Abstract
Background
A somatic mutation in the FOXL2 gene is reported to be present in almost all (97%; 86/89) morphologically defined, adult-type, granulosa-cell tumors (A-GCTs). This FOXL2 c.402C>G mutation changes a highly conserved cysteine residue to a tryptophan (p.C134W). It was also found in a minority of other ovarian malignant stromal tumors, but not in benign ovarian stromal tumors or unrelated ovarian tumors or breast cancers.
Methodology/Principal Findings
Herein we studied other cancers and cell lines for the presence of this mutation. We screened DNA from 752 tumors of epithelial and mesenchymal origin and 28 ovarian cancer cell lines and 52 other cancer cell lines of varied origin. We found the FOXL2 c.402C>G mutation in an unreported A-GCT case and the A-GCT-derived cell line KGN. All other tumors and cell lines analyzed were mutation negative.
Conclusions/Significance
In addition to proving that the KGN cell line is a useful model to study A-GCTs, these data show that the c.402C>G mutation in FOXL2 is not commonly found in a wide variety of other cancers and therefore it is likely pathognomonic for A-GCTs and closely related tumors.
Introduction
Malignant adult ovarian granulosa-cell tumors (A-GCTs) are malignant sex cord-stromal tumors known for their genomic stability and varied prognosis [1]. Until recently, there has been little insight into the molecular characteristics of A-GCTs. Using whole-transcriptome paired-end RNA sequencing, we identified a somatic missense mutation (c.402C>G, p. Cys134Trp) in the Forkhead transcription factor gene, FOXL2 [2]. This mutation was present in 97% of 89 morphologically identified A-GCTs [2]. Foxl2 has been shown to be crucial for granulosa-cell differentiation [3]. This was the first association of a somatic mutation in FOXL2 associated with cancer, however aberrant expression of Foxl2 has been reported in juvenile granulosa-cell tumor of the testis [4]. The mutation was also found at a lower frequency in two other related ovarian stromal tumors; 21% (3/14) thecomas and 10% (1/10) juvenile-type GCTs were mutation positive [2]. This single, recurrent mutation suggests that it is characteristic of granulosa-cell tumors, and its high frequency implies that it is potentially a driver in disease initiation.
To determine the specificity of this somatic mutation, high resolution melting or polymerase chain reaction (PCR) -based allelic discrimination was used to screen a diverse collection of tumors and ovarian tumor cell lines. Additional cytogenetic analysis was performed to demonstrate the stable karyotype of the A-GCT cell line, KGN [5].
Materials and Methods
Samples for the high resolution melt assay were purchased as either DNA or tissue blocks from vendors who provided unlinked anonymized specimens collected in accordance with applicable review boards approval, regulations and laws. Novartis does not require an ethical review committee for samples collected in this manner. Control DNA, used to validate the high resolution melt assay, was extracted from anonymized tumor specimens compiled by the frozen tumor bank, OvCaRe (Ovarian Cancer Research), under written informed consent. Approval for analysis of these samples for the FOXL2 mutation was obtained through the British Columbia Cancer Agency's research ethics board.
Seven hundred and fifty-two tumor DNA samples, of epithelial and mesenchymal origin (Table 1) were screened with a high resolution melting assay run on the LightScanner™ instrument (Idaho Technology Inc., Salt Lake City, Utah) [6], [7]. For each tumor block, malignant cells composed >50% of the cellularity and matched normal adjacent tissue was available for all cases. The assay was designed to detect sequence variants in the region from Ile102 to Phe138 in Foxl2 (NP_075555.1). Since FOXL2 is a single exon gene, PCR primers were placed in the coding region (forward primer 5′ AGAAGGGCTGGCAAAATAGC, reverse primer 5′ GCCGGTAGTTGCCCTTCT) resulting in a 150 base pair amplicon.
Table 1. Summary of tumor types screened by High Resolution Melt Curve Analysis (HRM).
| Total cases n = 752 (excluding controls) | Normal by HRM | Confirmed positive for FOXL2 c.402C>G mutation out of HRM positive cases | |
| Ovarian cancer negative controls | 14 | 11 | 0/3* |
| Ovarian A-GCT positive controls (including an unreported A-GCT case and the A-GCT cell line, KGN) | 13 | 0 | 13/13 |
| Bladder Cancer | 40 | 40 | |
| Breast Cancer | 74 | 71 | 0/3* |
| Carcinoid Cancer | 8 | 8 | |
| Cervical Cancer | 16 | 16 | |
| Colorectal Cancer | 77 | 75 | 0/2* |
| Endometrial Cancer | 12 | 12 | |
| Esophageal Cancer | 21 | 21 | |
| Gastric Cancer | 90 | 89 | 0/1* |
| Head & Neck Cancer | 28 | 26 | 0/2* |
| Hepatic (HCC & Cholangiocarcinoma) | 14 | 14 | |
| Lung Cancer (All types) | 125 | 123 | 0/2* |
| Melanoma | 31 | 31 | |
| Ovarian Cancer | 32 | 32 | |
| Pancreatic Cancer | 4 | 4 | |
| Prostate Cancer | 37 | 37 | |
| Renal Cancer | 52 | 51 | 0/1* |
| Leiomyosarcoma | 15 | 15 | |
| Malignant fibrous histiocytoma-pleomorphic sarcoma | 8 | 8 | |
| Rhabdomyosarcoma | 2 | 2 | |
| Liposarcoma | 4 | 4 | |
| Fibrosarcoma | 1 | 1 | |
| Testicular Cancer | 19 | 18 | 0/1* |
| Thyroid Cancer | 42 | 42 |
Sequence data is available for all screen positive samples.
Variants seen on HRM screen but not confirmed by sequencing (HRM false positive results).
The primary screen used whole genome amplified (Qiagen Repli-G kit) DNA derived from frozen tissue blocks of untreated primary tumors. All samples which had an aberrant melting curve or which failed to amplify in the initial screen were followed up with a repeat HRM assay using unamplified DNA prepared from tumor and adjacent normal tissue. Tumor samples which were repeat positive for an aberrant melting curve were sequenced in duplicate, and the resulting sequence trace files were analyzed for mutations using the phrap/phred/consed software package (www.phrap.org). DNA from 27 ovarian tumor samples previously genotyped for the mutation using a previously validated TaqMan real-time PCR-based allelic discrimination assay (Applied Biosystems, Foster City, CA) specific for the FOXL2 c.402C>G mutation [2] were used to validate the performance of the HRM assay. This included an unreported A-GCT case and the cell line KGN.
To establish the specificity of the FOXL2 c.402C>G mutation in ovarian cancer cell lines, we used the same TaqMan real-time PCR-based allelic discrimination assay to genotype 28 ovarian cancer cell lines and 52 cancer cell lines of different tissue origin for the FOXL2 c.402C>G mutation (Table S1).
To assess the cytogenetic profile of KGN, we utilized 24-color fluorescence in situ hybridization (FISH) (24XCyte, MetaSystems, Cat. D-0125-120-MC) and analyzed the results using the Axioplan 2, Zeiss,(MetaSystems, Isis), camera VAC-30054.
Results
All 11 previously reported FOXL2 c.402C>G mutation-positive A-GCT specimens as well as an unreported A-GCT case and the A-GCT cell line, KGN, validated the HRM assay by demonstrating a variant melt curve distinct from the common (wild-type) pattern. None of the 14 FOXL2 c.402C>G mutation negative samples exhibited this variant melt profile. However, three of the 10 high grade serous ovarian cancers showed an alternative variant profile; sequencing confirmed them to be false positives.
The primary screen of 752 whole genome amplified tumor DNA samples yielded 24 samples (∼4%) with a variant profile, distinct from that seen in association with the FOXL2 c.402C>G mutation-positive A-GCT specimens, as well as 41 with an indeterminate profile and 29 samples that failed. The secondary screen was performed on this set of 94 samples using unamplified genomic DNA derived from tumors and matching normal specimens. Eighty-two of the samples were found to be false positives where there was no variant profile seen between the tumor and normal DNA. Twelve of the samples remained indeterminate and were subsequently sequenced and confirmed to be false positives.
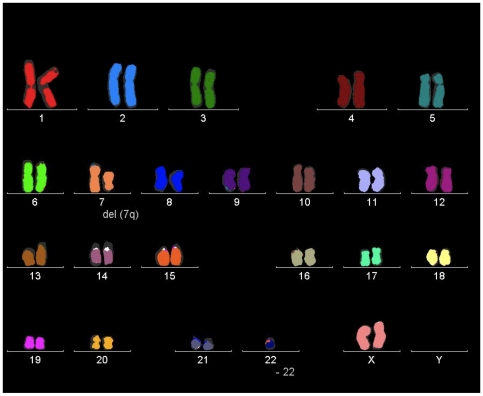
The granulosa-cell line KGN, which was derived through long-term passage of a recurrent A-GCT [5], was the only cell line found to harbor the mutation. The mutation was not present in an SVOG granulosa-cell line, immortalized by SV40 [8] or 26 other ovarian cancer derived cell lines. Unlike most ovarian cancer derived cell lines KGN shows relative genomic stability (Figure 1). In addition to deletion of 7q, it is monosomic for chromosome 22 which is the most frequent cytogenetic abnormality seen in A-GCTs [9]; another feature demonstrating its similarities to A-GCTs.
Figure 1. Cytogenetic analysis of the KGN cell line.
24-color fluorescence in situ hybridization (FISH) demonstrates the tumor cell line's stable karyotype 45, XX, 7q-, -22 consistent with the original publication [5]. FOXL2 is located at 3q23. Images were obtained using the Axioplan 2, Zeiss, (MetaSystems, Isis), camera VAC-30054.
Discussion
Loss-of-function germline mutations in FOXL2 are associated with blepharophimosis–ptosis–epicanthus–inversus syndrome [BPES;OMIM#110100]; an autosomal dominant developmental disorder characterized by eyelid malformations and premature ovarian failure due to a dysfunction of granulosa-cells [10]. The FOXL2 c.402C>G mutation is seen in the heterozygous state in most A-GCTs. Unlike in BPES, where germline FOXL2 mutations are spread across the gene [11], the somatic FOXL2 mutation in A-GCTs involves the same base pair in all cases. This favors a specific functional consequence such as a dominant negative effect or a change or gain of function as opposed to a generic loss of function and the ultimate impact of this mutation is oncogenic. Additionally, immunohistochemical data indicating that Foxl2 expression is maintained in the nuclei in A-GCTs, that were heterozygous or appeared to be hemizygous or homozygous for the mutation, implies that this mutation does not affect protein localization [2].
Analysis of 28 various ovarian cancer-derived cell lines demonstrated that the mutation was only present in the granulosa-cell tumor cell line, KGN, suggesting that it is molecularly akin to A-GCTs. The presence of the missense mutation in the well-characterized A-GCT cell line, KGN, is in keeping with the high frequency of the somatic mutation in A-GCTs and supports the use of this cell model to study the properties of this ovarian sex cord stromal tumor. This cell line has been used in a number of elegant studies which have addressed the question of the function of Foxl2 [12] and the effects of FOXL2 missense, haploinsufficient or hypomorphic mutations associated with BPES [13]–[15]. Further dissection of these phenomena with attention to the possible confounding effects of this mutation in one copy of the endogenous gene may elucidate the function of this missense mutation in the granulosa-cell tumor.
The absence of the FOXL2 c.402C>G mutation in this large series of common epithelial malignancies such as lung, colorectal, breast, gastric, bladder, thyroid, prostate, melanoma and ovarian carcinoma, in addition to a range of less frequent tumors, implies a high specificity of this recurrent mutation for ovarian sex cord stromal tumors. This study does not exclude the possibility that the mutation could be found in other rare or related neoplasms such as testicular stromal tumors. As the mutation was not found in non-GCT ovarian tumor cell lines and the SV40 transformed granulosa-cell line, SVOG, provides further support of its likely role in A-GCT disease initiation. Considering the extremely high frequency of this mutation in morphologically selected A-GCTs (97%) [2], these data provide further evidence suggesting that the mutation is also specific for this tumor type and could be useful as a diagnostic test. Further studies will be required to determine the relevance of the mutation in other sex cord stromal tumors of the ovary, however, it is possible that all mutation positive tumors could ultimately be considered to be a single entity of which the major component would be A-GCTs.
Supporting Information
Cell lines screened by TaqMan real-time PCR-based allelic discrimination assay for the FOXL2 c. 402 C>G mutation. Ovarian cancer cell lines are italicized and ovarian granulosa-cell-derived lines are underlined.
(0.02 MB XLS)
Acknowledgments
We would like to thank the National Cancer Institute for providing us with the NCI-60 cell line panel from the Division of Cancer Treatment and Diagnosis (DCTD) Tumor Repository.
Footnotes
Competing Interests: The British Columbia Cancer Agency is the holder of a provisional patent on the use of the FOXL2 mutation as a potential diagnostic and therapeutic target. Bella Gorbatcheva and John Monahan are both employees of the Novartis Institutes for BioMedical Research. These competing interests do not impact the sharing of data related to this publication.
Funding: This work was supported by a grant from the Canadian Institutes of Health Research MOP 97735. CIHR had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The high resolution melt assay used to screen the solid tumors was internally funded and performed at the Novartis Institutes for BioMedical Research. John Monahan and Bella Gorbatcheva of the Novartis Institutes for BioMedical Research designed this portion of the study, collected and analysed the resulting data. The decision to publish and preparation of the manuscript was done in collaboration with John Monahan and Bella Gorbatcheva.
References
- 1.Koukourakis GV, Kouloulias VE, Koukourakis MJ, Zacharias GA, Papadimitriou C, et al. Granulosa cell tumor of the ovary: Tumor review. Integr Cancer Ther. 2008;7(3):204–215. doi: 10.1177/1534735408322845. [DOI] [PubMed] [Google Scholar]
- 2.Shah SP, Kobel M, Senz J, Morin RD, Clarke BA, et al. Mutation of FOXL2 in granulosa-cell tumors of the ovary. N Engl J Med. 2009 doi: 10.1056/NEJMoa0902542. [DOI] [PubMed] [Google Scholar]
- 3.Schmidt D, Ovitt CE, Anlag K, Fehsenfeld S, Gredsted L, et al. The murine winged-helix transcription factor Foxl2 is required for granulosa cell differentiation and ovary maintenance. Development. 2004;131(4):933–942. doi: 10.1242/dev.00969. [DOI] [PubMed] [Google Scholar]
- 4.Kalfa N, Fellous M, Boizet-Bonhoure B, Patte C, Duvillard P, et al. Aberrant expression of ovary determining gene FOXL2 in the testis and juvenile granulosa cell tumor in children. J Urol. 2008;180(4 Suppl):1810–1813. doi: 10.1016/j.juro.2008.03.097. [DOI] [PubMed] [Google Scholar]
- 5.Nishi Y, Yanase T, Mu Y, Oba K, Ichino I, et al. Establishment and characterization of a steroidogenic human granulosa-like tumor cell line, KGN, that expresses functional follicle-stimulating hormone receptor. Endocrinology. 2001;142(1):437–445. doi: 10.1210/endo.142.1.7862. [DOI] [PubMed] [Google Scholar]
- 6.Reed GH, Wittwer CT. Sensitivity and specificity of single-nucleotide polymorphism scanning by high-resolution melting analysis. Clin Chem. 2004;50(10):1748–1754. doi: 10.1373/clinchem.2003.029751. [DOI] [PubMed] [Google Scholar]
- 7.Herrmann MG, Durtschi JD, Wittwer CT, Voelkerding KV. Expanded instrument comparison of amplicon DNA melting analysis for mutation scanning and genotyping. Clin Chem. 2007;53(8):1544–1548. doi: 10.1373/clinchem.2007.088120. [DOI] [PubMed] [Google Scholar]
- 8.Lie BL, Leung E, Leung PC, Auersperg N. Long-term growth and steroidogenic potential of human granulosa-lutein cells immortalized with SV40 large T antigen. Mol Cell Endocrinol. 1996;120(2):169–176. doi: 10.1016/0303-7207(96)03835-x. [DOI] [PubMed] [Google Scholar]
- 9.Mayr D, Hirschmann A, Marlow S, Horvath C, Diebold J. Analysis of selected oncogenes (AKT1, FOS, BCL2L2, TGFbeta) on chromosome 14 in granulosa cell tumors (GCTs): A comprehensive study on 30 GCTs combining comparative genomic hybridization (CGH) and fluorescence-in situ-hybridization (FISH). Pathol Res Pract. 2008;204(11):823–830. doi: 10.1016/j.prp.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 10.Crisponi L, Deiana M, Loi A, Chiappe F, Uda M, et al. The putative forkhead transcription factor FOXL2 is mutated in blepharophimosis/ptosis/epicanthus inversus syndrome. Nat Genet. 2001;27(2):159–166. doi: 10.1038/84781. [DOI] [PubMed] [Google Scholar]
- 11.Beysen D, De Jaegere S, Amor D, Bouchard P, Christin-Maitre S, et al. Identification of 34 novel and 56 known FOXL2 mutations in patients with Blepharophimosis syndrome. Hum Mutat. 2008;29(11):E205–19. doi: 10.1002/humu.20819. [DOI] [PubMed] [Google Scholar]
- 12.Batista F, Vaiman D, Dausset J, Fellous M, Veitia RA. Potential targets of FOXL2, a transcription factor involved in craniofacial and follicular development, identified by transcriptomics. Proc Natl Acad Sci U S A. 2007;104(9):3330–3335. doi: 10.1073/pnas.0611326104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benayoun BA, Batista F, Auer J, Dipietromaria A, L'Hote D, et al. Positive and negative feedback regulates the transcription factor FOXL2 in response to cell stress: Evidence for a regulatory imbalance induced by disease-causing mutations. Hum Mol Genet. 2009;18(4):632–644. doi: 10.1093/hmg/ddn389. [DOI] [PubMed] [Google Scholar]
- 14.Beysen D, Moumne L, Veitia R, Peters H, Leroy BP, et al. Missense mutations in the forkhead domain of FOXL2 lead to subcellular mislocalization, protein aggregation and impaired transactivation. Hum Mol Genet. 2008;17(13):2030–2038. doi: 10.1093/hmg/ddn100. [DOI] [PubMed] [Google Scholar]
- 15.Moumne L, Dipietromaria A, Batista F, Kocer A, Fellous M, et al. Differential aggregation and functional impairment induced by polyalanine expansions in FOXL2, a transcription factor involved in cranio-facial and ovarian development. Hum Mol Genet. 2008;17(7):1010–1019. doi: 10.1093/hmg/ddm373. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cell lines screened by TaqMan real-time PCR-based allelic discrimination assay for the FOXL2 c. 402 C>G mutation. Ovarian cancer cell lines are italicized and ovarian granulosa-cell-derived lines are underlined.
(0.02 MB XLS)