Abstract
6×His tag is one of the most widely used affinity fusion tags that facilitates detection and purification of recombinant proteins. However, the location of this tag within a particular type of protein may influence the expression, solubility, and bioactivity of the protein, and the optimal location needs to be determined experimentally. To provide a tool for rapid generation of 6× His tags at the N- or C-terminus of any recombinant protein, we have constructed a pair of Escherichia coli expression vectors—pLIC-NHis and pLIC-CHis—based on the pET30a vector, for ligation-independent cloning (LIC). Construction of this new pair of LIC vectors was accomplished by replacement of the multiple cloning site of pET30a with two specifically designed LIC cloning sites. A target gene derived by PCR with a pair of predesigned primers can be inserted into the LIC site of pLIC-NHis for expression of recombinant proteins fused with the N-terminal sequence MHHHHHHG or into that of pLIC-CHis for expression of recombinant proteins with the C-terminal sequence THHHHHH. Successful expression of two normal mammalian prion proteins and five bacterial proteins in E. coli using this pair of LIC vectors reveals that these vectors are valuable tools for the production of recombinant His-tagged proteins in E. coli.
Keywords: His-tagged fusion protein, cloning, protein expression
The increasing demand for recombinant proteins for applications such as biomedical research, therapeutics, diagnostics, and vaccine development has driven many improvements in protein expression and purification technology. For example, fusion of a 6× His affinity tag to the N- or C-terminus of target proteins through recombinant DNA techniques has greatly simplified the detection of protein expression using antibodies specific for the tag and the purification of recombinant proteins using immobilized metal affinity chromatography (IMAC).1–4 Although a number of other affinity tags, including FLAG and GST, have been described in the literature,5 6× His tag is still the most widely used tag for production and high-throughput purification of affinity-tagged recombinant proteins, as it exerts a low metabolic burden on expression hosts and offers flexible conditions for IMAC, such as mild, nondenaturing conditions or denaturing conditions.2,4, 6× His tag is particularly suitable for purifying proteins that often accumulate as insoluble aggregates (inclusion bodies) in the cytoplasm when overexpressed in Escherichia coli and require solubilization and purification under denaturing conditions.4 As a result of its relative small size and charge, as well as poor antigenicity, 6× His tag rarely has any effects on the structure and function of target proteins.6,7. As such, removal of the 6× His tag from fusion proteins is not always necessary for practical applications, such as the development of recombinant protein-based vaccines8 and immunoassays9,10 and for protein characterization and structure determination.11,12
It has been shown that the solubility and expression level of a fusion protein in an E. coli cell-free system can sometimes be influenced by the location of a 6× His tag.13 Woestenenk et al.14 also reported that N- and C-terminal His tags generally affect protein solubility negatively, but N-terminal 6× His tags have less negative effect on solubility than C-terminal 6× His tags. In spite of these generalizations, the effect of the 6× His tag location on protein solubility is target protein-specific14 and thus, needs to be determined experimentally for each individual protein to achieve the optimal result. The availability of plasmid expression vectors that allow rapid cloning of any target genes in frame with the N- or C-termini 6× His tag coding sequence of the vector would facilitate a quick assessment of the effect of the 6× His tag position on protein expression. Insertion of a gene or DNA fragment into a selected vector is achieved traditionally by a time-consuming and less cost-effective restriction-ligation cloning approach15 or by recombinase-mediated cloning,16–18 which introduces unwanted vector-derived residues into target proteins. A ligation-independent cloning (LIC) approach has been developed that improves the efficiency of cloning PCR-amplified DNA fragments into a plasmid vector by eliminating the restriction-ligation steps.19 The method involves the generation of 12–15 bp protruding overhangs at both ends of a PCR-amplified DNA product and annealing of the DNA fragment specifically to complementary, single-stranded tails of a linearized vector, followed by transformation of E. coli. Compared with the conventional restriction-ligation-dependent cloning method, LIC is fast, efficient, and independent of the target gene sequence.19
A number of LIC expression vectors have been reported in the literature20–22 or are available from a commercial source such as Novagen (Madison, WI, USA).23 The majority of these vectors was designed to contain the coding sequences for N-terminal tags (6× His tag, S tag, GST tag, or Nus tag) immediately upstream of a protease-cleavage site. The tags may be removed from the expressed proteins by specific proteolysis using enterokinase, factor Xa, or tobacco etch virus (TEV) protease if desired. C-terminal 6× His-tagged proteins may be generated with the pT7LIC series of LIC plasmids.21 However, during the generation of recombinant constructs to express His-tagged proteins for diagnostic applications, we noted that it was not possible to select, from all of the LIC vectors described to date, a pair of vectors for expression of a target protein fused only with the N- or C-termini 6× His tag. This limitation prompted us to create a pair of LIC expression vectors for the rapid generation of expression constructs to place the 6× His tag at the N- or C-terminus of recombinant proteins. In this work, we modified the T7-based vector pET30a to create a pair of LIC vectors that contain a short sequence, coding for 7–8 aa, including a 6× His tag at the N- or C-termini of target proteins. The two LIC vectors were evaluated further by cloning and expression of several selected genes of prokaryotic and eukaryotic sources and shown to be useful for the production of various recombinant proteins in E. coli.
MATERIALS AND METHODS
Materials and Bacterial Culture
The pET30a vector was obtained from Novagen. Restriction and modifying enzymes for molecular cloning were purchased from New England Biolabs (Pickering, Ontario, Canada), unless stated otherwise. PfuUltra Hotstart DNA polymerase was from Stratagene (Cedar Creek, TX, USA). QIAquick Gel Extraction and QIAprep Spin Miniprep kits, Penta-His mAb (anti-His mAb), and nickel-nitrilotriacetic acid (Ni-NTA) agarose resin were obtained from Qiagen (Mississauga, Ontario, Canada). Luria Bertani (LB) medium was purchased from VWR (Mississauga, Ontario, Canada). All chemicals were purchased from Sigma-Aldrich (Oakville, Ontario, Canada), unless noted otherwise. E. coli strains used for cloning or protein expression are DH5α from Invitrogen (Burlington, Ontario, Canada) and BL21(DE3) and Rosetta 2(DE3) from Novagen. Transformants of E. coli strains were cultured in LB medium, supplemented as required with 30 μg/ml kanamycin [DH5α, BL21(DE3)] or 30 μg/ml kanamycin plus 34 μg/ml chloramphenicol [Rosetta 2(DE3)].
Construction of LIC Plasmids
Standard DNA manipulation procedures were followed as described.15 For the construction of pLIC-NHis, to be used for expression of N-terminal 6× His tagged proteins, PCR was conducted to synthesize a DNA fragment from the pET30a template using PfuUltra Hotstart DNA polymerase and a pair of 5′-phosphorylated primers P698 (5′P-GTGATGATGATGATGGTGCATATG3′) and P699 (5′P-GTGGAAGTGGATAACTGAGATCCGGCTGCTAACAAAGC3′). Sequences of P698 and P699 annealing to nt 328–350 and nt 117–139 of pET30a, respectively, are underlined. The PCR product was gel-purified using a QIAquick Gel Extraction kit (Qiagen) and then self-ligated with T4 DNA ligase to create pLIC-NHis, in which a new sequence, 5′CGTGGAAGTGGATAAC3′, replaced a 188-bp segment (nt 140–327) of pET30a (Figure 1A).The pLIC-NHis plasmid (5250 bp), propagated in and prepared using a QIAprep Spin Miniprep kit (Qiagen) from E. coli DH5α, was analyzed by restriction digestion and DNA sequencing to confirm the presence of the new sequence containing a unique PmlI site (i.e., LIC site) and the loss of the original multiple cloning site (MCS), including the sequence coding for a thrombin site, an S-Tag, and a C-terminal 6× His tag. Similarly, for the construction of pLIC-CHis to be used for expression of C-terminal 6× His-tagged proteins, PCR was performed to obtain a DNA fragment with a set of 5′-phosphorylated primers P723 (5′P-ACTTATATCTCCTTCTTAAAGTTAAACAAAATTA3′) and P724 (5′P-ACTCACCACCACCACCACCACTG3′). The underlined sequences of P723 and P724 are those annealing to nt 351–381 and nt 138–157 of pET30a, respectively. Self-ligation of the PCR product with T4 DNA ligase resulted in a circularized plasmid pLIC-CHis (5235 bp), in which a unique ScaI site, 5′AGTACT3′ (i.e., LIC site), was introduced to replace a 193-bp sequence segment (nt 158–350) of pET30a (Fig. 1B). The presence of a new sequence in pLIC-CHis was confirmed by restriction digestion and DNA sequencing.
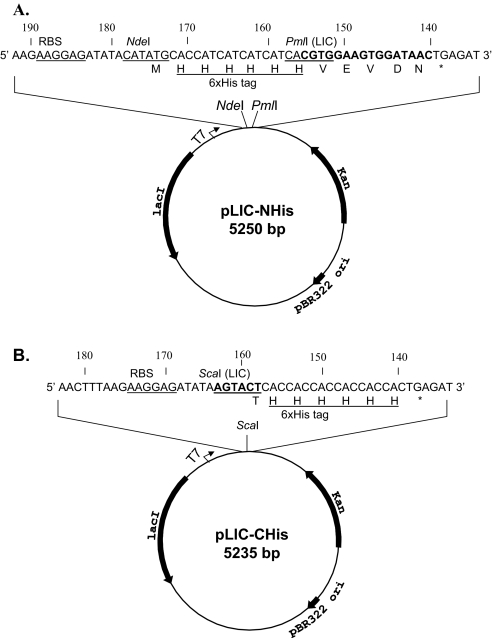
FIGURE 1.
Schematic illustration of the pET30a-derived LIC vectors pLIC-NHis and pLIC-CHis, which are derived from the parent plasmid vector pET30a (Novagen), as described in Materials and Methods. The nucleotide sequences of the derived plasmids are numbered with the first nucleotide corresponding to that of the parent plasmid pET30a. (A) pLIC-NHis is shown in a circularized plasmid DNA (5250 bp) with the locations of the T7 promoter (T7), lac repressor gene (lacI), kanamycin-resistance gene (Kan), and the pBR322 origin of replication (ori). The T7 expression portion is expanded and annotated with the positions of the ribosomal binding site (RBS), relevant restriction sites (NdeI and PmlI), and a 16-bp nucleotide sequence (boldface type), replacing the original 188-bp segment (nt 140–327) of pET30a. A unique PmlI site CACGTG (underlined) was created for LIC cloning. The corresponding amino acid sequence, including a 6× His tag (underlined), is depicted in a single-letter code. (B) pLIC-CHis (5235 bp) is shown and annotated similarly as pLIC-NHis. A unique ScaI site (boldface type and underlined) for LIC cloning was introduced into the T7 expression region to replace a 193-bp sequence segment (nt 158–350) of pET30a.
Linearization of LIC Expression Vectors for LIC Cloning
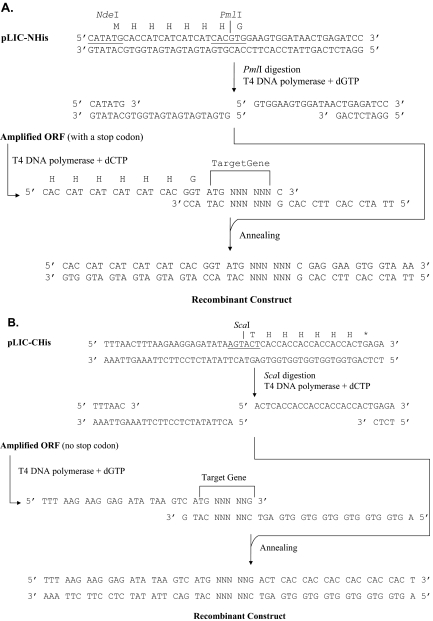
LIC plasmids were cut with a unique LIC restriction enzyme, followed by digestion with T4 DNA polymerase, as described elsewhere.22 Briefly, pLIC-NHis DNA was digested with PmlI, gel-purified as described above, and treated with T4 DNA polymerase in the presence of dGTP to create the cloning-ready vector with 5′ overhangs (18 and 14 nt, respectively) at both ends (Figure 2A). Similarly, pLIC-CHis DNA was linearized with ScaI, followed by digestion with T4 DNA polymerase in the presence of dCTP, to generate the cloning-ready vector with 5′ overhangs (20 and 22 nt, respectively) at both ends.
FIGURE 2.
Ligation-independent cloning of PCR-amplified open-reading frames (ORFs) into pLIC-NHis or pLIC-CHis. Procedure details are given in Materials and Methods. (A) PmlI-cut pLIC-NHis is treated with T4 DNA polymerase in the presence of dGTP. An ORF of interest, generated by PCR, using a specifically deigned primer set, is treated similarly in the presence of dCTP. Subsequently, the T4 DNA polymerase-treated PCR product is annealed to the treated vector at room temperature and used to transform E. coli DH5α for propagation of recombinant plasmids. (B) ScaI-cut pLIC-CHis is treated with T4 DNA polymerase in the presence of dCTP. An ORF of interest, generated by PCR, using a specifically designed primer set, is treated similarly in the presence of dGTP. Recombinant plasmids were propagated as stated above.
Primer Designs for LIC Cloning
To clone any target genes amplified by PCR in the correct orientation and reading frame into pLIC-NHis, sense primers were designed to begin with the sequence 5′CACCATCATCATCATCACGGT3′, followed by sequences encoding six or more N-terminal residues; antisense primers begin with the sequence 5′TTATCCACTTCCACG3′, followed by sequences reverse-complementary to the stop codon and the sequences encoding six or more C-terminal residues. Sense primers for cloning the genes of interest in the proper orientation and reading frame into pLIC-CHis should begin with the 5′TTTAAGAAGGAGATATAAGTC3′, followed by a gene-specific sequence coding for six or more N-terminal amino acids; antisense primers should begin with the sequence 5′AGTGGTGGTGGTGGTGGTGAGTC3′, followed by sequences reverse-complementary to the sequence encoding six or more C-terminal residues with omission of the stop codon and the nucleotide at the third codon position of the last codon. Accordingly, primer sets for PCR amplification of several genes, which had been selected to evaluate the LIC expression vectors described in this study, were synthesized and listed in Table 1.
TABLE 1.
Primers for LIC Cloning of Seven Target Genes into pLIC-NHis or pLIC-CHis
| Source of DNA for cloning | Target gene (encoded protein) | Oligonucelotide primera (Genbank Access No. for primer designing) | Annealing sites (cloned gene sequence)b | LIC cloning vector | Recombinant product |
|---|---|---|---|---|---|
| Leptospira borgpetersenii serovar Hardjo | lipL32 | P700: 5′CACCATCATCATCATCACGGTATGAAAAAACTTTCGATTTTGGC3′ | nt 1–23 | pLIC-NHis | rLipL32 (N) |
| (outer membrane lipoprotein LipL32) | P701: 5′TTATCCACTTCCACGTTACTTAGTCGCGTCAGAAGC3′ (AF121192) | nt 799–819 (full-length) | |||
| L. borgpetersenii serovar Hardjo | lipL36 | P725: 5′CACCATCATCATCATCACGGTATGAGAAGAAACATAATGAAAATTGC3′ | nt 1–26 | pLIC-NHis | rLipL36 (N) |
| (outer membrane lipoprotein LipL36) | P726: 5′TTATCCACTTCCACGTTAGTATTTAGGATAAGTTG3′ (AF024626) | nt 1100–1119 (full-length) | |||
| L. borgpetersenii serovar Hardjo | sphA | P802: 5′CACCATCATCATCATCACGGTGATAGACAATCTTTGTATAAAGATTTAC3′ | nt 82–109 | pLIC-NHis | rSphA-m (N) |
| (sphingomyolinase C) | P803: 5′TTATCCACTTCCACGTCAACGATAAATTAATTTCTTACTCC3′ (X52176) | nt 1646–1671 (partial ORF, nt 82–1671) | |||
| L. borgpetersenii serovar Hardjo | ompL1 | P804: 5′CACCATCATCATCATCACGGTAAATCATACGCAATTGTAGGATTC3′ | nt 73–96 | pLIC-NHis | rOmpL1-m (N) |
| (outer membrane protein OmpL1) | P805: 5′TTATCCACTTCCACGTTAGAGTTCGTGTTTATAACCAAAT3′ (AY622669) | nt 939–963 (partial ORF, nt 73–963) | |||
| Listeria monocytogenes serotype 4b | lmo1941 | P729: 5′CACCATCATCATCATCACGGTATGGTAAAAACAAGAAAAGAAAAAC3′ | nt 1–25 | pLIC-NHis | rLmo1941 (N) |
| (hypothetical peptidoglycan-binding protein) | P730: 5′TTATCCACTTCCACGTTACTGTGGAATTGTTAGAACTGTACC3′ (NC 003210) | nt 694–720 (full-length) | |||
| Bos taurus | bovine PRNP (bovine prion protein gene) | P765: 5′TTTAAGAAGGAGATATAAGTCATGAAGAAGCGACCAAAACCTG3′ | nt 73–91 | pLIC-CHis | rbPrP-m (C) |
| P774: 5′AGTGGTGGTGGTGGTGGTGAGTCGCCCCTCGTTGGTAATAAGC3′ (AB001468) | nt 703–722 (partial ORF, nt 73–722) | ||||
| Ovis aries | ovine PRNP | P765: 5′TTTAAGAAGGAGATATAAGTCATGAAGAAGCGACCAAAACCTG3′ | nt 73–91 | pLIC-CHis | roPrP-m (C) |
| (ovine prion protein gene) | P764: 5′AGTGGTGGTGGTGGTGGTGAGTCCCTACTATGAGAAAAATGAGGA3′ (U67922) | nt 746–767 (partial ORF, nt 73–767) |
The underlined sequences within the primers are gene-specific sequences derived from lipL32 of Leptospira kirschneri,24 lipL36 of L. kirschneri,25 sphA of Leptospira interrogans,26 ompL1 of L. kirschneri (Genbank Access No. AY622669), Lmo1941 of L. monocytogenes strain EGD-e,27 bovine PRNP,28 and ovine PRNP.29 Genbank access numbers for the above gene sequences are indicated in parentheses.
Numbers refer to the sequence position of target ORFs, the first nucleotide of which is numbered 1. The cloned portions of target genes are shown in parentheses.
-m, Mature form; (N), N-terminal; (C), C-terminal.
Ligation-Independent Cloning of Target Genes
To evaluate the LIC expression vectors, the genes coding for the following proteins were amplified by PCR from respective genomic DNAs with PfuUltra Hotstart DNA polymerase (Stratagene) and the appropriate primer pairs (Table 1): a L. monocytogenes protein Lmo1941 (hypothetical peptidoglycan-binding protein), four L. borgpetersenii proteins (the lipoproteins LipL32 and LipL36, SphA, and the OmpL), and two prion proteins (PrPc; bPrPc and oPrPc). For a serotype 4b strain LI0521 of L. monocytogenes, the genomic DNA was isolated as described30 using the GenomicPrep Cells and Tissue DNA Isolation kit (Amersham Biosciences, Baie d’Urfe, Quebec, Canada); for a serovar Harjo strain 6.92 of L. borgpetersenii, its genomic DNA was extracted by using the methods described previously.31 DNA from bovine and ovine blood samples was isolated using a MagNA Pure LC DNA Isolation kit (Roche, Laval, Quebec, Canada). The PCR products corresponding to the bacterial genes were gel-purified and treated with T4 DNA polymerase in the presence of dCTP to generate 5′ overhangs complementary to those of the cloning-ready pLIC-NHis (Fig. 2A). The PCR products corresponding to the two PRNP were treated with T4 DNA polymerase in the presence of dGTP to produce 5′ overhangs complementary to those of the cloning-ready pLIC-CHis (Fig. 2B). After annealing of the pretreated PCR products (10 ng) to the corresponding cloning-ready LIC vectors (10 ng) via compatible overhangs at room temperature for 5 min, the mixture was used to transform E. coli DH5α. Bacterial colonies were screened by colony PCR as described30 with T7 promoter and T7 terminator primers to identify the clones containing the inserted gene of expected sizes and verified further by DNA sequencing. The recombinant plasmids were introduced into E. coli BL21(DE3) or Rosetta 2(DE3) for protein expression.
Protein Expression and Purification
E. coli cells harboring an expression construct were grown in 20 ml LB broth containing 30 μg/ml kanamycin [for BL21(DE3)] or 30 μg/ml kanamycin plus 34 μg/ml chloramphenicol [for Rosetta 2(DE3)] at 37°C overnight with constant shaking. Overnight cultures of bacterial cells were diluted 100-fold into 2 L LB broth, supplemented with appropriate antibiotics, cultured as above until an OD590 of 0.6–1.0 was reached and then induced to express the recombinant proteins for 3 h by adding isopropyl β-d-galactopyranoside (IPTG) to a final concentration of 1 mM. After harvesting by centrifugation at 10,000 g for 20 min, bacterial pellets were resuspended in PBS (pH 7.2) containing 1 mM PMSF, lysed with a French Press at 1500 psi, and centrifuged (27,000 g, 20 min) to collect the insoluble materials (inclusion bodies), from which His-tagged recombinant proteins were solubilized and purified by Ni-NTA agarose chromatography, as described previously.32
SDS-PAGE and Western Blotting
SDS-PAGE and Western blots were performed to analyze the E. coli lysate containing His-tagged recombinant proteins or purified recombinant proteins as described elsewhere.33 The proteins, after separation by SDS-PAGE, were stained with Coomassie blue or electrotransferred to nitrocellulose membranes using a Trans-Blot SD semi-dry transfer cell (Bio-Rad, Hercules, CA, USA) for Western blotting. The membrane-immobilized proteins were probed with anti-His mAb. Bound antibodies were detected by using HRP-conjugated goat anti-mouse IgG (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA) and a 4-chloro-1-naphthol/H2O2 substrate kit (Bio-Rad), according to the manufacturer's instructions.
RESULTS
Features of LIC Expression Vectors
Two LIC vectors, pLIC-NHis and pLIC-CHis, derived here from the parental vector pET30a, are presented schematically in Figure 1. Both vectors retain the backbone of pET30a, including the T7 promoter, lac repressor gene lacI, RBS, kanamycin-resistance gene kan, and the pBR322 ori. In pLIC-NHis, the original MCS of pET30a was essentially deleted and replaced with a 16-bp sequence (5′CGTGGAAGTGGATAAC3′) immediately downstream of the 6× His coding sequence to create a unique PmlI site (CACGTG) for LIC cloning. In pLIC-CHis, a unique ScaI restrition site (AGTACT) for LIC cloning was introduced immediately upstream of the C-terminal 6× His coding sequence to replace the original MCS of pET30a. Using the LIC cloning strategies outlined in Figure 2 (see Materials and Methods for details), a target gene derived by PCR with a pair of predesigned primers (Table 1) can be inserted into the LIC site of pLIC-NHis to express a recombinant protein fused with an N-terminal sequence MHHHHHHG or into that of pLIC-CHis to encode a recombinant protein with a C-terminal sequence THHHHHH.
Evaluation of LIC Expression Vectors
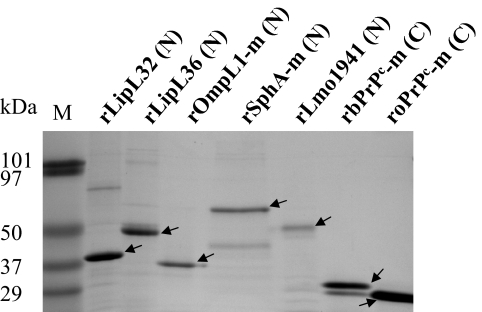
Cloning of the genes coding for L. monocytogenes Lmo1941 and L. borgpetersenii LipL32, LipL36, mature SphA (aa 28–556), and OmpL1 into pLIC-NHis and the genes coding for bPrPc and oPrPc into pLIC-CHis, using the LIC cloning procedure (Fig. 2), yielded seven respective expression constructs (Table 1). The expression constructs for all of these proteins were generated using one of the two LIC vectors. PCR analysis and DNA sequencing revealed that these constructs contained the correctly inserted genes. Western blot analysis of the total protein lysates from IPTG-induced E. coli, transformed with any one of these constructs, revealed protein bands recognized by anti-His mAb, in agreement with the predicted sizes of His-tagged fusion proteins (data not shown). This indicated that target proteins were expressed as N- or C-terminal His-tagged proteins from these constructs and that this set of LIC vectors is useful for expression of any target proteins. Each expressed protein described here was found predominantly in the insoluble fraction (presumably as a result of formation of protein aggregates referred to as inclusion bodies) of the cell lysates and was purified further after solubilization to 80–90% purity by Ni-NTA agarose affinity chromatography, as judged by SDS-PAGE (Figure 3).The additional minor bands observed most likely represent protein contaminants from E. coli (Figure 3). It is also possible that the minor lower molecular weight bands are a result of proteolyic degradation of the expressed target proteins.
FIGURE 3.
SDS-PAGE analysis of seven recombinant proteins expressed from the constructs generated with pLIC-NHis or pLIC-CHis. All recombinant proteins were produced from the expression constructs in E. coli BL21(DE3) with the exception of rSphA-m (N) in E. coli Rosetta 2(DE3), affinity purified by Ni-NTA agraose chromatography under denaturing conditions, and analyzed by SDS-PAGE. The position of 6× His tag at the N- and C-termini is indicated by (N) and (C), respectively. M, Prestained protein standards (Bio-Rad). rLipL32 (N), rLipL36 (N), rOmpL1-m (N), and rSphA-m (N) proteins are encoded by the corresponding genes from L. borgpetersenii serovar Harjo strain 6.92; rLmo1941 is a protein of L. monocytogenes serotype 4b strain LI0521. rbPrPc-m and roPrPc are mature bovine and ovine proteins, respectively. The amount of purified proteins loaded for each lane: rLipL32 (N), 4 μg; rLipL36 (N), 2 μg; rOmpL1-m (N), 1.5 μg; rSphA-m (N), 1.5 μg; rLmo1941, 1.0 μg; rbPrPc (C), 4 μg; roPrPc-m (C), 8 μg. Recombinant target proteins are indicted by arrows with calculated molecular masses of 33 kDa for LipL32 (N), 38 kDa for rLipL36 (N), 32.3 kDa for rOmpL1-m (N), 61 kDa for rSphA-m (N), 26 kDa for rLmo1941, 24.5 kDa for rbPrPc-m (C), and 23.8 kDa for roPrPc-m (C).
DISCUSSION
We have derived from the pET30a vector a pair of prokaryotic expression vectors, pLIC-NHis and pLIC-CHis, into which any target gene, regardless of its sequence makeup, can be inserted using a ligation-independent cloning strategy.19 The usefulness of these two vectors was demonstrated in this study by successful cloning and expression in E. coli of seven protein-coding genes. These genes were selected for protein expression using the LIC vectors for two main reasons: We are interested in producing these recombinant proteins to evaluate their diagnostic potential, and these rapidly available genes in our laboratory represent the genes of various sources (Gram-positive bacteria, Gram-negative bacteria, and eukaryotes) and may thus allow for assessing the ability of the two LIC vectors to express proteins of various organisms. As found commonly with expression of foreign proteins in E. coli, all seven recombinant proteins were expressed in an insoluble form (inclusion bodies). Given the fact that the location of a 6× His tag interferes with the expression level and/or solubility of fusion proteins,13,14 the availability of pLIC-NHis and pLIC-CHis would allow for expression of a target protein fused only with an N- or C-terminal 6× His tag and thus, facilitate the assessment of the effect of the tag position on protein expression. Although the mechanism by which the position of a His tag influences recombinant protein expression is not understood, it appears that the effect of the His tag position on fusion protein expression is dependent on the sequence of expressed proteins.
All of the previously reported LIC expression vectors20–23 do not have the flexibility of expressing two recombinant fusion proteins that are different, essentially only in the location of His tag. The majority of LIC vectors reported previously20–23 incorporates a number of non-native amino acid residues to fusion proteins. Although the pT7LIC series of LIC vectors described by Chanda et al.21 is similar to pLIC-CHis, in that all of these vectors express recombinant proteins, fused only to a C-terminal 6× His tag, there are no other LIC vectors partnered with them to express recombinant target proteins fused only to a N-terminal 6× His tag. To the best of our knowledge, pLIC-NHis, together with pLIC-CHis, represents a unique pair of LIC expression vectors having the same plasmid backbone for production of recombinant proteins fused only to a 6× His tag at the N- or C-terminus. The presence of a 6× His tag did not appear to produce unwanted interference with practical applications.8,9,11,12 Thus, incorporation of a specific protease cleavage site into recombinant proteins for the purpose of removal of the 6× His tag was not considered during the construction of the LIC expression vectors. If required, a specific protease cleavage site, such as enterokinase, TEV protease, or factor Xa, can be introduced between the 6× His tag and a target protein by synthesizing appropriate PCR primers and thus, allow for protease-guided removal of the tag from recombinant fusion proteins.
In summary, a detailed procedure for the preparation of cloning-ready vectors and the design of gene-specific primers, as reported in this communication, allows for rapid and easy cloning of any PCR-amplified genes into pLIC-NHis or pLIC-CHis. This new set of LIC vectors, like any other LIC expression vectors described previously,20–23 offers significant advantages over the conventional restriction-ligation cloning approach, including no need to digest vectors and PCR-amplified genes with restriction endonucleases to create compatible ends for cloning, the elimination of the T4 DNA ligase step, and the avoidance of interference with cloning from any restriction sites within target sequences. Thus, generation of protein expression constructs using the LIC expression vectors described in this study and elsewhere20–23 is efficient and cost-effective for high-throughput recombinant protein expression.
ACKNOWLEDGMENT
We gratefully thank Jasmine Rendulich for preparation of the bovine and ovine genomic DNA from blood samples.
REFERENCES
- 1.Hochuli E, Dobeli H, Schacher A. New metal chelate adsorbent selective for proteins and peptides containing neighboring histidine residues. J Chromatogr 1987; 411: 177– 184 [DOI] [PubMed] [Google Scholar]
- 2.Porath J. Immobilized metal ion affinity chromatography. Protein Expr Purif 1992; 3: 263– 281 [DOI] [PubMed] [Google Scholar]
- 3.Porath J, Carlsson J, Olsson I, Belfrage G. Metal chelate affinity chromatography, a new approach to protein fractionation. Nature 1975; 258: 598– 599 [DOI] [PubMed] [Google Scholar]
- 4.Waugh DS. Making the most of affinity tags. Trends Biotechnol 2005; 23: 316– 320 [DOI] [PubMed] [Google Scholar]
- 5.Arnau J, Lauritzen C, Petersen GE, Pedersen J. Current strategies for the use of affinity tags and tag removal for the purification of recombinant proteins. Protein Expr Purif 2006; 48: 1– 13 [DOI] [PubMed] [Google Scholar]
- 6.Mohanty AK, Wiener MC. Membrane protein expression and production: effects of polyhistidine tag length and position. Protein Expr Purif 2004; 33: 311– 325 [DOI] [PubMed] [Google Scholar]
- 7.Terpe K. Overview of tag protein fusions: from molecular and biochemical fundamentals to commercial systems. Appl Microbiol Biotechnol 2003; 60: 523– 533 [DOI] [PubMed] [Google Scholar]
- 8.Kaslow DC, Shiloach J. Production, purification and immunogenicity of a malaria transmission-blocking vaccine candidate: TBV25H expressed in yeast and purified using nickel-NTA agarose. Biotechnology (N Y) 1994; 12: 494– 499 [DOI] [PubMed] [Google Scholar]
- 9.Flannery B, Costa D, Carvalho FP, et al. Evaluation of recombinant Leptospira antigen-based enzyme-linked immunosorbent assays for the serodiagnosis of leptospirosis. J Clin Microbiol 2001; 39: 3303– 3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin M, Trottier E, Mallory M. Enzyme-linked immunosorbent assay based on a chimeric antigen bearing antigenic regions of structural proteins Erns and E2 for serodiagnosis of classical swine fever virus infection. Clin Diagn Lab Immunol 2005; 12: 877– 881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Derewenda ZS. The use of recombinant methods and molecular engineering in protein crystallization. Methods 2004; 34: 354– 363 [DOI] [PubMed] [Google Scholar]
- 12.Lesley SA, Kuhn P, Godzik A, et al. Structural genomics of the Thermotoga maritima proteome implemented in a high-throughput structure determination pipeline. Proc Natl Acad Sci USA 2002; 99: 11664– 11669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Busso D, Kim R, Kim SH. Expression of soluble recombinant proteins in a cell-free system using a 96-well format. J Biochem Biophys Methods 2003; 55: 233– 240 [DOI] [PubMed] [Google Scholar]
- 14.Woestenenk EA, Hammarstrom M, van den Berg S, Hard T, Berglund H. His tag effect on solubility of human proteins produced in Escherichia coli: a comparison between four expression vectors. J Struct Funct Genomics 2004; 5: 217– 229 [DOI] [PubMed] [Google Scholar]
- 15.Sambrook J, Russel DW. Molecular Cloning: A Laboratory Manual, 3rd ed, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 2001 [Google Scholar]
- 16.Walhout AJ, Temple GF, Brasch MA, et al. GATEWAY recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol 2000; 328: 575– 592 [DOI] [PubMed] [Google Scholar]
- 17.Brasch MA, Hartley JL, Vidal M. ORFeome cloning and systems biology: standardized mass production of the parts from the parts-list. Genome Res 2004; 14: 2001– 2009 [DOI] [PubMed] [Google Scholar]
- 18.Busso D, Delagoutte-Busso B, Moras D. Construction of a set Gateway-based destination vectors for high-throughput cloning and expression screening in Escherichia coli. Anal Biochem 2005; 343: 313– 321 [DOI] [PubMed] [Google Scholar]
- 19.Aslanidis C, de Jong PJ. Ligation-independent cloning of PCR products (LIC-PCR). Nucleic Acids Res 1990; 18: 6069– 6074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cabrita LD, Dai W, Bottomley SP. A family of E coliexpression vectors for laboratory scale and high throughput soluble protein production. BMC Biotechnol 2006; 6: 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chanda PK, Edris WA, Kennedy JD. A set of ligation-independent expression vectors for co-expression of proteins in Escherichia coli. Protein Expr Purif 2006; 47: 217– 224 [DOI] [PubMed] [Google Scholar]
- 22.Stols L, Gu M, Dieckman L, Raffen R, Collart FR, Donnelly MI. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr Purif 2002; 25: 8– 15 [DOI] [PubMed] [Google Scholar]
- 23.Novy RE, Yaeger KW, Kolb KM. Ligation independent cloning: efficient directional cloning of PCR products. InNovations 1996; 5: 1– 3 [Google Scholar]
- 24.Haake DA, Chao G, Zuerner RL, et al. The leptospiral major outer membrane protein LipL32 is a lipoprotein expressed during mammalian infection. Infect Immun 2000; 68: 2276– 2285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haake DA, Martinich C, Summers TA, et al. Characterization of leptospiral outer membrane lipoprotein LipL36: downregulation associated with late-log-phase growth and mammalian infection. Infect Immun 1998; 66: 1579– 1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Segers RP, van der Drift A, de Nijs A, Corcione P, van der Zeijst BA, Gaastra W. Molecular analysis of a sphingomyelinase C gene from Leptospira interrogans serovar hardjo. Infect Immun 1990; 58: 2177– 2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glaser P, Frangeul L, Buchrieser C, et al. Comparative genomics of Listeria species. Science 2001; 294: 849– 852 [DOI] [PubMed] [Google Scholar]
- 28.Yoshimoto J, Iinuma T, Ishiguro N, Horiuchi M, Imamura M, Shinagawa M. Comparative sequence analysis and expression of bovine PrP gene in mouse L-929 cells. Virus Genes 1992; 6: 343– 356 [DOI] [PubMed] [Google Scholar]
- 29.Lee IY, Westaway D, Smit AF, et al. Complete genomic sequence and analysis of the prion protein gene region from three mammalian species. Genome Res 1998; 8: 1022– 1037 [DOI] [PubMed] [Google Scholar]
- 30.Yu WL, Dan H, Lin M. Novel protein targets of humoral immune response to Listeria monocytogenes infection in rabbits. J Med Miccrobiol 2007; 56: 888– 895 [DOI] [PubMed] [Google Scholar]
- 31.Lin M, Li Y. PCR genome walking identifies a genetic locus comprising two heat shock genes (hslV and hslU) from Leptospira borgpetersenii serovar Hardjobovis. Curr Microbiol 2001; 43: 452– 456 [DOI] [PubMed] [Google Scholar]
- 32.Lin M, Trottier E, Pasick J. Antibody responses of pigs to defined Erns fragments after infection with classical swine fever virus. Clin Diagn Lab Immunol 2005; 12: 180– 186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang L, Lin M. Identification of IspC, an 86-kilodalton protein target of humoral immune response to infection with Listeria monocytogenes serotype 4b, as a novel surface autolysin. J Bacteriol 2007; 189: 2046– 2054 [DOI] [PMC free article] [PubMed] [Google Scholar]