Abstract
Sensitive molecular methods, such as the PCR, can detect low-level contamination, and careful technique is required to reduce the impact of contaminants. Yet, some assays that are designed to detect high copy-number target sequences appear to be impossible to perform without contamination, and frequently, personnel or laboratory environment are held responsible as the source. This complicates diagnostic and research analysis when using molecular methods. To investigate the air specifically as a source of contamination, which might occur during PCR setup, we exposed tubes of water to the air of a laboratory and clean hood for up to 24 h. To increase the chances of contamination, we also investigated a busy open-plan office in the same way. All of the experiments showed the presence of human and rodent DNA contamination. However, there was no accumulation of the contamination in any of the environments investigated, suggesting that the air was not the source of contamination. Even the air from a busy open-plan office was a poor source of contamination for all of the DNA sequences investigated (human, bacterial, fungal, and rodent). This demonstrates that the personnel and immediate laboratory environment are not necessarily to blame for the observed contamination.
Keywords: real-time PCR, qPCR, Alu repeat, B1 element, 16S rDNA
As one of the most sensitive methods available for detecting nucleic acids, the PCR is at risk of being affected by low levels of contamination. This susceptibility is compounded by the fact that PCR functions by generating billions of copies of the DNA sequence that is being analyzed. Consequently, conducting the PCR reaction generates products, which if not handled carefully, may contaminate later reactions.1 As PCR has developed over the last 20 years, specific practices have been introduced to reduce laboratory contamination, including physical separation of the different stages of the procedure,2 incorporating enzymatic3 or irradiation4 steps to remove contaminating molecules, and generating a careful approach to identify contamination risk during sampling, sample processing, and analysis.5 Despite this, PCR can detect low levels of extraneous template DNA, which may be described as a contaminant, and the targeting of certain sequences are more likely to be affected by contamination than others.
Detecting the 16S ribosomal sequences for broad species bacterial detection can be difficult to perform without detecting low levels of background contamination.6–8 This is partly because the enzymes involved in the reaction are generated using recombinant techniques in bacteria, and it is difficult to purify the enzyme completely free of DNA.8 The laboratory environment has also been blamed for the source of contaminating 16S rDNA.9 As the ribosomal sequences are frequently multicopy, their increased abundance compounds the problem. Bacterial and fungal DNA has also been found to contaminate the reaction during sampling and extraction procedures necessary prior to DNA analysis.10–13
High-copy DNA targets are also problematic when detecting human DNA, for example, Alu14 or mitochondrial15 sequences. It has been assumed frequently that the contamination source is the laboratory environment inhabited by humans performing the work.16,17 This type of contamination has often caused problems, especially when trying to detect low copy-number DNA.15 Human DNA contamination has proved a major source of criticism in specialist fields such as ancient DNA detection.18
Here, we ask whether the laboratory working environment is likely to be the source of high copy-number DNA contamination. To address this question specifically, we investigated contamination by high copy-number human DNA sequences from the air of different environments over a 24-h period.
MATERIALS AND METHODS
Exposure Experiment
UltraPure DNase/RNase-free distilled water (1 ml; Invitrogen, Carlsbad, CA, USA) was added to 63 1.5 ml nonstick microtubes (Alpha Laboratories, Eastleigh, UK). The tubes were placed at three different locations: a UV-sterilized/hepa-filtered PCR hood; a bench in the pre-PCR laboratory; and on a desk at the entrance to an open-plan office shared by 14 people. Three tubes were left open for each time-point: 0 min, 5 min, 15 min, 30 min, 60 min, 360 min (6 h), and 1440 min (24 h). At the end of each time-point, aliquots of the samples were made for all subsequent reactions and stored at −20°C for no more than 2 weeks. Aliquots of 5 μl of the exposed water were added to the respective real-time PCR reactions.
DNase/RNase-free distilled water (1 ml; Invitrogen, Carlsbad, CA, USA) was added to 63 1.5 ml nonstick microtubes (Alpha Laboratories, Eastleigh, UK). The tubes were placed at three different locations: a UV-sterilized/hepa-filtered PCR hood; a bench in the pre-PCR laboratory; and on a desk at the entrance to an open-plan office shared by 14 people. Three tubes were left open for each time-point: 0 min, 5 min, 15 min, 30 min, 60 min, 360 min (6 h), and 1440 min (24 h). At the end of each time-point, aliquots of the samples were made for all subsequent reactions and stored at −20°C for no more than 2 weeks. Aliquots of 5 μl of the exposed water were added to the respective real-time PCR reactions.
Real-Time PCR
Four real-time PCR reactions (Tables 1 and 2, Supplemental Material) were used in this study to examine if the level of contamination increases with the length of exposure time in different environments. Reaction designations are author-denoted or taken from the appropriate original reference. Reactions were designed to follow the Minimum Information for Publication of Quantitative Real-Time PCR Experiments guidelines (see Table 3, Supplemental Material).(19) Reactions targeted human high copy-number DNA (Alu-J; Alu J short interspersed nuclear element), mouse high copy-number DNA (B1; B1 short interspersed nuclear element), bacterial DNA (16S rDNA; bacterial 16S ribosomal sequence), or fungal DNA (ITS2; fungal ribosomal internal-transcribed spacer 2 region). Primers were synthesized by Sigma-Aldrich (Cambridge, UK) and purified using the manufacturer’s salt-free purification. All real-time PCR reactions were set up manually and conducted in 12.5 μl vol using a Rotorgene 6000 thermocycler and associated 100 μl tubes (Corbett Research, Cambridge, UK). Reactions were optimized by varying temperatures, times, and primer concentrations, and PCR amplification efficiencies were estimated using dilution series (described below), according to the formula E = 10(−1/slope)− 1. Table 1 in the Supplemental Material refers to the different Taq polymerase sources used in the different reactions, which were run for 45 cycles using assay-specific optimal parameters (Table 2, Supplemental Material) and were considered negative if no DNA accumulated before Cycle 40. All reagents—primer, water, tubes, and pipette tips—were newly purchased and not opened before performing the experiments.
Generation of Standard Curves
Human genomic DNA, Balb/c mouse cDNA, and Candida dubliniensis DNA were used as templates for PCR reactions with the Alu-J, B1, and ITS2 primers, respectively (Table 1, Supplemental Material). The PCR products of the Alu-J, B1, and ITS2 reactions were cloned into plasmid vectors [Alu-J: pGEMTeasy vector (Promega, Southampton, UK); B1 and ITS2: pCR4 TOPO (Invitrogen, Paisley, UK)], following the manufacturers’ instructions. Recombinant plasmids were isolated using the QIAprep Spin Miniprep kit (Qiagen, Crawley, UK), quantified with the NanoDrop Spectrophotometer ND-1000 (NanoDrop Technologies, Wilmington, DE, USA) at OD260, and sequenced using the 3730XL genetic analyzer along with the AB SeScanner software (Applied Biosystems, Warrington, UK). The resulting sequences were compared with those stored in the Genbank database using the BLAST alignment software to confirm the presence of the Alu-J, B1, and ITS2 sequences. Tenfold dilutions of the linearized plasmids (500–5×107) copies/reaction) were used as a standard curve in all real-time PCR reactions. The lowest dilution in these reactions was higher than we normally perform as a result of the endogenous contamination being >50 or >5 copy standards that we would usually include. This is an inherent problem when investigating contamination of this kind, and thus, copy-number estimation must be made that is outside of the range of the standard curve.
Escherichia coli DNA was used as the template for the standard curve in the 16S real-time PCR assay by preparing a fivefold dilution series from 80–5×105 copies/reaction.
Statistical Analysis
Distribution of contaminating DNA results was assessed by D’Agostino and Pearson Omnibus Normality test, and distributions were considered not normal when P = <0.05.
RESULTS
Real-Time PCR
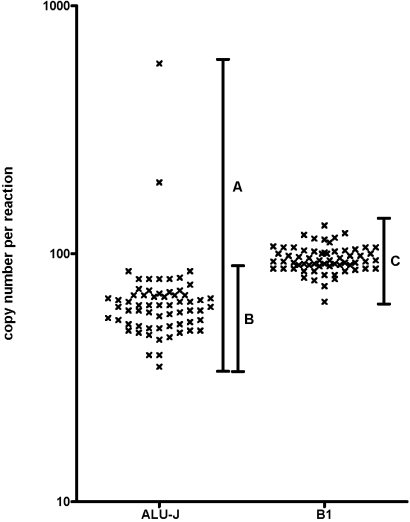
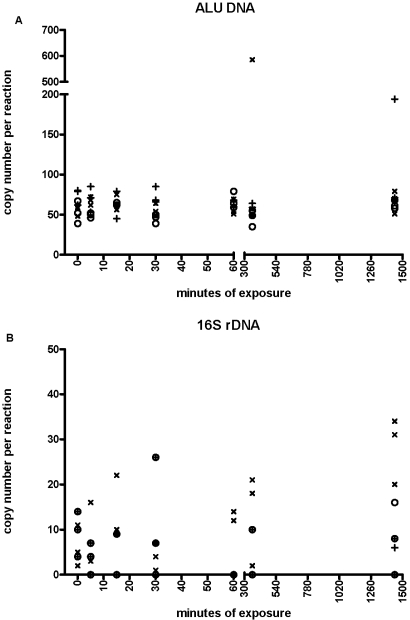
Alu-J reactions, performed with water, exposed to different environments for different periods of time, produced a product in all time-points, including Time 0. When all data were compared, there was a mean copy number of 71 copies/reaction, but the data were not distributed normally (Figure 1).There was no associated increase in the amount of Alu-J DNA detected with time, although there were two increased results: one at 6 h from the laboratory bench and one at 24 h from the open-plan office (Figure 2A), which were outside of the 3.09 sd from the mean (99.8% contained in interval). If we omit these two outliers, the distribution becomes normal with a mean of 61 copies/reaction and a coefficient of variation (CV) of 18.79% (Fig. 1).
FIGURE 1.
All Alu-J and B1 experimental data plotted from all exposure experiments. (A) Total spread of Alu-J results; (B) spread of Alu-J results minus outliers; (C) spread of B1 results.
FIGURE 2.
Effect of exposure to air from clean hood (○), laboratory (×), and open-plan office (+) on the detection of (A) Alu-J DNA sequences and (B) 16S rDNA.
Detection of 16S rDNA bacterial contamination was less frequent than with the Alu-J reaction (Fig. 2, B). The 38 exposure samples that did yield a result had a mean of eight copies/reaction. 16S rDNA contamination did not increase over time in any of the environments. When the same samples were analyzed using the mouse B1 element primers, all time-points yielded a result that had a normal distribution with a mean and CV of 95 copies/reaction and 12.29%, respectively (Fig. 1). Furthermore, as with the Alu-J and 16S rDNA reactions, there was no increase in mouse DNA contamination with time (data not shown).
Contamination assessment using the broad-spectrum fungal ITS reaction demonstrated that where it occurred, it was sporadic, occurring four times in the 63 reactions performed for all exposure samples. Approximate copy numbers in the four contaminated cases were approximately one, four, four, and seven copies/reaction, and no trend with replicates or time was observed (data not shown).
Sequencing Analysis
Sequence analysis confirmed that the molecules being amplified by the respective primer sets were Alu or B1 sequences (Figure 3).
FIGURE 3.
Sequences derived from PCR reactions from 0- and 1440-min exposure reactions with Alu-J (A) and B1 (B) primers. Boxes depict location of the primers.
DISCUSSION AND CONCLUSION
In this study, we investigated if the air of three different environments was a contamination source of four DNA sequences. Frequently, it is assumed that contamination from these selected high-abundance sequences, such as the human Alu element, are from the environment. If the environment were the source of observed contamination, then the experiment would have measured an accumulation of contaminants over time so that tubes left open for longer would be contaminated with more DNA than those left open for a shorter period of time.
There was no detectable increase over time with the 16S reaction. Because of the recognized problem of rDNA contamination,6–8 we chose the Amplitaq Gold enzyme from Applied Biosystems, as it is reported to have only ∼10 copies of 16S template/5 units Taq polymerase, so it is likely that this is what was being picked up by the data presented in Figure 2, B. Where our assay detected nothing in 25 of 63 samples, it is likely that we were at the limit of detection of this particular reaction. These data demonstrate that it is actually quite difficult to contaminate an open tube with bacterial DNA from the air, even if it is left open for 24 h in a busy office.
The same phenomenon was observed when measuring human DNA. The Alu sequence is a repeat that comprises 10% of the human genome, so within every cell, there are over 1 million copies of this sequence.14 It is generally assumed and has been reported that the inability to perform a PCR reaction negative for this amplicon is because this sequence is so numerous that ambient DNA is present in the air or water.(16) Furthermore, this has been suggested to be “an intrinsic phenomenon of an environment inhabited by humans that includes laboratory area, reagents and equipment.”(17) Our data suggest that at least in our hands, this is not correct. If the air from the laboratory area (or open-plan office with 14 people in it) were the source, then we would have seen an accumulation in the amount of this contamination over time. This was not the case, leading us to conclude that the air in these environments was not the source of most of the reported contamination.
We reported two occasions of an Alu measurement, which fell outside the 99.8% interval: one at 6 h and one at 24 h, where the results were ∼585 and ∼194 copies/reaction, respectively. None of the other respective replicate samples displayed comparable quantities of contaminating DNA. These two examples of higher copy-number contamination represent the only cases of environmental-sourced contamination in our hands. This would have occurred during the experiment or from the user and/or equipment during the manipulation of the tubes. Either way, the infrequency of this increased measurement demonstrates that contamination of human DNA was rare. If these two statistical outliers are omitted, the findings of contamination were very consistent (Fig. 1), with a mean of 61 copies/reaction and a CV of 18.79%. This CV represents a highly reproducible measurement with low error that further supports the notion that the source of the “effective laboratory background” was not derived from the air, the users, or their equipment. This is because the variation associated from DNA falling from a pipette during tube manipulation (for example) would be considerably more variable. To obtain replicates deliberately with such a CV by quantitative PCR requires careful experimental procedure (skilled pipetting, freshly thawed samples, etc.), and thus, we speculate that the low error of the observed effective laboratory background can only be explained if this contamination were already present in one or more of the reaction components that made up the master mix.
Identification of the source of this contamination would be a complicated and expensive set of experiments to perform, as a result of the fact that there are a vast number of suppliers of various reagents (which would need to be investigated to avoid prejudice), and any method used to remove contaminating DNA (enzymatic digestion, UV treatment, etc.) will have a detrimental effect on some of the constituent components of the PCR reagents (e.g., primers, dNTPs). We have performed preliminary experiments using UV radiation, which suggest the molecular grade UltraPure DNase/RNase-free distilled water (Invitrogen, Carlsbad, CA, USA) was not the source of the observed contamination (data not shown), but we feel that responsibility for identification and subsequent publication of the presence of contaminating DNA from molecular grade reagents rests with the suppliers.
DNase/RNase-free distilled water (Invitrogen, Carlsbad, CA, USA) was not the source of the observed contamination (data not shown), but we feel that responsibility for identification and subsequent publication of the presence of contaminating DNA from molecular grade reagents rests with the suppliers.
Our final observation was the presence of the rodent B1 element in all samples. There was no occasion of an increase above the effective laboratory background during the experiment. The CV was even less variable than with the Alu at 12.29%, with a mean of 95 copies/reaction. As with the Alu data, both observations support the reagents as the source of contaminating DNA, and we predicted that the mouse B1 element contamination was most likely a result of the hot-start mAb used to inactivate the Taq polymerase used by this reaction. Again, information about contaminating mouse DNA should be provided by the suppliers.
In conclusion, the absence of accumulation of contaminating DNA over time demonstrates that the air is unlikely to be the source of the observed high copy DNA sequence contamination, as speculated previously.(16,17) Additionally, the low copy-number variation observed by the ubiquitous contamination supports the finding further that this must have been present in one or more components of the master mix. Further work would be required to identify which component(s) contained contaminants; we did not do this, as the remit of our study was to investigate the air specifically as a contamination source and whether this was the cause of the contamination by high copy-number sequences, as has been assumed frequently. Laboratory air was not the source of the reported contamination in our hands, and we would advise researchers to include their reagents as a potential contaminant source when investigating PCR contamination.
ACKNOWLEDGMENTS
This work was funded by the European Commission through the Framework Six Program (Project LSHP-CT-2006-037785) and the Marie Curie Early Stage Training Site GALTRAIN (Project MEST-CT-2005-020524). We thank Tanya Novak and Clare Green for their assistance with experimentation and critical review of the manuscript; Tania Nolan, of Sigma Genosys, for her assistance with assay design and optimization; and Martin Lee, of Enigma Diagnostics Ltd., for his useful insights and discussion.
Footnotes
The authors declare no financial support or associations that may pose a conflict of interest within this manuscript.
REFERENCES
- 1.Stals A, Werbrouck H, Baert L, et al. Laboratory efforts to eliminate contamination problems in the real-time RT-PCR detection of Noroviruses. J Microbiol Methods 2009; 77: 72– 76 [DOI] [PubMed] [Google Scholar]
- 2.Kwok S, Higuchi R. Avoiding false positives with PCR. Nature 1989; 339: 237– 238 [DOI] [PubMed] [Google Scholar]
- 3.Longo MC, Berninger MS, Hartley JL. Use of uracil DNA glycosylase to control carry-over contamination in polymerase chain reactions. Gene 1990; 93: 125– 128 [DOI] [PubMed] [Google Scholar]
- 4.Tamariz J, Voynarovska K, Prinz M, Caragine T. The application of ultraviolet irradiation to exogenous sources of DNA in plasticware and water for the amplification of low copy number DNA. J Forensic Sci 2006; 51: 790– 794 [DOI] [PubMed] [Google Scholar]
- 5.Millar BC, Xu J, Moore JE. Risk assessment models and contamination management: implications for broad-range ribosomal DNA PCR as a diagnostic tool in medical bacteriology. J Clin Microbiol 2002; 40: 1575– 1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corless CE, Guiver M, Borrow R, et al. Contamination and sensitivity issues with a real-time universal 16S rRNA PCR. J Clin Microbiol 2000; 38: 1747– 1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lyons SR, Griffen AL, Leys EJ. Quantitative real-time PCR for Porphyromonas gingivalis and total bacteria. J Clin Microbiol 2000; 38: 2362– 2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meier A, Persing DH, Finken M. Bottger EC. Elimination of contaminating DNA within polymerase chain reaction reagents: implications for a general approach to detection of uncultured pathogens. J Clin Microbiol 1993; 31: 646– 652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rand KH, Houck H. Taq polymerase contains bacterial DNA of unknown origin. Mol Cell Probes 1990; 4: 445– 450 [DOI] [PubMed] [Google Scholar]
- 10.Kaul K, Luke S, McGurn C, Snowden N, Monti C, Fry WA. Amplification of residual DNA sequences in sterile bronchoscopes leading to false-positive PCR results. J Clin Microbiol 1996; 34: 1949– 1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loeffler J, Hebart H, Bialek R, et al. Contaminations occurring in fungal PCR assays. J Clin Microbiol 1999; 37: 1200– 1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rimek D, Garg AP, Haas WH, Kappe R. Identification of contaminating fungal DNA sequences in Zymolyase. J Clin Microbiol 1999; 37: 830– 831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van der Zee A, Peeters M, de Jong C, et al. Qiagen DNA extraction kits for sample preparation for legionella PCR are not suitable for diagnostic purposes. J Clin Microbiol 2002; 40: 1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Price AL, Eskin E, Pevzner PA. Whole-genome analysis of Alu repeat elements reveals complex evolutionary history. Genome Res 2004; 14: 2245– 2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yao YG, Bandelt HJ, Young NS. External contamination in single cell mtDNA analysis. PLoS ONE 2007; 2: e681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicklas JA, Buel E. Simultaneous determination of total human and male DNA using a duplex real-time PCR assay. J Forensic Sci 2006; 51: 1005– 1015 [DOI] [PubMed] [Google Scholar]
- 17.Urban C, Gruber F, Kundi M, Falkner FG, Dorner F, Hammerle T. A systematic and quantitative analysis of PCR template contamination. J Forensic Sci 2000; 45: 1307– 1311 [PubMed] [Google Scholar]
- 18.Melchior L, Kivisild T, Lynnerup N, Dissing J. Evidence of authentic DNA from Danish Viking Age skeletons untouched by humans for 1,000 years. PLoS ONE 2008; 3: e2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bustin SA, Benes V, Garson JA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 2009; 55: 611– 622 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.