Why Is Light Important for Pathogenic Microbes?
Light is an abundant signal that many organisms use to assess the status of their environment. Species from all kingdoms have evolved the capacity to sense and respond to wavelengths across the visible spectrum. Light has long been linked to disease (Figure 1); however, the mechanisms behind many of these observations are not well understood. Recently, a direct link has been established between specific protein photosensors and the ability to cause disease in both pathogenic bacteria and fungi [1]–[3]; thus, certain pathogens require these photosensors for full virulence. A role for photoperception is likely to emerge as a common theme in microbial pathogenesis.
Figure 1. Effects of light on microbial pathogenesis.
Light (or its absence) represents an environmental signal that is known to regulate many properties of a microbial cell, which may indirectly or directly influence the development of disease.
What Photosensors Are Known in Microbes That Could Regulate Virulence?
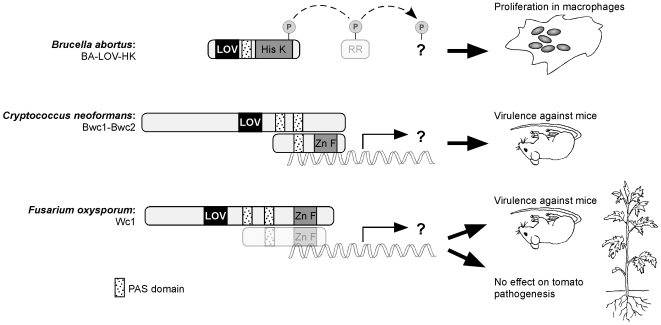
Microbes have sensory proteins that can perceive a range of energies across the visible spectrum, as well as the far ultraviolet (UV) and infrared wavelengths [4]. These protein photosensors fall into distinct classes depending on the chromophore that they bind, i.e., the cofactor that actually does the light sensing. Microbial photosensors include the phytochromes, cryptochromes, rhodopsins, photoactive yellow protein, and flavoproteins with BLUF (blue light sensing using FAD; [5]) or LOV (light, oxygen or voltage; [6]) domains. At present it is the blue light photoreceptors that are known to be required for virulence in microbes. In these cases, the photosensor proteins all contain a LOV domain that in the dark non-covalently binds a flavin cofactor (Figure 2). Light absorption by the flavin initiates formation of a covalent bond between a conserved cysteine residue in the LOV domain and the 4a carbon of the flavin [7]. Formation of this bond alters the conformation of the LOV domain, resulting in signal transmission. The output affected by the LOV domain can vary: in the bacterial systems discussed below, absorption of blue light via the LOV domain modulates the activity of a histidine kinase domain [2],[8], whereas in the fungi the LOV domain is predicted to affect gene expression via allosteric regulation of a zinc finger domain that binds DNA [9] (Figure 2). While it is only the LOV domain proteins that are currently established as regulators of microbial virulence, it will be of interest to test other photosensors for their role in pathogenesis.
Figure 2. LOV domain photosensors are required for pathogenesis.
Structure of the photosensors from Brucella abortus, Cryptococcus neoformans, and Fusarium oxysporum. The signal transduction pathway is unknown in all three cases, but is likely to involve a phosphorylation cascade including downstream signaling partners in B. abortus and a transcriptional response for the two fungal species. In the case of B. abortus, mutation of the photoreceptor gene affects proliferation in macrophages, while photoreceptor mutation in the two pathogenic fungi species modulates the rate at which mice succumb to infection. A wc-2 homlog of F. oxysporum is predicted (FOXG_01037.2) but has yet to be characterized from this species.
What Do We Currently Know about Photoregulation of Bacterial Pathogenesis?
Probably the most surprising example of photoregulation of bacterial pathogenesis was discovered by Swartz and colleagues, who demonstrated that proliferation of the Gram-negative pathogen Brucella abortus in a macrophage infection model requires exposure to visible light [2]. Moreover, this research team showed that the light dependence of cellular proliferation in macrophages required a blue light photosensory histidine kinase, which they named LOV-HK (Figure 2). Why virulence is regulated by visible light in this pathogen remains unresolved, as is the molecular/cellular mechanism underlying the light-dependent regulation of virulence. However, work on a photosensory LOV histidine kinase in the related species Caulobacter crescentus may provide some insight into light-regulated virulence in B. abortus. Specifically, activation of the photosensory two-component system LovK-LovR in C. crescentus results in modulation of the adhesive capacity of the cell [8]. Certainly, the ability of bacterial pathogens to adhere to host cells is often a critical determinant of virulence, and it will be interesting to see if the adhesive response observed in C. crescentus is conserved in its relative B. abortus, or in other pathogens that encode LOV histidine kinases (e.g., the plant pathogen Pseudomonas syringae). Bioinformatic analysis of over 600 bacterial genomes reveals that at least one LOV domain protein is present in 13% of species, including a number of pathogens [10]. The observations from B. abortus may therefore be widely relevant. Finally, it was recently reported that white light has a repressive effect on both expression of the flagellar genes flaA, flaB, and flaC and on cell adhesion and virulence in the plant pathogen Agrobacterium tumefaciens [11]. This photoresponse was independent of the three photoreceptors that can be identified in the genome sequence of A. tumefaciens, including two phytochromes and one photolyase/cryptochrome. An exciting possibility is that novel visible light photoreceptors are regulating these processes.
Does Light Also Influence Fungal Pathogenesis?
Sequencing projects reveal that fungal genomes encode putative photosensory proteins of the rhodopsin, phytochrome, cryptochrome, and LOV domain classes. A fungal LOV domain photosensor, first identified and named WHITE COLLAR 1 in the non-pathogen Neurospora crassa, is present throughout most of the kingdom, suggesting an ancient origin. In all species examined, WC-1 physically interacts with a second protein, WC-2, that contains a zinc finger DNA-binding domain such that the complex can act as a light-sensitive transcription factor (Figure 2). One fungus in which a wc-1 homolog is required for virulence is Cryptococcus neoformans, a cause of fatal meningitis in humans. Light regulates the filamentous mating process of the fungus via the WHITE COLLAR homologs Bwc1 and Bwc2. Mutation of these genes causes a reduction in resistance to UV light, and also renders the strains less virulent in a mouse inhalation model of the disease [1],[12]. Fusarium oxysporum is a filamentous species best known as a plant pathogen but is also a cause of human disease: its wc-1 homolog in a tomato isolate is required for full virulence in a mouse tail-vein injection model, but surprisingly has no effect on virulence in tomato roots [3]. Finally, there is tantalizing evidence suggesting that exposure to light influences virulence in several insect pathogens and the human pathogen Histoplasma capsulatum [13]. The cellular/molecular mechanism of light regulation of virulence in all fungal species is unknown.
How Can I Shed Light on the Photobiology of My Favorite Pathogen?
Clearly, changes in light quantity (i.e., fluence) or quality (i.e., wavelength) represent a cue in the regulation of virulence in select microbial pathogens. As described above, there are only three reports of photoreceptors that specifically affect virulence and the underlying mechanisms are unknown; thus, the field is wide open for new investigations. Grow your favorite microbe in the light and dark and see if it does anything differently. Or search the genome sequence for candidate photosensors, mutate these genes, and test the ability of the mutant strains to cause disease. There are several caveats to consider in microbial photobiology experiments: 1) it is likely that certain photosensory genes or pathways may have accumulated mutations over years of laboratory cultivation and are no longer functional, 2) the chemical properties of growth media can change on being exposed to bright visible light, and 3) proteins other than bona fide photosensory receptors also bind cofactors that absorb in the visible region of the spectrum and their function may be affected by light [14]. Other things one should consider when measuring the effects of light on virulence include logistical difficulties in controlling the light environment in certain plant and animal facilities, and the possibility that light also affects host defenses against microbes [15],[16]. Despite these experimental challenges, the continued impact of diseases worldwide, the rise in antibiotic resistance, and the emergence of new pathogens underscores the need for better understanding the basis of microbial pathogenesis. It is time to put microbial light sensing under the spotlight.
Footnotes
The authors have declared that no competing interests exist.
Supported in part by the NIH-NIAID, a Beckman Young Investigator Award, and the Mallinckrodt Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Idnurm A, Heitman J. Light controls growth and development via a conserved pathway in the fungal kingdom. PLoS Biol. 2005;3:e395. doi: 10.1371/journal.pbio.0030095. doi: 10.1371/journal.pbio.0030095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Swartz TE, Tseng T-S, Frederickson MA, Paris G, Comerci DJ, et al. Blue-light-activated histidine kinases: two-component sensors in bacteria. Science. 2007;317:1090–1093. doi: 10.1126/science.1144306. [DOI] [PubMed] [Google Scholar]
- 3.Ruiz-Roldán MC, Garre V, Guarro J, Mariné M, Roncero MIG. Role of the white collar 1 photoreceptor in carotenogenesis, UV resistance, hydrophobicity, and virulence of Fusarium oxysporum. Eukaryot Cell. 2008;7:1227–1230. doi: 10.1128/EC.00072-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Briggs WR, Spudich JL, editors. Weinheim: Wiley-VCH; 2005. Handbook of photosensory receptors. [Google Scholar]
- 5.Gomelsky M, Klug G. BLUF: a novel FAD-binding domain involved in sensory transduction in microorganisms. Trends Biochem Sci. 2002;27:497–500. doi: 10.1016/s0968-0004(02)02181-3. [DOI] [PubMed] [Google Scholar]
- 6.Christie JM, Salomon M, Nozue K, Wada M, Briggs WR. LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): binding sites for the chromophore flavin mononucleotide. Proc Natl Acad Sci USA. 1999;96:8779–8783. doi: 10.1073/pnas.96.15.8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salomon M, Christie JM, Knieb E, Lempert U, Briggs WR. Photochemical and mutational analysis of the FMN-binding domains of the plant blue light receptor, phototropin. Biochemistry. 2000;39:9401–9410. doi: 10.1021/bi000585+. [DOI] [PubMed] [Google Scholar]
- 8.Purcell EB, Siegal-Gaskins D, Rawling DC, Fiebig A, Crosson S. A photosensory two-component system regulates bacterial cell attachment. Proc Natl Acad Sci USA. 2007;104:18241–18246. doi: 10.1073/pnas.0705887104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Froehlich AC, Liu Y, Loros JJ, Dunlap JC. White Collar-1, a circadian blue light photoreceptor, binding to the frequency promoter. Science. 2002;297:815–819. doi: 10.1126/science.1073681. [DOI] [PubMed] [Google Scholar]
- 10.Losi A, Gärtner W. Bacterial bilin- and flavin-binding photoreceptors. Photochem Photobiol Sci. 2008;7:1168–1178. doi: 10.1039/b802472c. [DOI] [PubMed] [Google Scholar]
- 11.Oberpichler I, Rosen R, Rasouly A, Vugman M, Ron EZ, et al. Light affects motility and infectivity of Agrobacterium tumefaciens. Environ Microbiol. 2008;10:2020–2029. doi: 10.1111/j.1462-2920.2008.01618.x. [DOI] [PubMed] [Google Scholar]
- 12.Liu OW, Chun CD, Chow ED, Chen C, Madhani HD, et al. Systematic genetic analysis of virulence in the human fungal pathogen Cryptococcus neoformans. Cell. 2008;135:174–188. doi: 10.1016/j.cell.2008.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campbell CC, Berliner MD. Virulence differences in mice of type A and B Histoplasma capsulatum yeasts grown in continuous light and total darkness. Infect Immun. 1973;8:677–678. doi: 10.1128/iai.8.4.677-678.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hug DH. Photoactivation of enzymes. In: Smith KC, editor. Photochemical and photobiological reviews. New York and London: Plenum Press; 1981. pp. 87–138. [Google Scholar]
- 15.Roberts MR, Paul ND. Seduced by the dark side: integrating molecular and ecological perspectives on the influence of light on plant defence against pests and pathogens. New Phytol. 2006;170:677–699. doi: 10.1111/j.1469-8137.2006.01707.x. [DOI] [PubMed] [Google Scholar]
- 16.Sleijffers A, Garssen J, Van Loveren H. Ultraviolet radiation, resistance to infectious diseases, and vaccination responses. Methods. 2002;28:111–121. doi: 10.1016/s1046-2023(02)00215-3. [DOI] [PubMed] [Google Scholar]