Abstract
Proteolytic processing events are essential to physiological processes such as reproduction, development, and host responses, as well as regulating proteins in cancer; therefore, there is a significant need to develop robust approaches for characterizing such events. The current mass spectrometry (MS)-based proteomics techniques employs a “bottom-up” strategy, which does not allow for identification of different proteolytic proteins since the strategy measures all the small peptides from any given protein. The aim of this development is to enable the effective identification of specific proteolytic fragments. The protocol utilizes an acetylation reaction to block the N-termini of a protein, as well as any lysine residues. Following digestion, N-terminal peptides are enriched by removing peptides that contain free amines, using amine-reactive silica-bond succinic anhydride beads. The resulting enriched sample has one N-terminal peptide per protein, which reduces sample complexity and allows for increased analytical sensitivity compared to global proteomics.1 We initially compared the peptide identification and efficiency of blocking lysine using acetic anhydride (a 42 Da modification) or propionic anhydride (a 56 Da modification) in our protocol. Both chemical reactions resulted in comparable peptide identifications and ∼95 percent efficiency for blocking lysine residues. However, the use of propionic anhydride allowed us to distinguish in vivo acetylated peptides from chemically-tagged peptides.2 In an initial experiment using mouse plasma, we were able to identify >300 unique N-termini peptides, as well as many known cleavage sites. This protocol holds potential for uncovering new information related to proteolytic pathways, which will assist our understanding about cancer biology and efforts to identify potential biomarkers for various diseases.
Challenge (Fig. 1): Current liquid chromatography/mass spectrometry (LC-MS)-based “bottom-up” proteomics is relatively ineffective for detecting specific proteolytic modifications, despite the fact that they are highly prevalent in higher eukaryotic organisms.
FIGURE 1.
Objective: To enrich N-terminal peptides to identify:
proteolytic modifications
exact cleavage sites
natural protein N-termini
post-translationally modified N-termini
MATERIALS AND METHODS
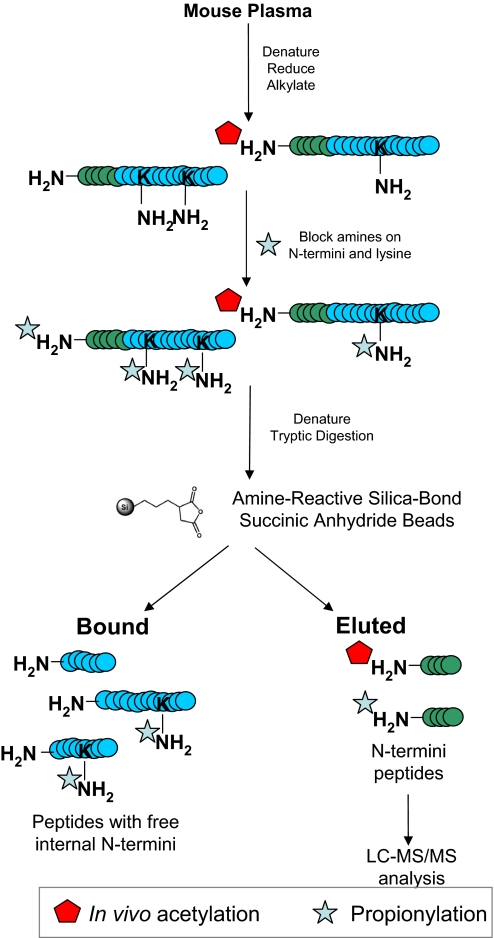
See Figure 2.
FIGURE 2.
RESULTS
TABLE 1.
Comparison between acetic anhydride and propionic anhydride to block amines and lysine
| Acetic anhydride | Propionic anhydride |
|---|---|
| ∼95% efficiency for blocking amines and lysines | ∼95% efficiency for blocking amines and lysines |
| 707 unique peptides identified | 766 unique peptides identified |
| +42 Da modification | +56 Da modification |
Previous studies have used acetic anhydride, but the use of propionic anhydride allows in vivo acetylation to be distinguished from the chemical modification.
TABLE 2.
LC-MS/MS identified N-terminal peptides from relatively low-abundance proteins
| Protein name | Identified N-terminal peptide sequence |
|---|---|
| Extracellular matrix protein 1 | A#SEGAFK*ASDQR |
| IGFBP3 | G#AGAVGAGPVVR |
| β-2-Microglobulin | I#QK*TPQIQVYSR |
| Plasma kallikrein | G#CMTQLYK*NTFFR |
| Glycoslyphosphatidylinositol | G#K*YHDVSER |
| C#GLSTHVEIGHR | |
| Leukemia inhibitory factor receptor | G#VQDLK*CTTNNMR |
| CD5 | E#SPTK*VQLVGGAHR |
#, Tagged N-terminal residue by propionylation or acetylation.
*, Propionylation of lysine residues.
IGFBP3, Insulin-like growth factor-binding protein 3.
TABLE 3.
LC-MS/MS identified N-terminal peptides from complement C3 and EGFR
| Complement C3 | Identified N-terminal peptide sequence | Propionylation (56 Da) | Acetylation (42 Da) | Event annotation |
|---|---|---|---|---|
| Complement C3 | I#PMYSIITPNVLR | 7 | Signal peptide removed | |
| S#VQLMER | 8 | C3 α chain | ||
| S#ELEEDIIPEEDIISR | 1 | C3b α chain | ||
| V#DVPAADLSDQVPDTDSETR | 6 | Trimmed (5AA) C3g fragment | ||
| S#EETK*QNEAFSLTAK | 3 | C3c α chain fragment 2 | ||
| T#DPGHEAK*IR | 3 | Internal cleavage | ||
| EGFR | G#ALEEK*K*VCQGTSNR | 4 | Trimmed proEGFR | |
| A#LEEK*K*VCQGTSNR | 1 | 2 | Signal peptide removed | |
| L#EEK*KVCQGTSNR | 4 | Trimmed |
#, Tagged N-terminal residue by propionylation or acetylation.
*, Propionylation of lysine residues.
EGFR, Epidermal growth factor receptor.
FIGURE 3.
SUMMARY
- Identified N-terminal peptides (unique) from 10 μL mouse plasma, using one-dimensional LC-MS/MS:
- —211 with propionlyation tags
- —40 acetylated
- —four identified with both forms
- Future experiments include:
- —applying this method toward additional sample types
- —further optimization
- Identification of different proteolytic products can lead to:
- —novel drug targets or biomarkers
- —understanding disease biology
ACKNOWLEDGMENTS
This research was supported by the National Institutes of Health National Center for Research Resources (RR18522) and Laboratory Directed Research Funds at Pacific Northwest National Laboratory (PNNL). Work was performed in the Environmental Molecular Sciences Laboratory, a U.S. Department of Energy (DOE) national scientific user facility on the PNNL campus in Richland, WA, USA. PNNL is operated for the DOE by Battelle under Contract DE-AC05-76RLO.
REFERENCES
- 1.Gevaert K, Goethals M, Martens L, et al. Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides. Nat Biotechnol 2003; 21: 566– 569 [DOI] [PubMed] [Google Scholar]
- 2.McDonald L, Robertson DH, Hurst JL, Beynon RJ. Positional proteomics: selective recovery and analysis of N-terminal proteolytic peptides. Nat Methods 2005; 2: 955– 957 [DOI] [PubMed] [Google Scholar]