Abstract
We have developed a sequencing-based gene expression profiling assay at single-cell resolution by combining a modified single-cell whole transcriptome amplification method with the next generation sequencing technique, the SOLiD™ system. Using this assay, we have shown that blastomeres in a four-cell stage embryo have similar gene expression, which is compatible with the fact that they have similar developmental potential. We proved that compared with cDNA microarray technique, our single-cell cDNA SOLiD sequencing assay can detect expression of thousands of more genes. Moreover, for the genes detected by microarray and SOLiD sequencing, our assay detected new transcript variants for a large proportion of them, which confirms unambiguously at single-cell resolution that the transcriptome complexity is higher than expected traditionally. Finally, by using our assay to Dicer knockout (KO) and Ago2 KO oocytes, we showed that a significant amount of transposons were up-regulated abnormally in Dicer/Ago2 KO mature oocytes compared with wild-type controls.
Keywords: gene expression, Ago2, Dicer
By analyzing the transcriptome at spectacular and unprecedented depth and accuracy, thousands of new transcript variants/isoforms were found expressed unambiguously in mammalian tissues or organs. These advances greatly accelerate our understanding of the complexity of gene expression regulation and networks for mammalian cells. The technique usually needs milligram amounts of total RNA for analysis, which corresponds to hundreds of thousands of cells. However, under certain conditions, it is practically not possible to get such amounts of materials for analysis, e.g., for early embryonic development. In fact, during mouse early development, when the founder population of germ line primordial germ cells (PGCs) just specified and emerged, there are only approximately 30 PGCs in an embryo. On the other hand, even for in vitro-cultured stem cells, for which the cell amount available for analysis is unlimited, there are limitations. For example, mouse embryonic stem cells, probably the most thoroughly analyzed type of stem cells during the past 27 years, were found to contain multiple subpopulations with strong differences of gene expression and physiological function. Altogether, a more sensitive, next-generation sequencing assay, ideally an assay working to single-cell resolution, is needed for these crucial developmental processes and stem cell biology.
Here, we modified a single-cell whole transcriptome amplification method to make it permissive to amplify cDNAs as long as 3 kb in an efficient and unbiased manner (see Figs. 8 and 9). We combined this modified single-cell cDNA amplification method with Applied Biosystems’ next generation sequencing technology, the SOLiD™ system, to set up a single-cell whole transcriptome assay. We proved that it is feasible to obtain gene expression profiles at single-cell resolution, which enables us to ask fundamental biological questions previously not possible, especially in the field of early embryonic development, and to understand biology at the single-cell resolution, which is the uniform, functional unit of any organism.
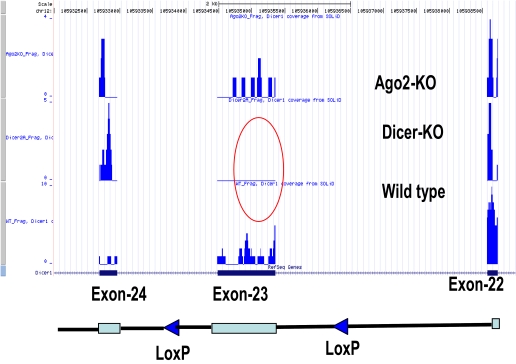
FIGURE 8.
Dicer locus on Chromosome 12 and coverage of Dicer Exon 23. LoxP, Locus of crossover in P1.
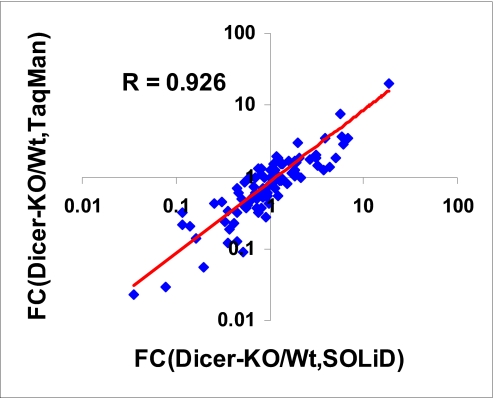
FIGURE 9.
The correlation plots of the fold changes that are determined by SOLiD reads and real-time PCR. FC, fold changes.
MATERIALS AND METHODS
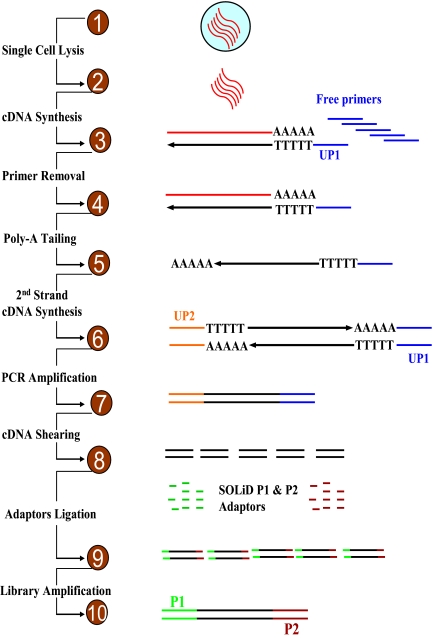
FIGURE 1.
Reaction schemes. UP1, universal primer 1; UP2, universal primer 2.
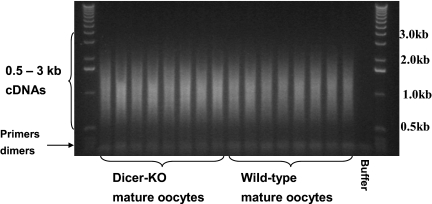
FIGURE 2.
cDNA products. KO, Knockout.
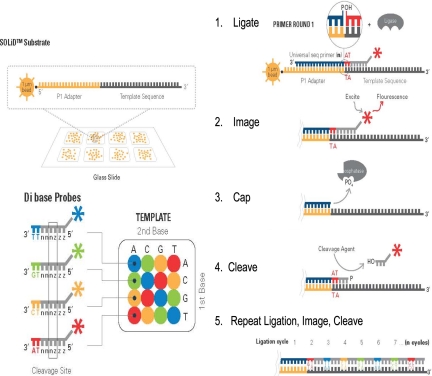
FIGURE 3.
SOLiD technology.
RESULTS
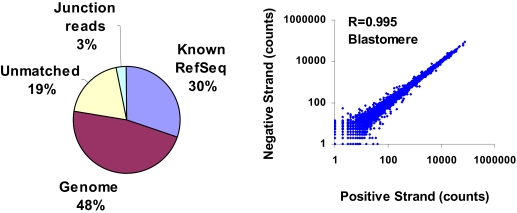
FIGURE 4.
SOLiD matching summary for mouse blastomeres in four-cell embryos.
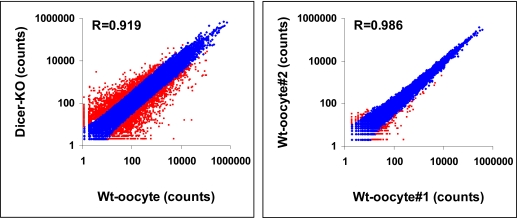
FIGURE 5.
SOLiD results for wild-type (Wt) and Dicer-KO oocytes.
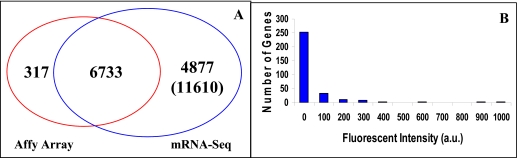
FIGURE 6.
Comparison of SOLiD sequencing data with those from microarrays of ∼80 pooled four-cell stage embryos (320 blastomeres) found that 6733 genes detected by Affymetrix GeneChip Mouse Genome 430 2.0 array (Affy Array) were also detected by SOLiD sequencing. The 317 genes, detected by microarray but missed by SOLiD, were relatively low-expressed genes and could result from cross-hybridization. SOLiD sequencing detects 4877 more expressed genes compared with microarray.
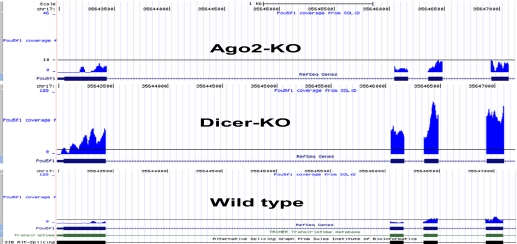
FIGURE 7.
Coverage of exons for Pou5f1 (Oct4).
CONCLUSIONS
In summary, we have established a SOLiD sequencing-based gene expression profiling assay at single-cell resolution. We proved that thousands of genes express two or more transcript variants in a same cell. We also proved that in Dicer KO mature oocytes, the transcripts of a lot of transposons and repeat elements are up-regulated abnormally. This single-cell sequencing assay will greatly facilitate understanding the transcriptome complexity during mammalian development, especially in the fields of stem cells and early embryonic development.
ACKNOWLEDGMENT
We thank Caroline Lee, Melissa A. Barker, Roland Wicki, Cinna Monighetti, Francisco M. De La Vega, and Neil A. Straus for excellent technical help.
Footnotes
Trademarks/Licensing: Applied Biosystems and AB (Design) are registered trademarks, and SOLiD is a trademark of Applied Biosystems or its subsidiaries in the United States and/or certain other countries.