 2) indicated that this protein was not used by B. cereus as an iron source.
IlsA binds heme
To determine whether IlsA in solution binds to heme or another component of hemoglobin, we incubated purified GST-IlsA and GST (negative control) with hemin (10−4 M) and analyzed the binding reaction on a non-denaturing polyacrylamide gel system (Figure 5A). This allows the separation of free heme, purified proteins and heme-loaded proteins without dissociating heme from the proteins. Proteins were transferred to a nitrocellulose filter, and heme peroxidase activity was detected by chemiluminescence. This method is the most sensitive test available for detecting heme bound to proteins [36]. Our results showed that GST-IlsA could bind heme-iron, whereas GST alone cannot. This is in agreement with the result of the multiple sequence alignment of the NEAT domain of IlsA with those found in Isd proteins of S. aureus (Figure 5B). Indeed, this comparison indicates that the three conserved residues (Ser82, Tyr166 and Tyr170) present in IsdA, which are essential for heme interaction [11],[12], are also present in IlsA. Furthermore, the residues Val157, Ile159 and Val161 that form the hydrophobic heme-pocket in IsdA are either conserved in IlsA or replaced with similar hydrophobic amino acids. The conservation in IlsA NEAT domain of amino acid residues known to interact with heme, suggests that IlsA binds heme through the NEAT domain.
Figure 5. Hemin binding analysis of purified IlsA.
(A) Chemoluminescent detection showing binding of GST-IlsA to hemin (He). Purified GST-IlsA or GST incubated either with hemin (+) or without (−) as indicated on the top of the lane, were migrated on non-denaturing PAGE and transferred to a nitrocellulose membrane. Phosphate buffer alone was also incubated with hemin (+) as a control. After reaction with an ECL reagent system, proteins with hemin-binding properties are detected on chemoluminescent-sensitive film. Upper arrow corresponds to GST-IlsA binding to hemin and lower arrows to hemin alone. (B) Multiple sequence alignments of NEAT domains of IlsA from B. cereus and Isd proteins from S. aureus. Positions shaded in yellow are identical in 100% of the aligned sequences, blue and green are identical or similar, respectively, in at least 50% of the aligned sequences. The first four letters of the sequence represent the protein identity. Asterisks represent the amino acids required for heme binding. Numbers 1 to 3 represent the NEAT domain numbered from the N-terminus out of the total number of NEAT domains in that protein. The accession numbers of Isd proteins from S. aureus are as follows: IsdH (Q99TD3), IsdB (Q7A656), IsdA (Q7A655), IsdC (Q7A654). The accession number of IlsA from B. cereus ATCC 14579 is NP_831113. NEAT domains were identified in the Pfam database. Alignments were generated in vector NTI advance 10 (Invitrogen), using a BLOSUM matrix with gap opening and extension penalties of 15 and 2, respectively.
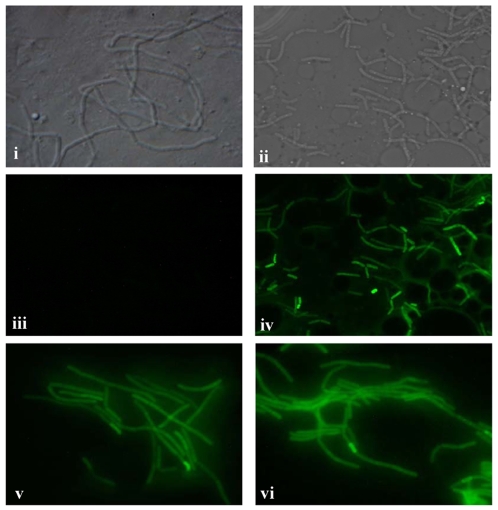
ilsA is strongly expressed in the hemocoel of infected larvae
The IVET screen showed that ilsA was strongly expressed during infection of the insect larvae [32]. However, it was not determined at which stage of the infection and in which tissue ilsA expression occurred. To determine where and when ilsA is expressed in vivo, larvae were infected with B. cereus harbouring a plasmid that contained a transcriptional fusion of the promoter pilsA and the gfp reporter gene. Bacteria recovered from the larvae at various times post-infection were examined under brightfield (Figure 6i–ii) and epifluoresence microscopy (Figure 6iii–vi). No fluorescence was observed in bacteria isolated from the intestine of living larvae after nine hours of infection (FIGURE 6iii). In sharp contrast, B. cereus bacteria isolated from the hemocoel of dying larvae after 24 hours of infection, showed strong fluorescence (FIGURE 6iv), thus indicating that ilsA is highly expressed in the hemocoel. Indeed, iron is not accessible to bacteria in the hemocoel due to the presence of proteins that bind iron such as transferrin and ferritin [43],[44]. B. cereus containing the plasmid (pHT315-paphA3'-gfp) with the constitutively expressed gfp gene, showed a strong expression in both the intestine and hemocoel of the infected larvae (Figure 6v–vi).
Figure 6. Analysis of ilsA expression in vivo.
Microscopic observations by Bright-field DIC (panels i,ii) and epifluorescence (panels iii–vi) were performed on B. cereus carrying pHT315-pilsA'gfp isolated at 9 hours after infection from the intestine of alive larvae (panels i,iii) and at 24 hours after infection from the hemocoel of dead larvae (panels ii,iv). Green bacteria indicate expression of gfp due to the activation of the ilsA promoter. B. cereus carrying pHT315-papha3'gfp was used as a control for its constitutive expression of GFP (panels v,vi).
IlsA is important for bacterial growth and adaptation of B. cereus in the hemocoel
Growth of the wild-type and ΔilsA mutant strains was assessed by homogenization of whole larvae at several time points after injection into the hemocoel of G. mellonella (Figure 7A). A dose of ∼500 mid-log phase bacteria (estimated as colonies forming units (cfu)) was injected and used as the start point. After 2 hours of injection, the number of the ΔilsA bacteria in the insect larvae decreased drastically (75.5 cfu), while the number of the wild-type bacteria remains stable (530 cfu). After 6 hours, B. cereus ΔilsA had a growth of about 10-fold less (39.4×103 cfu) than the B. cereus wild-type strain (50.3×104 cfu). After 24 hours, the growth of the mutant strain (18.5×105 cfu) was about 2,5-fold less than the B. cereus wild-type strain (50.7×105 cfu). The growth of B. cereus ΔilsA was significantly different (p<0.05) from the B. cereus wild-type strain at 2 hours and 24 hours after injection.
Figure 7.-. IlsA is required for B. cereus infection in the model G. mellonella.
(A) Effect of ilsA mutation on the growth of B. cereus after intrahemocoelic injection in G. mellonella. Wild-type and the mutant ΔilsA strains were injected separately into the hemocoel of last-instar larvae at a dose of 5×102 mid-log phase bacteria. Bacteria were counted at various times after injection (2, 6 or 24 h) from living larvae. Time 0 h is the real dose of each strain injected in the hemocoel. Results are the mean of bacterial counts from nine different larvae and error bars indicate standard errors of the means. (B) Effect of ilsA mutation on the virulence after intrahemocoelic injection in G. mellonella. Last-instar larvae were injected with various doses (102 to 103) of mid-log phase bacteria. Mortality was evaluated after 24 h of injection of B. cereus wild-type (bold histogram) and B. cereus ΔilsA (grey histogram). Results are mean values of four independent experiments and error bars indicate the standard errors of the means.
To test if the growth defect of the mutant strain affects the virulence of B. cereus in the hemocoel, we analyzed virulence tests where bacteria were directly injected into the hemocoel of G. mellonella. After 24 hours of infection with various doses of mid-log phase bacteria (102 to 103), the number of dead larvae was recorded (Figure 7B). The 50% lethal doses (LD50s) of the wild-type and of the ΔilsA mutant strains were assessed by Probit analysis and are shown as histograms. The wild-type strain had an LD50 of 2.2×102 (95% confidence limits between 1.8×102 and 2.6×102) bacteria per larva and the mutant showed an LD50 value of 4.8×102 (95% confidence limits between 4.1×102 and 5.7×102) bacteria per larva. A decrease of 2.2-fold (range 1.59 to 3.07 at the 5% confidence level) was determined by comparing the two dose-response regression lines. However, an additional statistical analysis (Student T-test) performed on each dose, indicates only a significant difference (p<0.05) at the lowest dose (100 bacteria/larvae). Then, the importance of IlsA by this route of infection seems less than by the oral route, as earlier reported [32], although IlsA helps B. cereus to survive in the hemocoel, as shown in the above in vivo growth analysis. These results indicate that IlsA is important for the growth and the survival of B. cereus in the hemocoel following injection or natural injury.
Discussion
The ability of living organisms to sequester free iron is not only necessary for protection against iron toxicity but it is also an innate resistance mechanism of hosts (mammals or insects) to fight infections [45],[46]. Iron incorporated into heme is the most abundant iron source in the mammalian host, being mainly associated with the oxygen-carrying molecule, hemoglobin, sequestered within erythrocytes. Ferritin, the major iron storage protein is a large molecule, composed of several subunits, containing about 4500 ferric ions per molecule, is present not only in mammals but also in almost all living organisms including insects, plants (phytoferritin) and bacteria (bacterioferritin) [47]. In insects, notably in G. mellonella, ferritin is present in high amounts in hemolymph and in hemocytes and may play a role in iron transport in addition to iron storage [44],[48]. Thus, free iron is not available and bacteria are iron restricted whatever their hosts. Therefore, they must be able to chelate and actively take up any available iron in order to grow and successfully infect their hosts. It has been shown that expression of the majority of genes involved in iron acquisition and metabolism, such as those coding for surface receptors, transport proteins, some hemolysins and cytotoxins, are controlled by the repressor Fur [1],[3]. In this study, we show that B. cereus can use hemoglobin, hemin and ferritin as iron sources through the surface receptor IlsA that also belongs to the Fur regulon. We show that B. cereus strain ATCC 14579 is not able to use lactoferrin or transferrin for its growth, which is in accordance with Sato et al. 1999 [30],[31] and in disagreement with Park et al. 2005 [29]. This result suggests that there might be strain variations, related to iron acquisition, but also that this B. cereus strain might be unable to cope with the antibacterial activity of lactoferrin [49], since higher concentrations of lactoferrin affected its growth. Hemoglobin and ferritin are intracellular iron rich proteins and must be released from cells in order to be used by B. cereus for its growth. It has been shown that B. cereus produces a large variety of cytotoxic proteins (Hbl, Nhe, CytK, Clo and HlyII) able to lyse various eukaryotic cells including erythrocytes [50]–[52]. Expression of these cytotoxic proteins with the exception of hemolysin II (HlyII), is regulated by the pleiotropic regulator PlcR [24],[53]. Then, all these factors may contribute to release hemoglobin, heme and ferritin from cells following cell lysis. These iron-binding molecules can then be used for B. cereus growth after direct binding to the surface receptor, IlsA. Our results show that purified recombinant IlsA can bind hemoglobin and ferritin in vitro. In addition, IlsA also binds heme alone, suggesting that the interaction between IlsA and hemoglobin is due to its interaction with heme. How IlsA binds heme is currently unknown; however, studies from other NEAT proteins have identified conserved tyrosine residues and hydrophobic residues that form a heme pocket, which mediates heme binding [11],[12]. These residues are also present in the IlsA NEAT domain, suggesting that IlsA mediates heme binding through this NEAT. The interaction with ferritin is much less evident, although it has previously been reported that the NEAT protein IsdA in S. aureus binds to several non-heme host proteins, including the ferric iron carrier transferrin [54], and several matrix and plasma proteins [55]. IlsA is a more complex protein than IsdA because of the LRRs domains. These domains are found in a large range of proteins with different functions and are known to bind structurally unrelated protein ligands [56],[57]. In Gram positive bacteria, only few LRR-containing proteins have been characterized to date, especially in Listeria monocytogenes, Streptococcus pyogenes and Enterococcus faecalis, where they are involved in the interaction with the host [57]. Thus, it is tempting to speculate that the LRR domains of IlsA are involved in the binding to ferritin. To our knowledge, IlsA is the first LRRs protein that has a function in iron uptake. A recent study revealed that a surface protein of Candida albicans, Als3, is essential for iron acquisition from ferritin by binding to the fungal hypha [58]. However, there is, no obvious structural homology, between Als3 (family of agglutinin like adhesion molecules) and IlsA. Moreover, the authors did not report any direct in vitro binding between ferritin and Als3. Thus, in depth structural studies related to the interaction between IlsA and ferritin are needed to elucidate this aspect.
Our results suggest that IlsA is a key protein of a new iron/heme acquisition system in B. cereus. To achieve the transport of ferric iron from ferritin and heme across the cell wall and the cytoplasmic membrane into the cytoplasm, IlsA has to interact with other bacterial partners. The iron-regulated surface determinants (Isd) involved in heme acquisition and transport has been studied in several Gram-positive bacteria especially in S. aureus and B. anthracis. In S. aureus, the Isd system is composed from cell wall anchored proteins acting as hemoprotein receptors that are in vicinity of an iron transporter ABC systems [9],[10]. Unlike S. aureus, the heme uptake system Isd in B. anthracis depends on secreted NEAT domains proteins in the extracellular environment and also on those located in the bacterial envelope which enable heme transfer to an ABC transporter system and then into the cytoplasm [13],[14,13].
The heme scavenging strategy evolved by B. cereus seems to be different from those described in S. aureus and B. anthracis, since it involves the surface protein IlsA, which contains SLH domains that enable its binding to peptidoglycane [59]. Homologs of IlsA are found in B. anthracis (belonging to Bsl surface proteins), but none of them possess the three conserved domains (NEAT, LRR, SLH) at the same time like IlsA. In addition to three SLH domains, BslK presents a NEAT domain, while BslL contains LLR domains [60]. Then, since B. anthracis can use heme as an iron source via the Isd uptake system [61], it might suggest that IlsA homologs in B. anthracis do not function in the same way as in B. cereus. Thus, on the basis of our studies and those from other Gram-positive bacteria, we propose a model (Figure 8) where IlsA interacts with NEAT proteins belonging to the Isd system to ensure heme transport through the bacterial envelope into the cytoplasm. In silico analysis of the B. cereus ATCC 14579 genome reveal, in addition to IlsA, several genes (Bc4547, Bc4548, Bc4549) encoding NEAT domain proteins. The protein encoded by Bc4548 contains a signal peptide, but lacks obvious anchoring motif. This protein, which has 98% identity with IsdX1 of B. anthracis [13],[14], may function as a secreted hemophore that captures heme from hemoglobin. The proteins encoded by Bc4547 and Bc4549 with the anchoring motifs NSKTA and NPKTG, respectively, may be the substrates of the putative sortase B (Bc4543) and being located in variable positions throughout the peptidoglycan. These proteins have 87% identity with IsdX2 [14] and 98% identity with IsdC of B. anthracis [61] respectively. Similar to S. aureus IsdC [62],[63] and B. anthracis IsdC [61], we propose that Bc4549 occupies a critical position enabling the transfer of heme to the iron permease system (Bc4544, Bc4545, Bc4546), followed by transport across the bacterial membrane. Finally, a putative intracellular monooxygenase, encoded by Bc4542 degrades heme to liberate iron to be used by B. cereus. The presence of IlsA is crucial for this iron-uptake system, since its disruption abolishes the ability of B. cereus to grow despite the presence of hemoglobin, hemin and ferritin. The mechanism by which IlsA permits the uptake of iron from ferritin is certainly not the same as from heme. We speculate that IlsA might be able to destabilize the ferritin structure, via a possible interaction with the IlsA LRR domains. The structural modification may permit other factors, such as reductases [64] or proteases [65] to liberate iron (Fe3+) that can then be captured by siderophores, such as petrobactin or bacillibactin [26] and transferred by an iron uptake-system into the cytosol. Studies are ongoing to investigate this hypothesis.
Figure 8. Model for heme iron acquisition in B. cereus.
In the host, B. cereus is in iron restrictive environment and the global iron regulator Fur is removed from its binding site upstream of ilsA and isd-like genes, leading to their expression. IlsA and Isd-like proteins are localized in their appropriate cellular locations. IlsA binds heme (H-Fe) directly from hemoglobin (Hb) or indirectly through Bc4548. Heme will then be transferred to Bc4547, Bc4549 and finally to the ABC transport system (Bc4544-45-46) located in the plasma membrane. Once heme is present in the cytoplasm, the monooxygenase (Bc4542) catalyzes the degradation of the heme and the released iron can be used for B. cereus growth. Arrows indicate the orientation and approximate sizes of the open reading frames. Potential transcription terminators are represented. The putative fur boxes in the promoter region upstream of ilsA (BC1331) and the Isd-like operon are indicated. Each cylinder indicates a NEAT domain. CW = cell wall, PM = plasma membrane, CY = cytoplasm.
To understand the contribution of IlsA in the pathogenesis of B. cereus, we analyzed its expression during infection using gfp transcriptional fusion. The results showed that ilsA is highly expressed in the hemocoel where free iron is not accessible to bacteria, being bound to proteins such as ferritin [44]. IlsA is also expressed in collected hemocoel and when bacteria are directly injected into the hemocoel (results not shown), indicating that the expression of ilsA is not depending on a dying insect. Then, expression of ilsA could be initiated in response to iron depleted conditions and/or due to bacterial sensing of ferritin, thus enabling capture of iron from this source and subsequent use by B. cereus for growth and survival within G. mellonella. Moreover, disruption of ilsA decreases the growth and the virulence of B. cereus after injection into the hemocoel. However, the effect of IlsA on the virulence of B. cereus was larger following infection by oral route [32], compared to what we found here, following injection into the hemocoel. This can be explained by a combination of several factors among which one might rely on the activation of the ferritin production. Assuming when the bacteria are entering the hemocoel from the intestinal lumen, the innate sensing (production of antimicrobial peptides, ferritin etc.) has been turned on for a while [66] and thus less free iron is available. In this situation, the ilsA mutant cannot grow as well as the wild-type. A second point is reliant to the dose dependent in vivo growth capacity of the ilsA mutant, reported here (Figure 7B). Indeed, unpublished histopathological studies (Nielsen-LeRoux et al.), have shown that bacteria, from the intestinal lumen, enter the hemocoel by small numbers which can be considered equivalent to a low dose of hemocoel injected bacteria.
Together, these observations indicate that IlsA is an important factor required for adaptation within an insect host especially during the early stages of infection, as soon as the bacteria enter the hemocoel and before iron ions are released from degraded tissues. Thus, IlsA is the first host iron receptor in B. cereus shown to contribute to in vitro growth by using hemoglobin, heme and ferritin as an iron source, as well as contributing to virulence in vivo . To our knowledge, it is also the first time that an extracellular pathogen has been shown to use ferritin for its growth in physiological conditions that are correlated with host infection. The intracellular bacteria Neisseria meningitidis has been reported to degrade ferritin by manipulating the cellular machinery and the lysosomal activity [67] and L. monocytogenes was reported to use ferritin in vitro via a reductase activity [64].
In this study, we provide evidence that IlsA is necessary and sufficient to mediate interaction and iron acquisition from several iron-binding proteins. The exceptional structure of IlsA (NEAT and LRRs) probably explains its ability to bind to these different ligands. Moreover, since IlsA-like proteins are restricted to B. cereus group bacteria, it may be a concrete example of how bacteria create molecules adapted to their particular surface structure. Indeed, the SLH domain of IlsA permits binding to the peptidoglycan via a mechanism often found in this bacterial group [68]. Further investigations are needed for a complete understanding of the mechanism of iron-uptake through IlsA. Detailed description of the mechanism by which IlsA allows B. cereus to acquire iron from hemoglobin, heme and ferritin will provide a better understanding not only of B. cereus pathogenesis but also for infections caused by other extracellular Gram-positive pathogenic bacteria.
In conclusion, our investigations show how a bacterial adaptation factor can be directly correlated with a certain level of virulence. Furthermore, they provide insights into the host adaptation and the evolutionary ecology of the B. cereus group bacteria, including the mammalian pathogen B. anthracis and the entomopathogen B. thuringiensis. Since identified homologs of IlsA are restricted to the B. cereus group, which is composed of closely related bacteria able to colonize diverse hosts, IlsA may be a key factor allowing colonization of these different environmental niches. B. cereus and B. thuringiensis can cause infection in insects, mice and other mammals [17],[25] while there is little evidence that B. anthracis can infect invertebrates [69]. Altogether, our results show the first steps of a new and unique mechanism of iron acquisition, which is specifically adapted for interaction with two different iron-rich host proteins found in vertebrates and invertebrates.
Supporting Information
Purification of IlsA. Coomassie-stained 10% SDS-PAGE analysis of the purified GST-IlsA and GST proteins from recombinant E. coli. Lane M, molecular weight markers in KDa. Purified GST-IlsA and GST with apparent molecular weights shown on the gel, was loaded in lane 1 and 2 respectively.
(0.51 MB PDF)
Footnotes
The authors are in the process of patenting IlsA, which may benefit from publication of this manuscript.
Financial support to N.D. is from the Conseil de la Recherche de l'Université Saint Joseph de Beyrouth, Lebanon, the Institut National de la Recherche Agronomique (INRA), and the French Embassy. This work was also supported by the Agence Nationale de la Recherche (ANR) through the project (PNRA 013-04). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Braun V. Iron uptake mechanisms and their regulation in pathogenic bacteria. Int J Med Microbiol. 2001;291:67–79. doi: 10.1078/1438-4221-00103. [DOI] [PubMed] [Google Scholar]
- 2.Wandersman C, Delepelaire P. Bacterial iron sources: from siderophores to hemophores. Annu Rev Microbiol. 2004;58:611–647. doi: 10.1146/annurev.micro.58.030603.123811. [DOI] [PubMed] [Google Scholar]
- 3.Ratledge C, Dover LG. Iron metabolism in pathogenic bacteria. Annu Rev Microbiol. 2000;54:881–941. doi: 10.1146/annurev.micro.54.1.881. [DOI] [PubMed] [Google Scholar]
- 4.Crosa JH, Walsh CT. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol Mol Biol Rev. 2002;66:223–249. doi: 10.1128/MMBR.66.2.223-249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stojiljkovic I, Larson J, Hwa V, Anic S, So M. HmbR outer membrane receptors of pathogenic Neisseria spp.: iron-regulated, hemoglobin-binding proteins with a high level of primary structure conservation. J Bacteriol. 1996;178:4670–4678. doi: 10.1128/jb.178.15.4670-4678.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewis LA, Sung MH, Gipson M, Hartman K, Dyer DW. Transport of intact porphyrin by HpuAB, the hemoglobin-haptoglobin utilization system of Neisseria meningitidis. J Bacteriol. 1998;180:6043–6047. doi: 10.1128/jb.180.22.6043-6047.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wandersman C, Stojiljkovic I. Bacterial heme sources: the role of heme, hemoprotein receptors and hemophores. Curr Opin Microbiol. 2000;3:215–220. doi: 10.1016/s1369-5274(00)00078-3. [DOI] [PubMed] [Google Scholar]
- 8.Genco CA, Dixon DW. Emerging strategies in microbial haem capture. Mol Microbiol. 2001;39:1–11. doi: 10.1046/j.1365-2958.2001.02231.x. [DOI] [PubMed] [Google Scholar]
- 9.Mazmanian SK, Ton-That H, Su K, Schneewind O. An iron-regulated sortase anchors a class of surface protein during Staphylococcus aureus pathogenesis. Proc Natl Acad Sci U S A. 2002;99:2293–2298. doi: 10.1073/pnas.032523999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mazmanian SK, Skaar EP, Gaspar AH, Humayun M, Gornicki P, et al. Passage of heme-iron across the envelope of Staphylococcus aureus. Science. 2003;299:906–909. doi: 10.1126/science.1081147. [DOI] [PubMed] [Google Scholar]
- 11.Pilpa RM, Fadeev EA, Villareal VA, Wong ML, Phillips M, et al. Solution structure of the NEAT (NEAr Transporter) domain from IsdH/HarA: the human hemoglobin receptor in Staphylococcus aureus. J Mol Biol. 2006;360:435–447. doi: 10.1016/j.jmb.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 12.Grigg JC, Vermeiren CL, Heinrichs DE, Murphy ME. Haem recognition by a Staphylococcus aureus NEAT domain. Mol Microbiol. 2007;63:139–149. doi: 10.1111/j.1365-2958.2006.05502.x. [DOI] [PubMed] [Google Scholar]
- 13.Maresso AW, Garufi G, Schneewind O. Bacillus anthracis secretes proteins that mediate heme acquisition from hemoglobin. PLoS Pathog. 2008;4:e1000132. doi: 10.1371/journal.ppat.1000132. doi: 10.1371/journal.ppat.1000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gat O, Zaide G, Inbar I, Grosfeld H, Chitlaru T, et al. Characterization of Bacillus anthracis iron-regulated surface determinant (Isd) proteins containing NEAT domains. Mol Microbiol. 2008;70:983–999. doi: 10.1111/j.1365-2958.2008.06460.x. [DOI] [PubMed] [Google Scholar]
- 15.Helgason E, Okstad OA, Caugant DA, Johansen HA, Fouet A, et al. Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis–one species on the basis of genetic evidence. Appl Environ Microbiol. 2000;66:2627–2630. doi: 10.1128/aem.66.6.2627-2630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kolsto AB, Lereclus D, Mock M. Genome structure and evolution of the Bacillus cereus group. Curr Top Microbiol Immunol. 2002;264:95–108. [PubMed] [Google Scholar]
- 17.Vilas-Boas GT, Peruca AP, Arantes OM. Biology and taxonomy of Bacillus cereus, Bacillus anthracis, and Bacillus thuringiensis. Can J Microbiol. 2007;53:673–687. doi: 10.1139/W07-029. [DOI] [PubMed] [Google Scholar]
- 18.Stenfors Arnesen LP, Fagerlund A, Granum PE. From soil to gut: Bacillus cereus and its food poisoning toxins. FEMS Microbiol Rev. 2008;32:579–606. doi: 10.1111/j.1574-6976.2008.00112.x. [DOI] [PubMed] [Google Scholar]
- 19.Miller JM, Hair JG, Hebert M, Hebert L, Roberts FJ, Jr, et al. Fulminating bacteremia and pneumonia due to Bacillus cereus. J Clin Microbiol. 1997;35:504–507. doi: 10.1128/jcm.35.2.504-507.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Callegan MC, Booth MC, Jett BD, Gilmore MS. Pathogenesis of gram-positive bacterial endophthalmitis. Infect Immun. 1999;67:3348–3356. doi: 10.1128/iai.67.7.3348-3356.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hilliard NJ, Schelonka RL, Waites KB. Bacillus cereus bacteremia in a preterm neonate. J Clin Microbiol. 2003;41:3441–3444. doi: 10.1128/JCM.41.7.3441-3444.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoffmaster AR, Ravel J, Rasko DA, Chapman GD, Chute MD, et al. Identification of anthrax toxin genes in a Bacillus cereus associated with an illness resembling inhalation anthrax. Proc Natl Acad Sci U S A. 2004;101:8449–8454. doi: 10.1073/pnas.0402414101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Slamti L, Lereclus D. A cell-cell signaling peptide activates the PlcR virulence regulon in bacteria of the Bacillus cereus group. EMBO J. 2002;21:4550–4559. doi: 10.1093/emboj/cdf450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gohar M, Faegri K, Perchat S, Ravnum S, Okstad OA, et al. The PlcR virulence regulon of Bacillus cereus. PLoS ONE. 2008;3:e2793. doi: 10.1371/journal.pone.0002793. doi: 10.1371/journal.pone.0002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salamitou S, Ramisse F, Brehelin M, Bourguet D, Gilois N, et al. The plcR regulon is involved in the opportunistic properties of Bacillus thuringiensis and Bacillus cereus in mice and insects. Microbiology. 2000;146 (Pt 11):2825–2832. doi: 10.1099/00221287-146-11-2825. [DOI] [PubMed] [Google Scholar]
- 26.Wilson MK, Abergel RJ, Raymond KN, Arceneaux JE, Byers BR. Siderophores of Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis. Biochem Biophys Res Commun. 2006;348:320–325. doi: 10.1016/j.bbrc.2006.07.055. [DOI] [PubMed] [Google Scholar]
- 27.Zawadzka AM, Abergel RJ, Nichiporuk R, Andersen UN, Raymond KN. Siderophore-mediated iron acquisition systems in Bacillus cereus: Identification of receptors for anthrax virulence-associated petrobactin. Biochemistry. 2009;48:3645–3657. doi: 10.1021/bi8018674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sato N, Kurotaki H, Watanabe T, Mikami T, Matsumoto T. Use of hemoglobin as an iron source by Bacillus cereus. Biol Pharm Bull. 1998;21:311–314. doi: 10.1248/bpb.21.311. [DOI] [PubMed] [Google Scholar]
- 29.Park RY, Choi MH, Sun HY, Shin SH. Production of catechol-siderophore and utilization of transferrin-bound iron in Bacillus cereus. Biol Pharm Bull. 2005;28:1132–1135. doi: 10.1248/bpb.28.1132. [DOI] [PubMed] [Google Scholar]
- 30.Sato N, Ikeda S, Mikami T, Matsumoto T. Bacillus cereus dissociates hemoglobin and uses released heme as an iron source. Biol Pharm Bull. 1999;22:1118–1121. doi: 10.1248/bpb.22.1118. [DOI] [PubMed] [Google Scholar]
- 31.Sato N, Kurotaki H, Ikeda S, Daio R, Nishinome N, et al. Lactoferrin inhibits Bacillus cereus growth and heme analogs recover its growth. Biol Pharm Bull. 1999;22:197–199. doi: 10.1248/bpb.22.197. [DOI] [PubMed] [Google Scholar]
- 32.Fedhila S, Daou N, Lereclus D, Nielsen-LeRoux C. Identification of Bacillus cereus internalin and other candidate virulence genes specifically induced during oral infection in insects. Mol Microbiol. 2006;62:339–355. doi: 10.1111/j.1365-2958.2006.05362.x. [DOI] [PubMed] [Google Scholar]
- 33.Dower WJ, Miller JF, Ragsdale CW. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lereclus D, Arantes O, Chaufaux J, Lecadet M. Transformation and expression of a cloned delta-endotoxin gene in Bacillus thuringiensis. FEMS Microbiol Lett. 1989;51:211–217. doi: 10.1016/0378-1097(89)90511-9. [DOI] [PubMed] [Google Scholar]
- 35.Arantes O, Lereclus D. Construction of cloning vectors for Bacillus thuringiensis. Gene. 1991;108:115–119. doi: 10.1016/0378-1119(91)90495-w. [DOI] [PubMed] [Google Scholar]
- 36.Vargas C, McEwan AG, Downie JA. Detection of c-type cytochromes using enhanced chemiluminescence. Anal Biochem. 1993;209:323–326. doi: 10.1006/abio.1993.1127. [DOI] [PubMed] [Google Scholar]
- 37.Fortinea N, Trieu-Cuot P, Gaillot O, Pellegrini E, Berche P, et al. Optimization of green fluorescent protein expression vectors for in vitro and in vivo detection of Listeria monocytogenes. Res Microbiol. 2000;151:353–360. doi: 10.1016/s0923-2508(00)00158-3. [DOI] [PubMed] [Google Scholar]
- 38.Fedhila S, Nel P, Lereclus D. The InhA2 metalloprotease of Bacillus thuringiensis strain 407 is required for pathogenicity in insects infected via the oral route. J Bacteriol. 2002;184:3296–3304. doi: 10.1128/JB.184.12.3296-3304.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guerout-Fleury AM, Shazand K, Frandsen N, Stragier P. Antibiotic-resistance cassettes for Bacillus subtilis. Gene. 1995;167:335–336. doi: 10.1016/0378-1119(95)00652-4. [DOI] [PubMed] [Google Scholar]
- 40.Bouillaut L, Ramarao N, Buisson C, Gilois N, Gohar M, et al. FlhA influences Bacillus thuringiensis PlcR-regulated gene transcription, protein production, and virulence. Appl Environ Microbiol. 2005;71:8903–8910. doi: 10.1128/AEM.71.12.8903-8910.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Finney DJ. London: Cambridge University Press; 1971. Probit Analysis. [Google Scholar]
- 42.Raymond M, Prato G, Ratsira D. Saint Georges d'Orgue, France: PRAXEME; 1993. PROBIT analysis of mortality assays displaying quantal response. [Google Scholar]
- 43.Locke MN, H. Iron economy in insects: transport, metabolism, and storage. Annu Rev Entomol. 1992;37:195–215. [Google Scholar]
- 44.Yuk JE, Seo DH, Choi CW, HAN J, Choi HK, PARK JB, Hwang SY, Koh SK, Yun CY. Characterization of Tissue-Ferritin Purified from Wax Moth. Entomol Reaserch. 2005;35:227–234. [Google Scholar]
- 45.Dunphy GB, Niven DF, Chadwick JS. Iron contributes to the antibacterial functions of the haemolymph of Galleria mellonella. J Insect Physiol. 2002;48:903–914. doi: 10.1016/s0022-1910(02)00159-2. [DOI] [PubMed] [Google Scholar]
- 46.Bullen JJ, Rogers HJ, Spalding PB, Ward CG. Natural resistance, iron and infection: a challenge for clinical medicine. J Med Microbiol. 2006;55:251–258. doi: 10.1099/jmm.0.46386-0. [DOI] [PubMed] [Google Scholar]
- 47.You SA, Wang Q. Ferritin in atherosclerosis. Clin Chim Acta. 2005;357:1–16. doi: 10.1016/j.cccn.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 48.Kim BS, Lee CS, Yun CY, Yeo SM, Park WM, et al. Characterization and immunological analysis of ferritin from the hemolymph of Galleria mellonella. Comp Biochem Physiol A Mol Integr Physiol. 2001;129:501–509. doi: 10.1016/s1095-6433(01)00287-2. [DOI] [PubMed] [Google Scholar]
- 49.Valenti P, Antonini G. Lactoferrin: an important host defence against microbial and viral attack. Cell Mol Life Sci. 2005;62:2576–2587. doi: 10.1007/s00018-005-5372-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lindback T, Okstad OA, Rishovd AL, Kolsto AB. Insertional inactivation of hblC encoding the L2 component of Bacillus cereus ATCC 14579 haemolysin BL strongly reduces enterotoxigenic activity, but not the haemolytic activity against human erythrocytes. Microbiology. 1999;145 (Pt 11):3139–3146. doi: 10.1099/00221287-145-11-3139. [DOI] [PubMed] [Google Scholar]
- 51.Andreeva ZI, Nesterenko VF, Yurkov IS, Budarina ZI, Sineva EV, et al. Purification and cytotoxic properties of Bacillus cereus hemolysin II. Protein Expr Purif. 2006;47:186–193. doi: 10.1016/j.pep.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 52.Fagerlund A, Lindback T, Storset AK, Granum PE, Hardy SP. Bacillus cereus Nhe is a pore-forming toxin with structural and functional properties similar to the ClyA (HlyE, SheA) family of haemolysins, able to induce osmotic lysis in epithelia. Microbiology. 2008;154:693–704. doi: 10.1099/mic.0.2007/014134-0. [DOI] [PubMed] [Google Scholar]
- 53.Agaisse H, Gominet M, Okstad OA, Kolsto AB, Lereclus D. PlcR is a pleiotropic regulator of extracellular virulence factor gene expression in Bacillus thuringiensis. Mol Microbiol. 1999;32:1043–1053. doi: 10.1046/j.1365-2958.1999.01419.x. [DOI] [PubMed] [Google Scholar]
- 54.Taylor JM, Heinrichs DE. Transferrin binding in Staphylococcus aureus: involvement of a cell wall-anchored protein. Mol Microbiol. 2002;43:1603–1614. doi: 10.1046/j.1365-2958.2002.02850.x. [DOI] [PubMed] [Google Scholar]
- 55.Clarke SR, Wiltshire MD, Foster SJ. IsdA of Staphylococcus aureus is a broad spectrum, iron-regulated adhesin. Mol Microbiol. 2004;51:1509–1519. doi: 10.1111/j.1365-2958.2003.03938.x. [DOI] [PubMed] [Google Scholar]
- 56.Kobe B, Kajava AV. The leucine-rich repeat as a protein recognition motif. Curr Opin Struct Biol. 2001;11:725–732. doi: 10.1016/s0959-440x(01)00266-4. [DOI] [PubMed] [Google Scholar]
- 57.Bierne H, Sabet C, Personnic N, Cossart P. Internalins: a complex family of leucine-rich repeat-containing proteins in Listeria monocytogenes. Microbes Infect. 2007;9:1156–1166. doi: 10.1016/j.micinf.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 58.Almeida RS, Brunke S, Albrecht A, Thewes S, Laue M, et al. the hyphal-associated adhesin and invasin Als3 of Candida albicans mediates iron acquisition from host ferritin. PLoS Pathog. 2008;4:e1000217. doi: 10.1371/journal.ppat.1000217. doi: 10.1371/journal.ppat.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kern JW, Schneewind O. BslA, a pXO1-encoded adhesin of Bacillus anthracis. Mol Microbiol. 2008;68:504–515. doi: 10.1111/j.1365-2958.2008.06169.x. [DOI] [PubMed] [Google Scholar]
- 61.Maresso AW, Chapa TJ, Schneewind O. Surface protein IsdC and Sortase B are required for heme-iron scavenging of Bacillus anthracis. J Bacteriol. 2006;188:8145–8152. doi: 10.1128/JB.01011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maresso AW, Schneewind O. Iron acquisition and transport in Staphylococcus aureus. Biometals. 2006;19:193–203. doi: 10.1007/s10534-005-4863-7. [DOI] [PubMed] [Google Scholar]
- 63.Muryoi N, Tiedemann MT, Pluym M, Cheung J, Heinrichs DE, et al. Demonstration of the iron-regulated surface determinant (Isd) heme transfer pathway in Staphylococcus aureus. J Biol Chem. 2008;283:28125–28136. doi: 10.1074/jbc.M802171200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Deneer HG, Healey V, Boychuk I. Reduction of exogenous ferric iron by a surface-associated ferric reductase of Listeria spp. Microbiology. 1995;141 (Pt 8):1985–1992. doi: 10.1099/13500872-141-8-1985. [DOI] [PubMed] [Google Scholar]
- 65.Whitby PW, Vanwagoner TM, Springer JM, Morton DJ, Seale TW, et al. Burkholderia cenocepacia utilizes ferritin as an iron source. J Med Microbiol. 2006;55:661–668. doi: 10.1099/jmm.0.46199-0. [DOI] [PubMed] [Google Scholar]
- 66.Vierstraete E, Verleyen P, Baggerman G, D'Hertog W, Van den Bergh G, et al. A proteomic approach for the analysis of instantly released wound and immune proteins in Drosophila melanogaster hemolymph. Proc Natl Acad Sci U S A. 2004;101:470–475. doi: 10.1073/pnas.0304567101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Larson JA, Howie HL, So M. Neisseria meningitidis accelerates ferritin degradation in host epithelial cells to yield an essential iron source. Mol Microbiol. 2004;53:807–820. doi: 10.1111/j.1365-2958.2004.04169.x. [DOI] [PubMed] [Google Scholar]
- 68.Fouet A, Mesnage S. Bacillus anthracis cell envelope components. Curr Top Microbiol Immunol. 2002;271:87–113. doi: 10.1007/978-3-662-05767-4_5. [DOI] [PubMed] [Google Scholar]
- 69.Jensen GB, Hansen BM, Eilenberg J, Mahillon J. The hidden lifestyles of Bacillus cereus and relatives. Environ Microbiol. 2003;5:631–640. doi: 10.1046/j.1462-2920.2003.00461.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Purification of IlsA. Coomassie-stained 10% SDS-PAGE analysis of the purified GST-IlsA and GST proteins from recombinant E. coli. Lane M, molecular weight markers in KDa. Purified GST-IlsA and GST with apparent molecular weights shown on the gel, was loaded in lane 1 and 2 respectively.
(0.51 MB PDF)