Abstract
The Antigen I/II (AgI/II) family of proteins are cell wall anchored adhesins expressed on the surface of oral streptococci. The AgI/II proteins interact with molecules on other bacteria, on the surface of host cells, and with salivary proteins. Streptococcus gordonii is a commensal bacterium, and one of the primary colonizers that initiate the formation of the oral biofilm. S. gordonii expresses two AgI/II proteins, SspA and SspB that are closely related. One of the domains of SspB, called the variable (V-) domain, is significantly different from corresponding domains in SspA and all other AgI/II proteins. As a first step to elucidate the differences among these proteins, we have determined the crystal structure of the V-domain from S. gordonii SspB at 2.3 Å resolution. The domain comprises a β-supersandwich with a putative binding cleft stabilized by a metal ion. The overall structure of the SspB V-domain is similar to the previously reported V-domain of the Streptococcus mutans protein SpaP, despite their low sequence similarity. In spite of the conserved architecture of the binding cleft, the cavity is significantly smaller in SspB, which may provide clues about the difference in ligand specificity. We also verified that the metal in the binding cleft is a calcium ion, in concurrence with previous biological data. It was previously suggested that AgI/II V-domains are carbohydrate binding. However, we tested that hypothesis by screening the SspB V-domain for binding to over 400 glycoconjucates and found that the domain does not interact with any of the carbohydrates.
Keywords: Streptococcus gordonii, X-ray crystallography, dental plaque, structural biology
Introduction
Dental plaque is a mixed-species biofilm that can be made up from of over 700 species of micro-organisms.1 Commensal bacteria such as gram-positive oral streptococci and Actinomyces spp. are primary colonizing bacteria that initiate the formation of dental plaque by adhering to the tooth surface. When bound, they constitute adhesion sites for secondary colonizers, usually gram-negatives.
Streptococcal species may comprise up to 78% of the bacteria in the early plaque2 and have the ability to interact with a wide variety of salivary proteins, bacteria, and host cells. Streptococcus gordonii is a commensal bacterium which colonizes multiple sites in the human oral cavity. Its interaction properties are important not only for initial adhesion to saliva-coated surfaces but also for interbacterial adhesion and secondary colonization by organisms such as Porphyromonas gingivalis.3 Therefore S. gordonii is believed to play an important role in the development of bacterial communities associated with dental caries, gingivitis, and periodontitis.4 Furthermore, the bacterium has a pathogenic potential at non-oral sites, for example, as an etiological agent in infective endocarditis.5 S. gordonii is a complex organism that expresses a number of cell surface proteins with which it interacts with its surroundings; CshA and CshB bind to fibronectin,6,7 Hsa binds to cell surface glycoproteins on leukocytes,8 and GspB binds to platelets.9 S. gordonii also expresses two cell surface proteins SspA and SspB that belong to the antigen I/II (AgI/I) family of proteins.10
AgI/II proteins are expressed by virtually all species of oral streptococci. The proteins are structurally well conserved within the family. The precursor proteins are 1310–1653 amino acids long and contain seven discrete regions based on primary sequence (see Fig. 1). These are a signal peptide, an N-terminal (N-) region, an alanine-rich repeat (A-) domain, a variable (V-) domain, a proline-rich repeat (P-) region, a C-terminal domain, and a cell wall anchoring region.10 The cell wall anchored adhesins SspA and SspB expressed by S. gordonii mediate a wide range of interactions with host proteins and other bacteria. For example, they mediate binding to salivary agglutinin glycoprotein (SAG),11 collagen type I12, β1-integrin,13 Actinomyces naeslundii,14,15 and P. gingivalis16. The N- and C-terminal regions of SspA and SspB are very similar in sequence, 84 and 98%, respectively, in sequence identity. The major difference between the proteins resides in the V-domains that only display 27% sequence identity (see Fig. 2). Overall the V-domain is the most variable region within all proteins of the AgI/II family, although the overall sequence identity for PAc (Streptococcus mutans NG8), SpaPG (S.mutans Guy's) and SpaPI (S. mutans Ingbritt), and Pas (Streptococcus intermedius) is 75% in the V-region.15 The V-domain of SspB is most different with only 26% identity to the other AgI/II proteins. In fact, despite the high sequence similarities between SspA and SspB, the variable region of SspA more closely resembles that of S. mutans PAc (78% sequence identity).11
Figure 1.
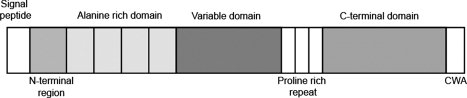
Domain architecture of Ag I/II proteins. The 1500 residue protein is comprised of seven regions (distinguished by shading): a signal peptide is followed by an N-terminal region, an alanine-rich repeat domain, a variable domain, a proline-rich repeat region, a C-terminal domain and a cell-wall anchoring region (CWA).
Figure 2.
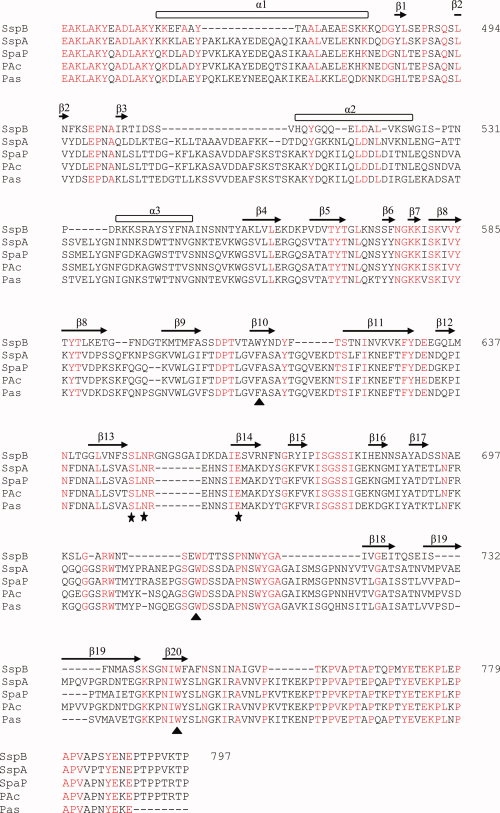
Sequence alignment. Multiple sequence alignment of SspB-V and related AgI/II variable domains. Sequences from the following organisms were compared; S. gordonii (SspB); S. gordonii (SspA), S. mutans NG5 (SpaP); S. mutans (PAc) and S. intermedius (Pas). Secondary structure elements of S. gordonii SspB-V are indicated above the sequence. Conserved residues are shown in red. The metal coordinating residues are marked with filled stars and the two conserved tryptophans and a third tryptophan/phenylalanine located in the binding pocket are marked with filled triangles. Numbering is according to the sequence of S. gordonii SspB-V.
Despite the sequence similarity of AgI/II proteins, evidence suggests that different members of the protein family have different binding specificity. Brooks et al.17 showed that AgI/II SspB of S. gordonii binds to P. gingivalis fimbrial protein Mfa1, whereas the related AgI/II PAc from Streptococcus mutans does not. It has also been shown that the substrate-binding properties of S. gordonii SspA and SspB expressed on the surface of Lactococcus lactis differ in affinity for SAG and collagen substrates.18 Furthermore, SspA and SspB also have independent functions in coaggregation with different A. naeslundii coaggregation groups.14 By studying two chimeric polypeptides, one comprising the A- and V-domains of SspA fused to the P- and C-domains of SspB and the other comprising the A- and V-domains of SspB fused to the P- and C-domains of SspA, for aggregation with A. naeslundii groups, Jakubovics et al.15 conclude that the A- and V-domains determine their coaggregation specificities.
Previously, one crystal structure of an AgI/II variable domain had been determined, the S. mutans SpaP V-domain (SpaP-V).19 From that crystal structure, it was proposed that the V-domain adopts a lectin-like fold that displays a putative preformed carbohydrate-binding site. The fact that SspB is most different regarding the AgI/II V-domains, and that substrates and function are unknown make it of interest to study this protein in more detail.
We undertook studies to determine the crystal structure of the S. gordonii SspB V-domain (SspB-V) and to compare its structure to the homologous S. mutans SpaP-V structure. Here we report the 2.3 Å resolution structure of the S. gordonii SspB V-domain, which shares the same overall fold with the S. mutans protein SpaP-V, albeit it has a significantly smaller presumed binding pocket. In the proposed ligand binding pocket of SspB-V a Ca2+ ion is bound, apparently crucial for the correct folding of the domain. A glycan array also suggests that SspB-V is possibly not carbohydrate binding, and thus the function of the protein remains to be elucidated.
Results and Discussion
Structure determination
The full length SspB protein consists of 1500 residues divided into seven domains. The crystal structure of the recombinant V-domain was solved by single anomalous dispersion (SAD) on a single crystal of Selenomethionine (SeMet)-substituted protein to a resolution of 3.2 Å. Native data to 2.3 Å resolution was subsequently used for refinement. The asymmetric unit contains two molecules of SspB-V (A and B). The molecules are very similar with an rms deviation (rmsd) of 0.3 Å for all observed Cα atoms. The final model is well ordered with an overall B-factor of 24.8 Å2 and a final crystallographic R factor of 0.18 (Rfree = 0.21). The structure consists of residues 462–763 in chain A and 468–760 in chain B as well as one Ca2+ ion per chain, and one K+ ion located at the interface between the two molecules. Four glycerol molecules and 281 water molecules have also been included in the model. The numbering is based on the full-length SspB protein. The histidine tag, the linker, and the first 13 residues, as well as the last 32 residues of the C-terminal are flexible and were not included in the final model. Diffraction quality crystals could only be obtained after subjecting the surface lysines to reductive methylation.20 Although the methylation reduced the surface entropy and was crucial for crystal growth, the electron density for the surface lysines was not of such quality that the methyl groups were modeled.
Overview of the SspB-V structure
The V-domain is organized into a central immunoglobulin-like β-sandwich flanked by two subdomains, SDA and SDB [Fig. 3(A,B)]. The β-sandwich consists of two anti-parallel sheets comprising eight (S1) and seven β-strands (S2), respectively. The sheets pack against each other adopting a β-supersandwich-like fold.21 The S1 and S2 sheets are connected by two helices, α2 and α3, bridging β3 and β4 that build up the major part of SDA. SDB is positioned on the other side of the β-sandwich and comprises a long loop connecting β13 with β14, and a stretch connecting β17 with β18 composed of turns and three shorter segments with 310 helix character. In addition, the structure comprises a small three-stranded anti-parallel sheet that lies almost perpendicular to the β-sandwich. The S2 sheet is broken between strands β20 and β13, separated by a metal ion, modeled as a Ca2+ ion. In connection to the Ca2+ ion, a putative ligand binding pocket resides, positioned between SDA and SDB and the top and bottom β-sheet. Furthermore, a long N-terminal helix (∼30 Å) and a C-terminal coiled peptide that runs anti-parallel to it, protrude from the core of the protein to form a hinge that connects the SspB-V with the neighboring domains of the protein, the A-domain on the N-terminal side, and the P-region on the C-terminal. It is unclear whether the C-terminal peptide is technically a part of the P-region or not but it indeed contains two prolines that pack against aromatic and hydrophobic residues in the N-terminal helix. Also, the N-terminal helix may belong to the A-domain; the helix contains six alanines. The hinge is anchored to the protein core by hydrogen bonds between the C-terminal peptide and α1. There is also a salt bridge between Asp483 in the N-terminal helix and Arg703 in SDB.
Figure 3.
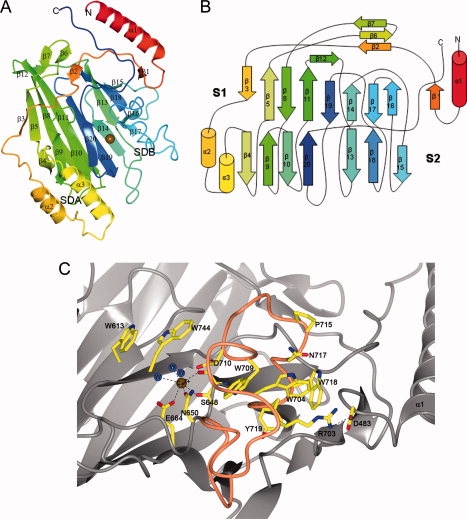
Overall structure of SspB-V. (A) Ribbon representation of SspB-V. The coloring blends through the model from red (N-terminus) to blue (C-terminus). The domain is organized into a central immunoglobulin-like β-sandwich flanked by two subdomains, subdomain A (SDA) and subdomain B (SDB). The β-sandwich consists of two anti-parallel β-sheets packed against each other in a β-supersandwich-like fold with a putative binding pocket stabilized by a Ca2+ ion, depicted as a gold sphere. (B) Topology diagram of SspB-V. β-strands are represented as arrows, helices as cylinders and loops as lines. Colors are the same as in (A). (C) The putative binding pocket. The Ca2+ ion (gold sphere) is coordinated by four atoms of the protein and three water molecules (blue spheres). The coiled region comprising the major part of SDB is colored orange. Amino acids in the binding pocket and in SDB are represented as ball-and-stick models and are labeled.
The putative binding pocket contains a tightly bound calcium ion
An open pocket is formed in association with the Ca2+ ion. The cavity is found between strands β13 and β20 of sheet S2 and strands β14 and β19 of the underlying sheet S1. Further, the turn after β20, which runs perpendicular to the upper part of S2, and a turn in the segment connecting β17 and β18 in SDB, contribute to the cavity. The Ca2+ ion is coordinated by four protein atoms: Ser648 O and OG, Asn650 OD1 from β13 in S2, and Glu664 OE1 from β14 in S1. The metal is further coordinated by three water molecules [Fig. 3(C)]. Although the crystals were grown in 0.2M KBr, the metal ion in the pocket is modeled as Ca2+ because the metal-protein distances refine to an average of 2.36 ± 0.12 Å, which is more consistent with Ca2+ (2.36–2.39 Å) than with K+ (2.74–2.82 Å).22
To verify if the SspB-V domain is stabilized by the bound metal, thermally induced melting analysis was performed on a set of mono-, and divalent ions using the thermofluor method.23 Results showed that the protein was most stable when supplemented with CaCl2 (Tm = 49°C). Other divalent metals, Mg2+ and Mn2+, also gave a relatively high melting point (∼45°C). Monovalent ions, Na+, K+, Li+, did not affect the thermostability when compared with the nonmetal supplemented protein (Tm = 41°C). The concentration of the metal did not alter the Tm. The presence of Ca2+ is also supported by the notion that Ca2+ is needed for aggregation to occur between salivary gp340 and lactococci expressing AgI/II on their surfaces15 and for the adherence of SspA and SspB proteins to β1-integrin.13
The putative binding pocket exhibits a predominantly negative electrostatic surface potential in close connection with the metal ion [Fig. 4(A)]. The opening of the pocket is also lined with two conserved tryptophans (709 and 744) and a third tryptophan (613) (phenylalanine in the other aligned sequences) [Figs. 2 and 3(C)] that may stack with, or form a lid over the bound ligand. Interestingly, in the coiled region that constitutes most of SDB, conserved residues cluster in a core of mostly hydrophobic and aromatic residues (Trp704, Trp709, Trp718, and Tyr719) [Figs. 3(C) and 4(A,B)]. The SDB coiled region is linked to the N-terminal helix via a salt bridge between Arg703 and Asp483 [Fig. 3(C)]. One can hypothesize that upon conformational changes in the A-domain, or when a ligand is bound, the salt bridge is broken and the structure of the coiled SDB is rearranged, perhaps to form a hydrophobic and aromatic lid over the binding pocket. Electron density in the opening of the binding pocket has been modeled as two glycerol molecules since glycerol was used for cryoprotection.
Figure 4.
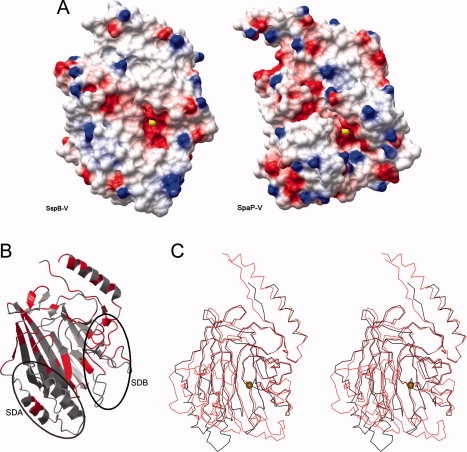
Structure comparison and analysis. (A) Electrostatic surface of SspB-V (left) and SpaP-V, pdb code 1JMM,19 (right). The molecular surfaces are colored blue and red according to positive and negative electrostatic potential, respectively. The Ca2+ ion is represented by a yellow sphere. (B) Ribbon representation of SspB-V in which the strictly conserved residues are colored red. SDA (Subdomain A) and SDB (Subdomain B) are indicated by oval lines. (C) Stereo view of SspB-V and SpaP-V. SspB-V is colored in black and SpaP-V in red. The Ca2+ ion is represented by a gold sphere. In S. gordonii and S. mutans V-domains, the core of the β-sandwich is highly conserved. The main structural differences reside in SDA resulting in a smaller and shallower pocket in SspB-V.
A homology search
A homology search using the DALI server24 identified the S. mutans SpaP-V domain (PDB code 1JMM19) as the closest structural relative (Z-score 31.2), as expected. A comparison between the structures will be discussed below. The DALI search also resulted in several distant relatives, mostly carbohydrate-binding proteins. The second closest structural relative is β-mannosidase (Z-score 4.6, PDB code 1OF325), a protein belonging to the Galactose binding domain superfamily. Comparison of the three-dimensional structure and topology of SspB-V with β-mannosidase shows that the overall fold of the β-sandwich is similar (rmsd 4.1 Å calculated on 127 aligned Cα atoms). However, the carbohydrate binding site of β-mannosidase is located on the opposite side of the β-sandwich when compared with the presumed ligand cleft in SspB-V. If we examine the surface of SspB-V equivalent to the β-mannosidase carbohydrate-interacting site, we find that the SspB-V surface constitutes mostly polar and positively charged amino acids and no aromatic residues. Hence, this surface contains residues normally not favored in carbohydrate binding. In addition, this area of the V-domain is not conserved among the AgI/II family. We therefore conclude that although the two proteins are distant structural relatives, SspB-V most likely does not share the mode of substrate binding.
Comparison between S. gordonii SspB-V and S. mutans SpaP-V
SspB-V is 61 residues shorter than SpaP-V and the sequence identity between them is 26%. Despite that, the core of the β-sandwich is structurally conserved (rmsd of 2.9 Å based on 281 aligned Cα atoms) [Fig. 4(C)]. The most conserved regions in the protein are the contact area between the N-terminal helix and the C-terminal coil and areas in connection with the ligand binding pocket [Fig. 4(B)]. The most significant differences reside in SDA which is among the least conserved regions in the V-domains. In SspB-V, SDA comprises two helices and the loop region that connects them, whereas SDA in SpaP-V is larger and constitutes a helix-turn-helix motif and a small β-sheet.
In contrast, SDB is well conserved, both in structure and sequence (discussed above). The main difference in SDB is that SpaP-V has a seven-residue turn, not present in SspB-V, inserted in the 310 helix region. This turn together with the larger SDA make the walls of the SpaP-V binding pocket significantly more pronounced, resulting in a larger and deeper cleft. In SspB-V the two strands β13 and β14 are longer than the corresponding strands in SpaP-V and the turn that connects those seals the bottom of the pocket and makes it smaller and less accessible than the SpaP-V pocket. As calculated by the CASTp server,26 the pocket volume of SspB-V is five times smaller than the SpaP-V pocket (418 and 2280 Å3, respectively). A narrower pocket indicates that smaller molecules fit into the putative binding site in SspB-V when compared with SpaP-V, but we can only speculate that this may result in different binding specificity.
Inside the pocket there is a metal-binding site present in both structures. This indicates that it is a structurally conserved binding site which may be required for conformational stability of the AgI/II V-domains. The residues coordinating the metal in the two structures are conserved and albeit the metal in SpaP-V is modeled as Na+, we expect that the native ion in all the V-domains is Ca2+. Troffer-Charlier et al.19 speculated that the metal-containing pocket constitutes a preformed carbohydrate-binding site. This is a likely possibility judging from the negative electrostatic surface potential along with aromatic residues in the pockets [Figs. 3(C) and 4(A)]. However, ligands to neither SpaP-V nor SspB-V have been determined and there is a possibility that molecules of another nature can bind into the pocket or to other binding surfaces of the domain. Additional minor pockets are identified by the CASTp server, but these are significantly smaller (<185 Å3). Nevertheless, other binding surfaces may be created when the full-length protein is folded.
Carbohydrate binding
To clarify whether SspB-V is carbohydrate binding or not a glycan array screening was performed at the Consortium for Functional Glycomics, Core H. A total of 406 glycans (of mammalian and pathogen origin) were screened but no binding was detected for any of the sugars. From this analysis, it can be concluded that SspB-V is probably not carbohydrate binding. Because the SspB-V is only one domain of several in the SspB protein, one must also consider the possibility that any binding may be enhanced in the presence of additional domains.
Interaction properties of SspB-V
SspB-V is unique among the AgI/II family and exhibits different interaction properties when compared with the other proteins. It has been shown that AgI/II proteins are responsible for distinguishing between different strains of Actinomyces naeslundii and that SspB-V dictates the aggregation to A. naeslundii group A and E but not any of the other A. naeslundii groups.15 Other studies have revealed that the N-terminal parts of SspA and SspB (including the NAV domains) are responsible for the adherence to β1-integrin.13 Because no difference is seen in binding to SspA and SspB, integrin recognition must be different from Actinomyces recognition. Therefore, the V-region may be versatile; it is the sole domain for discriminating between different Actinomyces strains but one out of several that has to fold correctly to bind integrin.
In SspB-V as well as in SpaP-V, the N- and C-termini form a stalk that protrudes from the V-domain. This implies that the N- and C-parts of the AgI/II proteins pack against each other and expose the V-domain to interacting microorganisms and other binding partners. In addition to the structures, this hypothesis is supported by the fact that Guy's monoclonal antibody recognize epitopes from both the A-domain and the P-region,27 thus they form tight interactions in the functional protein. For a deeper understanding of how AgI/II proteins are folded, and to elucidate the mechanism by which they bind to their interaction partners, emphasis has to be put on further structural and functional studies on all domains, either alone or in combination with each other. This type of knowledge is necessary for design of anti-adhesive substances to prevent detrimental bacteria–bacteria or bacteria–host cell interactions.
Methods
Cloning, protein expression, and purification
Plasmid pEB-528 encoding sspB was used as a template to amplify the S. gordonii sspB V-region. The gene fragment encoding the V-domain was PCR amplified using primers based on the gene sequence corresponding to sspB from S. gordonii M5 (accession code U40026). Forward and reverse primers were 5′AAA AAC CAT GGA AGC TAA ATT AGC AAA A3′ and 5′TTT TTG GTA CCT TAT GGA GTT TTT ACC GGT GGA GT-3′, respectively. The PCR product was cleaved with Acc65I and NcoI and then ligated into the equivalent sites of the pET-M11 expression vector (EMBL, Germany). The final plasmid encodes a hexa-histidine tag followed by a cloning vector linker (PMSNTNIPTTENLYPEGAM) and the V-domain, SspB449-797. The construct was verified with DNA sequencing. The expression construct was overexpressed in E. coli BL21 (DE3) cells at 37°C in Luria-Bertani media supplemented with 50 μg mL−1 kanamycin. When the culture reached an OD600 of 0.6, the temperature was lowered to 30°C and the expression was induced with 0.4 mM IPTG after which the culture was grown for an additional 5 h. Cells were harvested by centrifugation at 5000g and the pellet was frozen at −80°C. Cells were resuspended in 20 mM Tris pH 7.5, 150 mM NaCl, and 10 mM imidazole supplemented with EDTA-free protease inhibitor cocktail (Roche). The cells were lysed on ice by sonication, and cellular debris was removed by centrifugation. The supernatant was loaded onto a column packed with 3 mL Ni-NTA (Qiagen). SspB-V was eluted in 20 mM Tris pH 7.5, 150 mM NaCl, and 300 mM imidazole. The buffer was exchanged to 20 mM Tris pH 7.5, 150 mM NaCl, 0.5 mM EDTA, and 1 mM DTT, and the protein was further purified by gel filtration on a Superdex 200 16/60 PG column (Amersham Biosciences). Protein purity was judged by SDS-PAGE. For crystallization, the purified protein was subjected to reductive methylation.20 In short, SspB-V at 1 mg mL−1 in 50 mM HEPES pH 7.5 and 250 mM NaCl was incubated with 20 μL of dimethylamine-borane complex (ABC) and 40 μL of 1M formaldehyde per mL protein solution for 2 h at 4°C. Again, 20 μL of ABC and 40 μL of formaldehyde were added and the incubation continued for 2 h. After a final addition of 20 μL of ABC, the reaction was incubated over night. After concentration and buffer exchange to 20 mM Tris pH 7.5, 150 mM NaCl, 1 mM DTT, the protein was purified by another round of gelfiltration. The protein was concentrated to 18 mg mL−1 in 20 mM Tris pH 7.5 and 1 mM DTT using an Amicon Ultra centrifugal filter device (Millipore), frozen in liquid nitrogen and stored at −80°C. The His-tag was not removed before crystallization trials.
For expression of SeMet substituted protein, cells were grown in M9 media supplemented with glucose at 37°C. At an OD600 of 0.6, lysine, threonine, phenylalanine at 100 mg L−1, and leucine, isoleucine, valine, proline, and SeMet at 50 mg L−1 were added to down-regulate the synthesis of methionine.29 The culture was cooled to 20°C, protein expression induced with 0.4 mM IPTG, and the culture grown overnight. SeMet labeled SspB-V was purified as described above for the wild-type protein, except that tris (2-carboxyethyl)-phosphine hydrocholoride (TCEP) was used instead of DTT. The SeMet substituted protein was also subjected to reductive methylation before crystallization.
Thermal shift assay
SspB-V was screened for stabilizing metal-buffer conditions using the thermoflour method.23 In short, solutions of 7.5 μL 10× Sypro Orange (Molecular Probes), 12.5 μL 2× test compounds (buffer and metal), 2 μL 7.5 mg mL−1 SspB-V protein, and 3 μL water were added to 0.1 mL PCR tubes and heated in a qPCR detection system (Rotor-Gene 6000, Corbett Life Science) from 28 to 80°C in increments of 0.2°C. Changes in fluorescence were monitored and the melting temperature (Tm) was determined by calculating the derivative of the midpoint of the protein unfolding transition. The experiment was performed in triplicate in 100 mM Tris pH 7.5 supplemented with NaCl, KCl, LiCl, CaCl2, MgCl2, and MnCl2 at concentrations of 30, 15, and 7.5 mM.
Glycan array
SspB-V in 25 mM Tris pH 8.0 was concentrated to 10 mg mL−1 and lyophilized (total amount 0.9 mg). The protein was dissolved in 20 mM Tris-HCL pH 7.4, 150 mM NaCl, 2 mM CaCl2, 2 mM MgCl2, 0.05% Tween 20, and 1% BSA to a concentration of 200 μg mL−1. A total of 406 glycans in replicates of six were screened and any binding was detected with an anti-his antibody. The glycan array was performed by the Consortium for Functional Glycomics, Core H (https://www.functionalglycomics.org).
Protein crystallization and data collection
Crystals of native SspB-V were grown by vapor diffusion using the hanging-drop method at room temperature. Drops consisted of 1 μL of protein at 18 mg mL−1 mixed with 1 μL reservoir solution containing 25% (w/v) PEG3350 and 0.2M KBr. The crystals grew within 2 weeks in space group P65 with cell dimensions a = b = 110.2 Å, c = 121.5 Å, with two molecules in the asymmetric unit. Crystals of native protein were cryo-protected by soaking the crystals in mother liquid supplemented with 10% glycerol for a few seconds, then flash frozen in a stream of liquid nitrogen at 100 K. The SeMet-labeled protein crystals were grown in hanging-drops consisting of 1 μL protein at 13 mg mL−1 and 1 μL reservoir solution containing 20% (w/v) PEG 6000, 100 mM HEPES pH 7.3, and 0.2M LiCl. SeMet crystals were equilibrated in cryoprotectant solution containing 20% (w/v) PEG 6000, 100 mM Tris pH 7.5, and 10% glycerol. Native data to 2.3 Å resolution and a single-wavelength anomalous diffraction (SAD) data set to 3.2 Å were collected on a marmosaic 225 detector at beam line I911-3 at MAX-lab, Lund. Diffraction images were processed with MOSFLM30 and scaled with SCALA from the CCP4 program suit.31 Relevant processing statistics are presented in Table I.
Table I.
Data Collection, Refinement, and Model Quality Statistics for SspB-V
| Native | SeMet SAD | |
|---|---|---|
| Data processing | ||
| Space group | P65 | P65 |
| Cell dimensions a, b, c (Å) | 110.17, 110.17, 121.49 | 111.19, 111.19, 125.93 |
| Wavelength (Å) | 0.917 | 0.97855 |
| Resolution (Å) | 60.75–2.3 | 38.48–3.2 |
| Highest resolution shell (Å) | 2.42–2.3 | 3.37–3.2 |
| Total reflectionsa | 408,683 (56,049) | 167,609 (24,545) |
| Unique reflectionsa | 37,172 (5381) | 14,658 (2139) |
| I/σ (I)a | 24.4 (4.4) | 23.6 (12.7) |
| Rmergeab (%) | 9.6 (52.2) | 10.3 (16.2) |
| Completenessa (%) | 100 (99.9) | 99.9 (100) |
| Overall redundancya | 11.0 (10.4) | 11.4 (11.5) |
| Refinement | ||
| Rwork/Rfreec (%) | 17.6/20.9 | |
| Average B-factors (Å2) | ||
| Protein A-chain/B-chain | 23.9/24.9 | |
| Water | 30.1 | |
| Calcium ions | 21.2 | |
| No of protein atoms | 4638 | |
| No of metal ions | 3 | |
| No water molecules | 281 | |
| No of glycerol molecules | 4 | |
| RMSD from ideal | ||
| Bond lengths (Å) | 0.020 | |
| Bond angles (°) | 1.634 | |
| Ramachandran plot (most favored, allowed, generously allowed, disallowed) (%) | 86.3/12.8/0.6/0.4 | |
Values in parentheses indicate statistics for the highest resolution shell.
Rmerge = Σhkl Σi |Ii(hkl) − <I(hkl)>|/ Σhkl Σi Ii (hkl), where Ii(hkl) is the intensity of the ith observation of reflection hkl and <I(hkl)> is the average of all observations of reflection hkl.
Rwork = Σ | |Fobs| − | Fcalc| |/Σ|Fobs|, where Fobs and Fcalc are the observed and calculated structure factor amplitudes, respectively. Rfree is Rwork calculated using 5% of the data, randomly omitted from refinement.
Structure determination and refinement
The structure of the SspB-V was solved with SAD phasing using the program suite PHENIX.32 Locations of four Se atoms per monomer were determined and reflection phases to 3.2 Å resolution were calculated and improved with density modification and molecular averaging in SOLVE/RESOLVE (PHENIX). Automatic model building by the PHENIX routine AutoBuild resulted in a readily interpreted electron density map. Native data at 2.3 Å resolution was used for refinement with 5% of the data removed for calculation of Rfree. The full atomic model was built into the electron density using COOT33 and the model refined using REFMAC5.34 In the last rounds of refinement translational-liberation-screw (TLS) refinement was used, treating each molecule as an individual TLS group.34 The quality of the model was analyzed with WHATCHECK.35 Refinement statistics are presented in Table I. Figures were drawn with CCP4MG.36 The X-ray coordinates and structure factors have been deposited in the Protein Data Bank under accession code 2wd6.
Acknowledgments
We are grateful for the access to the beamline I911-3 at MAXlab, Lund and for the support of beamline scientists. We thank Gunter Stier, EMBL, Germany for cloning vectors.
Glossary
Abbreviations:
- Ag I/II
Antigen I/II
- SspB-V
SspB variable domain
- SpaP-V
SpaP variable domain
- SDA
Subdomain A
- SDB
Subdomain B
- SAG
Salivary agglutinin glycoprotein.
References
- 1.Jenkinson HF, Lamont RJ. Oral microbial communities in sickness and in health. Trends Microbiol. 2005;13:589–595. doi: 10.1016/j.tim.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Nyvad B, Kilian M. Comparison of the initial streptococcal microflora on dental enamel in caries-active and in caries-inactive individuals. Caries Res. 1990;24:267–272. doi: 10.1159/000261281. [DOI] [PubMed] [Google Scholar]
- 3.Jenkinson HF, Lamont RJ. Streptococcal adhesion and colonization. Crit Rev Oral Biol Med. 1997;8:175–200. doi: 10.1177/10454411970080020601. [DOI] [PubMed] [Google Scholar]
- 4.Rosan B, Lamont RJ. Dental plaque formation. Microbes Infect. 2000;2:1599–1607. doi: 10.1016/s1286-4579(00)01316-2. [DOI] [PubMed] [Google Scholar]
- 5.Douglas CW, Heath J, Hampton KK, Preston FE. Identity of viridans streptococci isolated from cases of infective endocarditis. J Med Microbiol. 1993;39:179–182. doi: 10.1099/00222615-39-3-179. [DOI] [PubMed] [Google Scholar]
- 6.McNab R, Holmes AR, Clarke JM, Tannock GW, Jenkinson HF. Cell surface polypeptide CshA mediates binding of Streptococcus gordonii to other oral bacteria and to immobilized fibronectin. Infect Immun. 1996;64:4204–4210. doi: 10.1128/iai.64.10.4204-4210.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McNab R, Jenkinson HF, Loach DM, Tannock GW. Cell-surface-associated polypeptides CshA and CshB of high molecular mass are colonization determinants in the oral bacterium Streptococcus gordonii. Mol Microbiol. 1994;14:743–754. doi: 10.1111/j.1365-2958.1994.tb01311.x. [DOI] [PubMed] [Google Scholar]
- 8.Urano-Tashiro Y, Yajima A, Takashima E, Takahashi Y, Konishi K. Binding of the Streptococcus gordonii DL1 surface protein Hsa to the host cell membrane glycoproteins CD11b, CD43, and CD50. Infect Immun. 2008;76:4686–4691. doi: 10.1128/IAI.00238-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bensing BA, Sullam PM. An accessory sec locus of Streptococcus gordonii is required for export of the surface protein GspB and for normal levels of binding to human platelets. Mol Microbiol. 2002;44:1081–1094. doi: 10.1046/j.1365-2958.2002.02949.x. [DOI] [PubMed] [Google Scholar]
- 10.Jenkinson HF, Demuth DR. Structure, function and immunogenicity of streptococcal antigen I/II polypeptides. Mol Microbiol. 1997;23:183–190. doi: 10.1046/j.1365-2958.1997.2021577.x. [DOI] [PubMed] [Google Scholar]
- 11.Demuth DR, Duan Y, Brooks W, Holmes AR, McNab R, Jenkinson HF. Tandem genes encode cell-surface polypeptides SspA and SspB which mediate adhesion of the oral bacterium Streptococcus gordonii to human and bacterial receptors. Mol Microbiol. 1996;20:403–413. doi: 10.1111/j.1365-2958.1996.tb02627.x. [DOI] [PubMed] [Google Scholar]
- 12.Love RM, McMillan MD, Jenkinson HF. Invasion of dentinal tubules by oral streptococci is associated with collagen recognition mediated by the antigen I/II family of polypeptides. Infect Immun. 1997;65:5157–5164. doi: 10.1128/iai.65.12.5157-5164.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nobbs AH, Shearer BH, Drobni M, Jepson MA, Jenkinson HF. Adherence and internalization of Streptococcus gordonii by epithelial cells involves beta1 integrin recognition by SspA and SspB (antigen I/II family) polypeptides. Cell Microbiol. 2007;9:65–83. doi: 10.1111/j.1462-5822.2006.00768.x. [DOI] [PubMed] [Google Scholar]
- 14.Egland PG, Du LD, Kolenbrander PE. Identification of independent Streptococcus gordonii SspA and SspB functions in coaggregation with Actinomyces naeslundii. Infect Immun. 2001;69:7512–7516. doi: 10.1128/IAI.69.12.7512-7516.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jakubovics NS, Kerrigan SW, Nobbs AH, Stromberg N, van Dolleweerd CJ, Cox DM, Kelly CG, Jenkinson HF. Functions of cell surface-anchored antigen I/II family and Hsa polypeptides in interactions of Streptococcus gordonii with host receptors. Infect Immun. 2005;73:6629–6638. doi: 10.1128/IAI.73.10.6629-6638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamont RJ, El-Sabaeny A, Park Y, Cook GS, Costerton JW, Demuth DR. Role of the Streptococcus gordonii SspB protein in the development of Porphyromonas gingivalis biofilms on streptococcal substrates. Microbiology. 2002;148:1627–1636. doi: 10.1099/00221287-148-6-1627. [DOI] [PubMed] [Google Scholar]
- 17.Brooks W, Demuth DR, Gil S, Lamont RJ. Identification of a Streptococcus gordonii SspB domain that mediates adhesion to Porphyromonas gingivalis. Infect Immun. 1997;65:3753–3758. doi: 10.1128/iai.65.9.3753-3758.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holmes AR, Gilbert C, Wells JM, Jenkinson HF. Binding properties of Streptococcus gordonii SspA and SspB (antigen I/II family) polypeptides expressed on the cell surface of Lactococcus lactis MG1363. Infect Immun. 1998;66:4633–4639. doi: 10.1128/iai.66.10.4633-4639.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Troffer-Charlier N, Ogier J, Moras D, Cavarelli J. Crystal structure of the V-region of Streptococcus mutans antigen I/II at 2.4 a resolution suggests a sugar preformed binding site. J Mol Biol. 2002;318:179–188. doi: 10.1016/S0022-2836(02)00025-6. [DOI] [PubMed] [Google Scholar]
- 20.Walter TS, Meier C, Assenberg R, Au KF, Ren J, Verma A, Nettleship JE, Owens RJ, Stuart DI, Grimes JM. Lysine methylation as a routine rescue strategy for protein crystallization. Structure. 2006;14:1617–1622. doi: 10.1016/j.str.2006.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lo Conte L, Ailey B, Hubbard TJ, Brenner SE, Murzin AG, Chothia C. SCOP: a structural classification of proteins database. Nucleic Acids Res. 2000;8:257–259. doi: 10.1093/nar/28.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harding MM. Small revisions to predicted distances around metal sites in proteins. Acta Crystallogr D Biol Crystallogr. 2006;62:678–682. doi: 10.1107/S0907444906014594. [DOI] [PubMed] [Google Scholar]
- 23.Ericsson UB, Hallberg BM, Detitta GT, Dekker N, Nordlund P. Thermofluor-based high-throughput stability optimization of proteins for structural studies. Anal Biochem. 2006;357:289–298. doi: 10.1016/j.ab.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 24.Holm L, Kaariainen S, Wilton C, Plewczynski D. Using Dali for structural comparison of proteins. Current Protocols in Bioinformatics; 2006 doi: 10.1002/0471250953.bi0505s14. Chapter 5:Unit 5.5. [DOI] [PubMed] [Google Scholar]
- 25.Boraston AB, Revett TJ, Boraston CM, Nurizzo D, Davies GJ. Structural and thermodynamic dissection of specific mannan recognition by a carbohydrate binding module, TmCBM27. Structure. 2003;11:665–675. doi: 10.1016/s0969-2126(03)00100-x. [DOI] [PubMed] [Google Scholar]
- 26.Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006;34:W116–W118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Dolleweerd CJ, Chargelegue D, Ma JK. Characterization of the conformational epitope of Guy's 13, a monoclonal antibody that prevents Streptococcus mutans colonization in humans. Infect Immun. 2003;71:754–765. doi: 10.1128/IAI.71.2.754-765.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Demuth DR, Davis CA, Corner AM, Lamont RJ, Leboy PS, Malamud D. Cloning and expression of a Streptococcus sanguis surface antigen that interacts with a human salivary agglutinin. Infect Immun. 1988;56:2484–2490. doi: 10.1128/iai.56.9.2484-2490.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J Mol Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 30.Leslie AG. The integration of macromolecular diffraction data. Acta Crystallogr D Biol Crystallogr. 2006;62:48–57. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- 31.Collaborative Computational Project No4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 32.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 33.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 34.Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr D Biol Crystallogr. 1999;55:247–255. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
- 35.Hooft RW, Vriend G, Sander C, Abola EE. Errors in protein structures. Nature. 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- 36.Potterton L, McNicholas S, Krissinel E, Gruber J, Cowtan K, Emsley P, Murshudov GN, Cohen S, Perrakis A, Noble M. Developments in the CCP4 molecular-graphics project. Acta Crystallogr D Biol Crystallogr. 2004;60:2288–2294. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]


