Abstract
The stringent response effector, guanosine tetraphosphate (ppGpp), adjust gene expression and physiology in bacteria, by affecting the activity of various promoters. RNA polymerase-interacting protein, DksA, was proposed to be the co-factor of ppGpp effects; however, there are reports suggesting independent roles of these regulators. Bacteriophage λ major lytic promoter, pR, is down-regulated by the stringent response and ppGpp. Here, we present evidence that DksA significantly stimulates pR-initiated transcription in vitro in the reconstituted system. DksA is also indispensable for pR activity in vivo. DksA-mediated activation of pR-initiated transcription is predominant over ppGpp effects in the presence of both regulators in vitro. The possible role of the opposite regulation by ppGpp and DksA in λ phage development is discussed. The major mechanism of DksA-mediated activation of transcription from pR involves facilitating of RNA polymerase binding to the promoter region, which results in more productive transcription initiation. Thus, our results provide evidence for the first promoter inhibited by ppGpp that can be stimulated by the DksA protein both in vivo and in vitro. Therefore, DksA role could be not only independent but antagonistic to ppGpp in transcription regulation.
INTRODUCTION
Gene expression control in bacteria is often exerted at the level of transcription by employing global regulatory networks. Such systems are designated for precise and suitable adaptations of unicellular organisms to rapid environmental changes and the availability of nutrient sources. The stringent response effector, a specific nucleotide, guanosine tetraphosphate (ppGpp) is rapidly produced in response to a variety of physico-chemical and nutritional stresses (1–3). ppGpp can directly and indirectly regulate expression of variety of genes, adjusting the survival of bacterial cells. It interacts with RNA polymerase (RNAP) near the active center (4); however, the exact binding site and determinants in the Escherichia coli enzyme remain controversial (5). Upon binding, ppGpp does not introduce any long-lasting conformational alterations to the enzyme (6). A major mechanism of ppGpp direct action at promoters was proposed to be based on the decreased stability of promoter–RNAP open complexes upon transcription initiation, for its both inhibitory (7) and stimulatory roles (8). In addition to the direct effect, ppGpp can influence the transcription capacity of RNAP by affecting the availability of core polymerase to σ factors in the cell (9,10) and controlling the synthesis of other transcription regulators (1,2,3,11). ppGpp is one of the most global and far-reaching regulators in bacteria: as shown by global transcription profiling in E. coli, stringent response involves alterations in expression of several hundred genes (12). However, in spite of over 40 years of studies, complete mechanism of the gene expression regulation by ppGpp remains not entirely solved (3). Significant and undisputable in vivo ppGpp effects, for example, were usually difficult to reproduce in purified in vitro system (3,11). The discovery of the DksA protein role resolved this discrepancy and was thus a mile-stone in understanding of the mechanism of the stringent response (13,14). DksA was shown to be a critical component for ppGpp-mediated inhibition of the ribosomal promoters in vivo (13,14). This 151 amino-acid protein interacts with RNAP binding in the enzyme’s secondary channel (13,14) The effect of the concerted action of DksA and ppGpp is promoter-specific, depending on the intrinsic properties of a given promoter (3,15). For all promoters examined so far, DksA destabilizes competitor-resistance complexes between RNAP and promoter region (10,16). Recent studies propose that the mechanism of DksA-mediated regulation of transcription relies on allosteric alteration to the RNAP at the promoter, affecting the transition from closed to intermediate complexes (17,18). As DksA potentiates ppGpp regulation in vivo and in vitro, it was proposed to be a co-factor of the stringent response, enhancing its negative (e.g. ribosomal promoters) and positive (amino acids biosynthesis promoters) effects (13–15,19). However, several lines of evidence indicate that the roles of these transcription regulators can be opposing in vivo, when DksA could regulate gene expression without ppGpp (20), and independent in vivo and in vitro for the fimB promoter (21). Also, the transcriptomic analysis of ppGpp- and DksA-deficient strains indicates that certain genes (e.g. involved in chemotaxis) could be independently or differentially regulated by these factors (22).
In the prokaryotic world, expression of many genes is regulated simultaneously or sequentially by multiple factors, to obtain optimal adaptation to environmental challenges. Promoters of such genes can serve as model systems to study complex transcription regulation. Bacteriophage λ major lytic promoter, pR, can be controlled both positively and negatively by variety of phage and host factors, including λ-encoded Cro and CI proteins and bacterial regulators, DnaA and SeqA (23). Moreover, DNA topology alterations (mediated by IHF and potentially by other chromosome-organizing proteins) could affect pR promoter’s activity (24). The stringent control alarmon, ppGpp, inhibits pR transcription both in vivo and in vitro (25–27). The correlation proposed by Barker et al. (7) and accepted in several other publications (13,16,28), between short life-time of competitor-resistance complexes and negative regulation by ppGpp, does not apply directly to pR, as its promoter-RNAP open complex is extremely stable (27). For pR, ppGpp was reported to affect the promoter escape step in transcription initiation, specifically, the first phospho-diester bond formation (27). At the time of publication of that work (2002), the DksA role in the stringent response was unknown, thus, the principle aim of this study was to elucidate a possible effect of DksA in pR-initiated transcription. Unexpectedly, we found that DksA does not act synergistically to the ppGpp effect; on the contrary, DksA directly stimulates pR-initiated transcription.
MATERIALS AND METHODS
Bacterial strains and plasmids
Escherichia coli MG1655 ΔlacZ strain (29), and its derivatives: ppGpp-null (relA spoT) ΔlacZ (from M. Cashel), dksA (RK201) (30) and the triple mutant relA spoT dksA lacZ were used in this work. Triple mutant relA spoT dksA was constructed by P1 transduction of the dksA::Tn5 (kanamycin resistance) allele to the relA spoT lacZ strain. Cultures were routinely grown at 30°C in Luria broth (LB) (31) supplemented with appropriate antibiotics (kanamycin, 50 μg/ml and ampicillin, 100 μg/ml). Plasmid pTAC3734 (32), bearing a promoter-less lacZ reporter gene, was used for construction of pR-lacZ fusions as described by Łyżeń et al. (33). For construction of supercoiled templates for in vitro transcription, plasmid pTE103 (34) was used as a vector, as described previously (33).
Nucleotides and proteins
Nucleotides were purchased from Roche Applied Science, (α-32P)UTP and (γ-32P)ATP from Hartmann Analytic GmbH, while ppGpp was synthesized and purified as described previously (34). The σ70-RNAP holoenzyme was purified according to a general protocol described in (35) with modification as in (36). N-terminal His-tagged E. coli DksA was purified according to (21).
Measurement of β-galactosidase activity
Activity of β-galactosidase in E. coli cells was measured according to a previously described method (37). Multicopy gene fusions (located on plasmids) were used, thus the results were normalized per amount of plasmid DNA in cells; plasmid copy number in E. coli was estimated by isolation of plasmid DNA from a known number of cells, linearization with restriction endonuclease, separation during agarose gel electrophoresis, staining with ethidium bromide and subsequent densitometric analysis of the bands relative to a known amount of plasmid DNA separated on the same gel, as described in ref. (38).
In vitro transcription assay
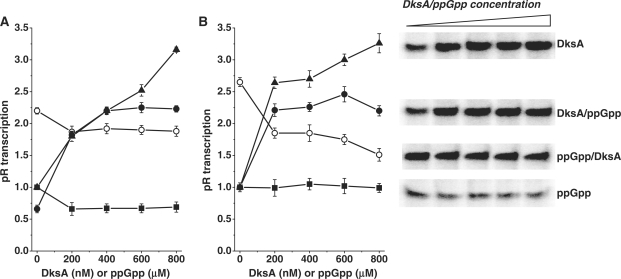
Supercoiled template was obtained by isolation of pTE103-derived plasmid from wild-type MG1655 strain, and purification of supercoiled plasmid DNA by ultracentrifugation in CsCl-ethidium bromide density gradient (31). All in vitro transcription reactions were performed in general as described previously (24), with some modifications. The reactions were performed in a total volume of 25 μl in the transcription buffer containing 50 mM Tris–HCl pH 8, 10 mM MgCl2, 10 mM β-mercaptoethanol, 10 μg/ml bovine serum albumin) with the final concentration of nucleotides: CTP and GTP of 150 μM, ATP 1 mM, UTP up to 15 μM and (α-32P)UTP up to 10 μCi with heparin (100 μg/ml) for single-round transcription. Reactions were terminated by addition of stop buffer (150 mM EDTA, 1.05 M NaCl, 7 M urea, 10% glycerol, 0.0375% xylene cyanol, 0.0375% bromophenol blue). The samples were separated by electrophoresis in 4.5% polyacrylamide gel containing 7 M urea in the TBE buffer (22) at 30 mA. The gel was dried, and RNA bands were visualized and quantified using the PhosphorImager system (BioRad). For experiments as described in Figure 1A, RNAP (15 nM) was pre-incubated with the indicated amounts of the ppGpp and/or DksA protein for 7 min at the room temperature prior mixing with KCl (140 mM), followed by 3 min incubation in 37°C. After the addition of DNA (5 nM), the samples were incubated at 37°C for 10 min. The reaction were started by the addition of nucleotides (see above) with heparin (100 μg/ml), the samples were incubated at 37°C for 10 min. The reaction was terminated by addition of 5 μl of the stop buffer and analyzed as described earlier. For experiments as in Figure 1B, single-round transcription reactions were performed in a total volume of 17 μl in transcription buffer (see above) containing 150 mM KCl. Template DNA (5 nM) with RNAP (15 nM), KCl (150 mM) and the indicated amounts of the ppGpp and/or DksA protein were incubated for 10 min at 37°C. After the addition of nucleotides (see above) with heparin (100 µg/ml), the samples were incubated at 37°C for 10 min. The reaction was terminated by addition of 3 µl of the stop buffer and analyzed as described earlier.
Figure 1.
In vitro transcription from pR promoter in the presence and absence of ppGpp and DksA. (A) Transcription in the presence of increasing concentrations of ppGpp (squares), DksA (triangles), DksA with addition of 200 μM ppGpp (closed circles) or ppGpp with addition of 400 nM DksA (open circles). RNAP and ppGpp/DksA were incubated 7 min at room temperature, then KCl was added (final concentration 140 mM) and temperature increased to 37°C prior to DNA addition. Transcription was initiated by adding of NTPs (see details in ‘Materials and Methods’ section). (B) Transcription in the presence of increasing concentrations of ppGpp (squares), DksA (triangles), DksA with addition of 200 μM ppGpp (closed circles) or ppGpp with addition of 400 nM DksA (open circles). RNAP, DNA, ppGpp/DksA were incubated in the buffer with KCl (150 mM) in 37°C then reaction was initiated by adding of NTPs. Transcription in the absence of ppGpp and DksA was set as 1. Data are from three independent experiments with error bars indicated. Inset: Autoradiogram of transcription illustrating the example experiment performed as described in (B), corresponding to the plots.
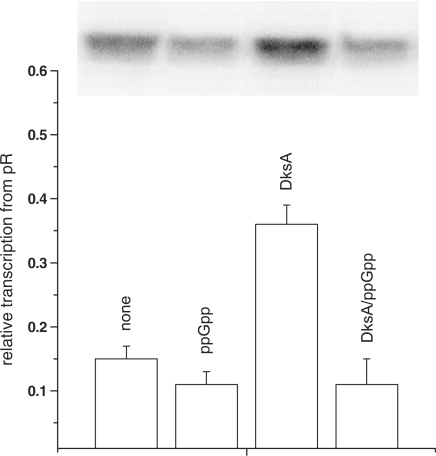
To analyze productive complex formation (Figure 4), the transcription reactions were carried out in transcription buffer (see above) supplemented by DNA template (5 nM) with KCl (150 mM) and the ppGpp (200 μM) and/or DksA (400 nM) at 20°C for 10 min. The reactions were started by addition of RNAP (15 nM) and nucleotides mix (see above) with heparin (100 μg/ml). At time-points, 17 μl aliquots were removed and added to 3 µl of the stop buffer and analyzed as described above. Figure 4 represents the transcription obtained at 3 min reaction time.
Figure 4.
The effect of ppGpp and DksA on the in vitro transcription initiation. Relative transcription levels at 3 min after the reaction start (in the time-course experiment) Autoradiogram of transcription products is inserted above the columns representing transcription from pR. DNA was incubated with ppGpp/DksA in buffer containing 150 mM KCl with no addition, ppGpp at 200 μM, DksA at 400 nM or DksA at 400 nM with addition of 200 μM ppGpp. The reaction was initiated by simultaneous addition of RNAP and NTP with heparin, and samples were withdrawn at indicated times. Values were normalized to the level of transcription for 30 min with no addition (set as 1). The results are from three independent experiments with standard errors.
Open complex stability assay
RNAP (15 nM), DNA template (5 nM), KCl (150 mM) and ppGpp (200 μM) and/or DksA (400 nM) were incubated for 10 min at 37°C in transcription buffer (see above). After heparin (100 µg/ml) addition, 15 μl aliquots were removed to a tube containing 2 µl of nucleotides (see above), at indicated times (0, 30, 60, 90, 120, 150, 180, 240 min). Reactions were carried out by 10 min at 37°C. The reaction was stopped by addition of 3 µl of the stop buffer and analyzed as above.
Electromobility shift assays
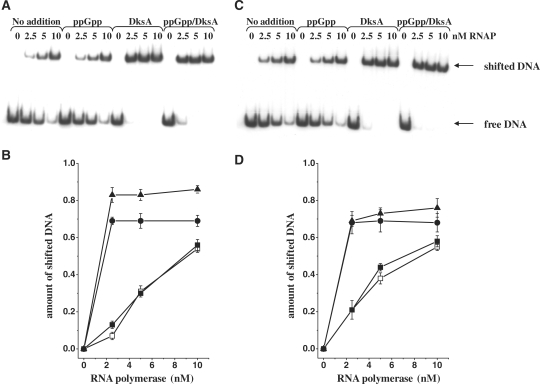
DNA linear fragments for RNAP in vitro binding studies were obtained by PCR using 5′-labeled (P32 primers: 5′-GCTCATACGTTAAATCTATCACCGCAAGGG and non-labeled 5′-GTAAGAGCGGGGTTATTTATGCTG followed by subsequent purification of the 323 nt DNA fragment. DNA (15 ng) was incubated in B-buffer (25 mM HEPES, pH 7.5, 0.1 mM EDTA, 5 mM DTT and 10% glycerol) supplemented with 0.1 mg/ml poly(dI-dC), 80 mM KCl, 200 μM ppGpp and/or 400 nM DksA and/or 1 mM ATP and UTP with increasing polymerase RNA concentration (0–10 nM) for 20 min in 37°C. The samples were separated on 3.5% Tris–glycine (pH 8.5) polyacrylamide gel at 120 V in 4°C. The DNA bands were visualized and quantified using PhosphorImager system (BioRad).
DNase I footprinting assay
End-labelled pR fragments (P32) were obtained as for EMSA. Purification of the PCR products and subsequent footprinting analysis was performed as described in ref. (40), using 500 nM DksA, 400 μM ppGpp and/or ATP and UTP at 1 mM and 40 nM RNAP.
RESULTS
DksA protein can activate transcription from pR in vitro
The stringent response and ppGpp overproduction inhibit pR activity; however, the mode of action of this effector is unusual comparing to other promoters (27). Generally, the in vivo effect of ppGpp on various promoters was usually difficult to reproduce in vitro. For both down-regulation of ribosomal RNA promoters and stimulation of amino acids biosynthesis promoters, DksA was shown to enhance the ppGpp effect in the reconstituted in vitro system. Thus, we aimed to test a hypothetic synergistic effect of DksA on ppGpp-mediated inhibition of the pR promoter. We employed the specific experimental set-up designed to study ppGpp effect in vitro (27), where ppGpp inhibited pR activity by about 30–40 % (Figure 1A). In all our in vitro transcription reactions, supercoiled template was used. This choice was justified by dependence of pR activity on the DNA topology (ref. 24 and our unpublished data). To our surprise, contrary to the anticipated results, DksA present in the in vitro transcription reaction stimulated pR activity over 3-fold at 800 nM DksA (Figure 1A). Moreover, the transcription was also enhanced when both regulators were present, at the constant level of ppGpp (200 μM). It is worth mentioning that in these experiments single-round transcription assay was performed, thus, the DksA activation did not involve the re-initiation events. The specific assay, where ppGpp inhibition was initially observed, requires the RNAP to be incubated with ppGpp in low-salt buffer and room temperature, before other components, including salt, are added and temperature increases to the regular reaction level (37°C) (27). For other experiments, the usual set-up requires the salt to be present already in the reaction from the initial complex formation step. Therefore, we tested ppGpp and DksA effect in pR-initiated transcription in this experimental system. All components, including 150 mM KCl and buffer conditions, were the same as in the previous assay. In this case, we observed similar DksA-mediated stimulation of pR-initiated transcription (up to 3.5-fold); however, ppGpp inhibitory effect was negligible (Figure 1B). In the presence of both regulators, activation of transcription was observed only when DksA concentration increased at constant ppGpp level (200 μM) (Figure 1). In the presence of increasing ppGpp concentrations, at constant DksA (400 nM), ppGpp has inhibitory effect on the transcription; however, one should note that the start-point is at 2.5-fold over the transcription with no addition and, even at the highest ppGpp concentrations, inhibited transcription was nevertheless 1.5-fold over the point where only ppGpp was present at the same concentration. The ability of DksA protein to inhibit transcription, as reported previously (13,14), was tested in the in vitro test employing the rrnB P1 promoter. In the experimental set-up as described in ref. (10), we observed a very similar inhibition of P1-initiated transcription (data not shown), indicating that this preparation of DksA protein retained its specific function.
Our results indicate that DksA can directly activate the pR promoter, alone and in the presence of ppGpp, in the experimental system allowing ppGpp-mediated inhibition of transcription. Thus, DksA can regulate pR transcription independently of ppGpp.
DksA is necessary for pR-initiated transcription in vivo
Elevated ppGpp level, due to the stringent response (e.g. by amino acid starvation) or RelA overproduction, resulted in the inhibition of pR-initiated transcription (25). Based on the finding that DksA can activate pR in vitro, we assessed the in vivo pR activity in the dksA mutant. A lack of DksA resulted in a significant decrease in pR activity, which was below 10% of that observed in the wild-type strain (Figure 2). The measurements were performed in the rich medium, in the exponential phase of bacterial growth. The pR activity in cells lacking both ppGpp and DksA was only slightly higher than in the dksA mutant, indicating that DksA role in pR-initiated transcription is prevailing at these growth conditions. The decrease in pR activity in cells lacking ppGpp (relA spoT), although surprising at first (given known ppGpp-mediated inhibition of pR), was already reported (26), where it was observed that low ppGpp levels are necessary for basal pR activity (Discussion section). These results suggest that DksA activity is indispensable for full pR activity in vivo. Taken together with the in vitro data, it can be concluded that DksA acts as an activator of pR.
Figure 2.
In vivo activity of pR promoter (β-galactosidase activity) in wild-type, relA spoT (ppGpp-null), dksA and relA spoT dksA strains. Activity in the wild-type (wt) strain was set as 1 (actual value was 12318 ± 1003 Miller units). The data are from six independent measurements with SD indicated.
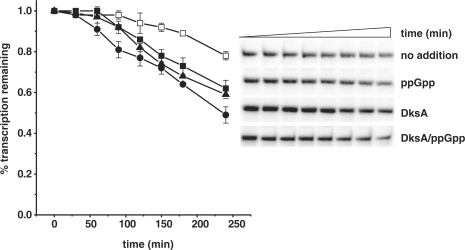
DksA stimulates pR transcription by enhancing the binding of RNAP to DNA
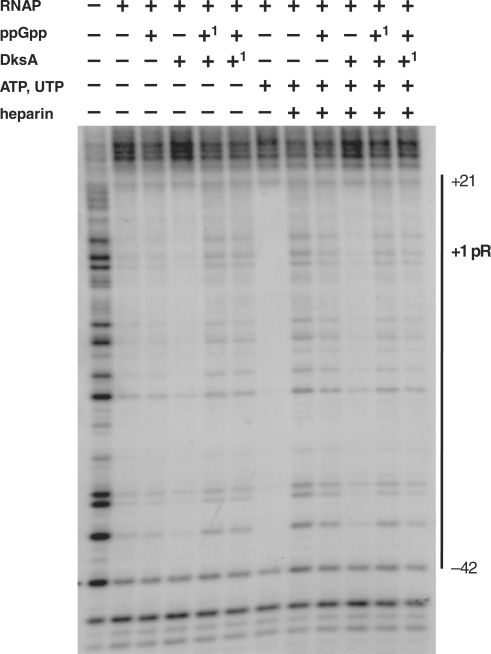
To address the question about the molecular mechanism of DksA-mediated activation of pR-initiated transcription, we tested DksA effects at various steps in the transcription initiation. DksA was reported to decrease the stability of the competitor-resistant RNAP–promoter complexes for its both positive and negative regulatory effects (13,14,19). Thus, we assessed the effect of ppGpp and DksA on complex stability in the same as previously employed (Figure 1B) in vitro transcription system. Generally, pR forms extremely stable open complexes, with the half-life time exceeding 6 h (extrapolation of the time-course plot and previous, unpublished observations). The presence of any of the regulators decreased the stability of the heparin-resistant complex down to about 4–4.5 h (Figure 3). However, the effect of DksA did not differ considerably from that of ppGpp on the complex stability. Moreover, the presence of both regulators did not result in further destabilization of complexes. Thus, we concluded that DksA does not act as a pR-transcription regulator by affecting the open complex half-life, although the protein itself destabilizes the complexes to the same extend as ppGpp. It was previously shown that ppGpp influences the promoter escape rate in transcription initiation (27). Therefore, we aimed to test a possible effect of DksA at this step of transcription. We found that DksA did not affect promoter escape, under the experimental conditions where ppGpp effect was observed (data not shown). At many bacterial promoters, RNAP produces certain amount of short transcripts, in addition to the full-length products, and this abortive transcription can influence the overall transcription efficiency. Thus, some regulators can affect this step in gene expression. However, the level of abortive transcripts measured in the different time points (including very short ones) in the presence of DksA was not altered, indicating that this cannot be a regulatory step involving DksA protein. The major effect of DksA in pR transcription was observed when in the in vitro transcription experiment RNAP was added at the same time as NTPs to the buffer–DNA reaction mixture containing also ppGpp/DksA (immediate start of transcription). In such experiments, the effects of regulators were evaluated at the steps of RNAP binding to DNA and productive initiation complex formation (the amount of the formed full-length transcript was assessed, Figure 4). The presence of DksA resulted in a significant increase in the amount of the transcript (2-fold effect) comparing to the transcription with no addition (Figure 4). Moreover, the time necessary for formation of full-length transcripts was shorter in the presence of DksA (the stimulatory effect was observed already after 3 min). The slight inhibitory effect of ppGpp visible also in the presence of both regulators in longer reaction time (over 5 min) disappeared and DksA showed overall stimulatory effect on pR also in the presence of ppGpp (data not shown). These results indicate that pR activation by DksA occurs at the first step of the transcription initiation, i.e. RNAP interaction with the promoter region, which results in forming of the productive transcription complexes. The direct DksA-mediated stimulation of the RNAP binding to DNA was shown previously (21,41). These findings and our observation described above suggested that DksA-mediated pR transcription activation could be based on related mechanism. To test this hypothesis, we performed electrophoretic mobility shift assay (EMSA) of a pR promoter DNA fragment with increasing concentrations of RNAP and constant amount of DksA protein and/or ppGpp. As presented in Figure 5, DksA significantly stimulated RNAP ability to bind the promoter. DksA alone did not cause DNA shift (control experiment, data not shown). The presence of ppGpp did not result in any alterations in RNAP-DNA binding. However, when ppGpp was added together with DksA, the enhancement of RNAP–DNA interaction was slightly reduced (Figure 5B). This corresponds to the results presented in Figure 4, where addition of ppGpp decreased DksA stimulatory effect. The presence of two initiating nucleotides of the transcript, ATP and UTP, resulted in abolishing of ppGpp effect at this step—at 2.5 nM RNAP majority of the DNA was visible as a complex with the protein (Figure 5C and D) for both DksA and DksA/ppGpp reactions. As EMSA experiment may have limitations due to the fragment linearity, problems in electrophoresis of the complexes and dissociation, we undertook another approach to study DksA role in RNAP binding to the pR promoter. The DNase I footprint analysis of the DNA fragment containing pR region showed more effective protection by RNAP in the presence of DksA. The binding was initiated by adding of RNAP and the effect was mostly visible at the short time—2 min (Figure 6); later, the footprint of RNAP could be too strong to visualize differences. The slightly weaker footprint of RNAP in the presence of both DksA and ppGpp occurred only when initiating nucleotides were absent; in the reaction where A+U were added RNAP bound very efficiently (data not shown, the protection was as strong as in the lane containing RNAP with A+U). The most pronounced DksA effect was visible when re-association of RNAP with pR was prevented by heparin. This set-up corresponds with single-round in vitro transcription, where DksA-mediated stimulation of pR was observed. However DksA increased the DNA protection by RNAP, the protection pattern and extent was not appreciably altered by DksA alone or with A+U.
Figure 3.
The effects of ppGpp and DksA on pR promoter half-life. (A) The time course of competitor-resistant open complexes was monitored by in vitro transcription challenged by heparin in the presence of none addition (DksA storage buffer, open squares), ppGpp at 200 μM (closed squares), DksA at 400 nM (triangles) or DksA at 400 nM with addition of 200 μM ppGpp (circles). The values found at 20 s after competitor addition were set as 1. Data are the average from three independent experiments with standard errors. (B) The autoradiogram from the experiment performed as shown in the (A).
Figure 5.
Binding of RNAP to pR DNA fragment. (A and C) Gel-shift assay using 15 ng of DNA and indicated amounts of the RNAP. (B and D) Quantification of the percentage of shifted (bound) DNA fragment. Symbols: none addition (DksA storage buffer, open squares), ppGpp at 200 μM (closed squares), DksA at 400 nM (triangles) or DksA at 400 nM with addition of 200 μM ppGpp (circles). In (C and D), ATP and UTP were present at 1 mM. The results are mean values from three measurements with error bars.
Figure 6.
Footprinting analysis of RNAP protection at pR promoter in the presence of ppGpp and/or DksA. Reactions contained double-stranded template with the 5′-end (P32)-labeled, 40 nM RNAP, 400 μM ppGpp, 500 nM DksA, ATP and UTP at 1 mM, heparin at 100 µg/ml where indicated. In the lanes containing both DksA and ppGpp ‘1’ indicates the regulator added as first.
We concluded from these in vitro data that the mechanism of DksA-mediated activation of pR-initiated transcription involves mainly facilitation of the association of RNAP with the pR promoter.
DISCUSSION
Discovery of DksA protein function in the stringent response, relatively recent comparing to the 40-years knowledge of ppGpp, elucidated its global role that initially was not expected when DksA was described as a suppressor of dnaKJ phenotype (30). From 2004, new aspects of DksA impact on gene expression as well as the approaches to explain the molecular mechanism of this regulation have been reported. Its importance for ppGpp-mediated transcription regulation has been documented in details (13,14,19), however unexpected roles of DksA are being recently revealed—as RNAP-binding protein DksA can affect transcription in the process independent on ppGpp (20,21).
Model promoter, bacteriophage λ-derived pR, is known to be down-regulated in vivo during stringent response and in vitro by its effector, ppGpp. In our work we aimed to elucidate the role of DksA in pR transcription. We present evidence that DksA does not facilitate ppGpp-mediated down-regulation of pR activity. In contrary, our in vitro and in vivo results indicate that DksA acts as an activator of pR-initiated transcription. Under the same transcription reaction conditions where ppGpp evidently inhibits pR activity DksA can stimulate transcription from pR nearly 3-fold (Figure 1). In the set-up resembling in vivo situation (both regulators are present) DksA could stimulate pR at the low levels of ppGpp. However, increased ppGpp concentrations diminished DksA-mediated activation (Figure 1). Total transcription level, though, exceeded that observed without any regulator or with ppGpp alone. This supports stimulatory role of DksA in pR transcription. Our observation brought two main questions: what is the molecular mechanism of DksA-mediated activation of pR, and what is a possible role of this stimulation for physiological activity of pR in the context of phage–host interaction. To address first question, we investigated DksA impact on pR transcription initiation. For pR, as for all promoters tested so far, DksA can decrease competitor-resistant RNAP–DNA complex half-life (Figure 3); nevertheless, for the same promoter, it can significantly activate the transcription. The explanation of this inconsistence is discussed below. The detailed analysis of the transcription initiation excluded DksA effect at promoter escape step and the abortive transcription. The results presented in this work indicate that the main stimulation occurs at the step of RNAP binding to DNA. This DksA effect was reported recently for the fimB promoter (21). However, in the case of fimB, opposite DksA- and ppGpp-mediated regulation was observed only in vivo. The facilitation of RNAP binding to pR region by DksA was observed in EMSA and DNase I footprint assays. Interestingly, the presence of both regulators, ppGpp and DksA resulted in less effective binding only in the absence of initiating nucleotides. When the ternary complexes were created by first two phosphodiester bonds in the presence of ATP and UTP, ppGpp did not affect DksA-mediated enhancement of the binding. It could be speculated that this may correspond to the in vivo situation occurring upon starvation and stress, facilitating ppGpp effect when nucleotides are limited. Although DksA stimulated the RNAP binding to pR, it did not shift the endpoints of the DNase I protection, suggesting that the extent of the contact site of RNAP at pR is not influenced by DksA.
The discrepancy between destabilization of open complexes and activation of transcription by facilitation of the RNAP binding by DksA for the same promoter could be confusing at first. It appears that even if DksA decreases competitor-resistant complex life-time shifting the equilibrium into the dissociation direction for pR, the activation of the overall promoter activity occurs at independent level and the DksA effect depends on the intrinsic properties of a promoter as suggested previously (17,28). In the contrary to the activation of transcription from amino acids biosynthesis promoters by ppGpp/DksA, when stable complexes need destabilization in order to start effective transcription (12), ppGpp inhibits pR. Notably, presence of both regulators did not cause synergistical destabilization of RNAP–pR complex (Figure 3). The formation of transcription-competent open complex of RNAP at pR involves at least two intermediates (42). The recent studies reported DksA-mediated allosteric alterations in RNAP during transcription initiation at rrnB P1, affecting the transition between intermediates (17). Whether DksA binding can result in the allosteric changes in RNAP at pR promoter, remains yet unsolved, however, one could speculate that if these alterations affect RNAP–DNA interface, this may enhance the effective binding of RNAP to promoter region. For the promoters forming stable complexes, this binding could be slow and require substantial energy, thus, enhancement at this step could facilitate overall promoter activity.
DksA can play a predominant role in pR regulation over ppGpp in the in vitro reconstituted system and in certain conditions of bacterial growth (discussed below). It is worth noting, that not only RNAP binding to DNA (which can be even inhibitory, if too strong), but also effective transcription (assessed by the formed transcript) is stimulated by DksA.
DksA is essential for pR transcription in vivo, with 10-fold decrease in pR activity in DksA-deficient E. coli. A lack of the anticipated effect of ppGpp-deficiency (an increase of the pR transcription in the absence of ppGpp would be expected), may be explained by the effects of cellular ppGpp concentrations. The reporter activity assays were performed in bacteria at exponential growth phase (at OD575 of 0.2–0.3). Under these conditions, ppGpp level is the lowest relative to other growth rates. As was reported previously (26), low concentration of ppGpp can be stimulatory for pR, while a complete deficiency of ppGpp results in a decrease in pR-initiated transcription. Bacteria lacking both ppGpp and DksA supported pR activity to the level comparable to this observed in dksA strain, indicating that at least in the exponential growth phase, DksA is indispensable for effective pR-initiated transcription. The in vivo dependence of pR activity on DksA strongly indicates that the in vitro stimulation documented here is not an artifact of the specific experimental conditions in the reconstituted system.
Based on our observations, we therefore concluded that DksA plays important stimulatory role in pR transcription. Hence, DksA can be added to the list of λ pR regulators as a host-encoded activator among other important ones, such as DnaA or SeqA. This appends the new factor to the already complex model promoter. The transformation of λ plasmid is a function of efficient pR transcription due to transcriptional activation of oriλ. The transformation efficiency of the dksA strain was impaired (data not shown) suggesting that DksA has significant impact on the consequences of pR transcription in λ plasmid DNA replication.
To address the second question about the physiological role of the opposite regulation of pR transcription by ppGpp and DksA, one could hypothesize about its relation of the adaptation of λ phage development to host condition. λ, as a temperate phage, upon infection, can follow one of two alternative developmental pathways: lytic or lysogenic. The proper decision, supposed to ensure optimal phage propagation, has to reflect the current physiological state of the host. Stringent response negatively regulates pR in vivo, indicating that ppGpp effect prevails in this specific situation. However, the absence of DksA results in the significant down-regulation of pR in the exponential growth. In addition to the situation when ppGpp is produced rapidly upon transition from nutrient availability (allowing fast growth) to starvation (stringent response), alarmon level increases significantly also in the situation of prolonged nutrient limitations (43). The latter situation is encountered by most bacteria (especially free-living prokaryotes) (44) and thus affects life cycle of bacteriophages infecting such host. Upon nutrient availability, phage lytic cycle would be preferable, thus, λ pR transcription responsible for synthesis of mRNA for replication proteins and crucial for transcriptional activation of oriλ, should be stimulated (26). This would explain multiple positive regulators of pR, and support the need for DksA while ppGpp level is low. In the situation when ppGpp concentration increases upon nutrient limitation, phage lytic development is inhibited (45); thus, pR transcription would be down-regulated, favoring lysogenic cycle. DksA-mediated up-regulation would be then taken over by ppGpp-mediated inhibition. Such a situation is mimicked in the in vitro experiment when ppGpp increases at the constant DksA level (Figure 1). However, the biological role of the sensitive balance of these regulators and their physiological functions leading to λ phage adjusting to environmental challenges of the host bacteria remain not entirely solved and are the subject of our current studies.
In summary, the results presented here shed a new light on the mechanisms of DksA and ppGpp effects during transcription regulation and give a new insight at pR transcription regulation. These data strongly indicate that although the stringent control requires DksA as a co-cofactor, this protein (by a different molecular mechanism) can play independent and antagonistic to ppGpp role in the regulation of gene expression.
FUNDING
Polish Ministry of Science and Higher Education (project grant no N N301 161635 to ASP). Funding for open access charge: The University of Gdańsk and the above mentioned project grant.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are grateful to Drs V. Shingler, A. Aberg and M. Cashel for important ideas and sharing essential reagents and results prior to publications; to Dr B. Kędzierska for encouragement; and to Dr K. Potrykus for discussion and critical reading of the article.
REFERENCES
- 1.Cashel M, Gentry D, Hernandez VJ, Vinella D. Escherichia coli and Salmonella: Cellular and Molecular Biology. I. Washington DC: American Society for Microbiology; 1996. The stringent response; pp. 1458–1496. [Google Scholar]
- 2.Magnusson LU, Farewell A, Nystrom T. ppGpp: a global regulator in Escherichia coli. Trends Microbiol. 2005;13:236–242. doi: 10.1016/j.tim.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Potrykus K, Cashel M. (p)ppGpp: still magical? Annu. Rev. Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 4.Artsimovitch I, Patlan V, Sekine S, Vassylyeva MN, Hosaka T, Ochi K, Yokoyama S, Vassylyev DG. Structural basis for transcription regulation by alarmone ppGpp. Cell. 2004;117:299–310. doi: 10.1016/s0092-8674(04)00401-5. [DOI] [PubMed] [Google Scholar]
- 5.Vrentas CE, Gaal T, Berkmen MB, Rutherford ST, Haugen SP, Vassylyev DG, Ross W, Gourse RL. Still looking for the magic spot: the crystallographically defined binding site for ppGpp on RNA polymerase is unlikely to be responsible for rRNA transcription regulation. J. Mol. Biol. 2008;377:551–564. doi: 10.1016/j.jmb.2008.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Szalewska-Pałasz A. Properties of Escherichia coli RNA polymerase from a strain devoid of the stringent response alarmone ppGpp. Acta. Biochim. Pol. 2008;55:317–323. [PubMed] [Google Scholar]
- 7.Barker MM, Gaal T, Josaitis CA, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J. Mol. Biol. 2001;305:673–688. doi: 10.1006/jmbi.2000.4327. [DOI] [PubMed] [Google Scholar]
- 8.Barker MM, Gaal T, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. II. Models for positive control based on properties of RNAP mutants and competition for RNAP. J. Mol. Biol. 2001;305:689–702. doi: 10.1006/jmbi.2000.4328. [DOI] [PubMed] [Google Scholar]
- 9.Bernardo LMD, Johansson L, Solera D, Skarfstad E, Shingler V. The guanosine tetraphosphate (ppGpp) alarmone, DksA and promoter affinity for RNA polymerase in regulation of sigma-dependent transcription. Mol. Microbiol. 2006;60:749–764. doi: 10.1111/j.1365-2958.2006.05129.x. [DOI] [PubMed] [Google Scholar]
- 10.Szalewska-Palasz A, Johansson LUM, Bernardo LMD, Skärfstad E, Stec E, Brännström K, Shingler V. Properties of RNA polymerase bypass mutants: implications for the role of ppGpp and its co-factor DksA in controlling transcription dependent on sigma54. J. Biol. Chem. 2007;282:18046–18056. doi: 10.1074/jbc.M610181200. [DOI] [PubMed] [Google Scholar]
- 11.Srivatsan A, Wang JD. Control of bacterial transcription, translation and replication by (p)ppGpp. Curr. Opin. Microbiol. 2008;11:100–105. doi: 10.1016/j.mib.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 12.Durfee T, Hansen AM, Zhi H, Blattner FR, Jin DJ. Transcription profiling of the stringent response in Escherichia coli. J. Bacteriol. 2007;90:1084–1096. doi: 10.1128/JB.01092-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paul BJ, Barker MM, Ross W, Schneider DA, Webb C, Foster JW, Gourse RL. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell. 2004;118:311–322. doi: 10.1016/j.cell.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 14.Perederina A, Svetlov V, Vassylyeva MN, Tahirov TH, Yokoyama S, Artsimovitch I, Vassylyev DG. Regulation through the secondary channel–structural framework for ppGpp-DksA synergism during transcription. Cell. 2004;118:297–309. doi: 10.1016/j.cell.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 15.Roberts JW. Promoter-specific control of E. coli RNA polymerase by ppGpp and a general transcription factor. Genes Dev. 2009;23:143–146. doi: 10.1101/gad.1770509. [DOI] [PubMed] [Google Scholar]
- 16.Haugen SP, Ross W, Gourse RL. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat. Rev. Microbiol. 2008;6:507–519. doi: 10.1038/nrmicro1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rutherford ST, Villers CL, Lee JH, Ross W, Gourse RL. Allosteric control of Escherichia coli rRNA promoter complexes by DksA. Genes Dev. 2009;23:236–248. doi: 10.1101/gad.1745409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blankschien MD, Lee JH, Grace ED, Lennon CW, Halliday JA, Ross W, Gourse RL, Herman C. Super DksAs: substitutions in DksA enhancing its effects on transcription initiation. EMBO J. 2009;28:1720–1731. doi: 10.1038/emboj.2009.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paul BJ, Berkmen MB, Gourse RL. DksA potentiates direct activation of amino acid promoters by ppGpp. Proc. Natl Acad. Sci. USA. 2005;102:7823–7828. doi: 10.1073/pnas.0501170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Magnusson LU, Gummesson B, Joksimović P, Farewell A, Nyström T. Identical, independent, and opposing roles of ppGpp and DksA in Escherichia coli. J. Bacteriol. 2007;189:5193–5202. doi: 10.1128/JB.00330-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aberg A, Shingler V, Balsalobre C. Regulation of the fimB promoter: a case of differential regulation by ppGpp and DksA in vivo. Mol. Microbiol. 2008;67:1223–1241. doi: 10.1111/j.1365-2958.2008.06115.x. [DOI] [PubMed] [Google Scholar]
- 22.Aberg A, Fernández-Vázquez J, Cabrer-Panes JD, Sánchez A, Balsalobre C. Similar and divergent effects of ppGpp and DksA deficiencies on transcription in Escherichia coli. J. Bacteriol. 2009;191:3226–3236. doi: 10.1128/JB.01410-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szalewska-Pałasz A, Węgrzyn G, Węgrzyn A. Mechanisms of physiological regulation of RNA synthesis in bacteria: new discoveries breaking old schemes. J. Appl. Genet. 2007;48:281–294. doi: 10.1007/BF03195225. [DOI] [PubMed] [Google Scholar]
- 24.Łyżeń R, Kochanowska M, Wegrzyn G, Szalewska-Pałasz A. IHF- and SeqA-binding sites, present in plasmid cloning vectors, may significantly influence activities of promoters. Plasmid. 2008;60:125–130. doi: 10.1016/j.plasmid.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 25.Szalewska-Pałasz A, Wegrzyn A, Herman A, Wegrzyn G. The mechanism of the stringent control of lambda plasmid DNA replication. EMBO J. 1994;13:5779–5785. doi: 10.1002/j.1460-2075.1994.tb06916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slomińska M, Neubauer P, Wegrzyn G. Regulation of bacteriophage lambda development by guanosine 5′-diphosphate-3′-diphosphate. Virology. 1999;262:431–441. doi: 10.1006/viro.1999.9907. [DOI] [PubMed] [Google Scholar]
- 27.Potrykus K, Wegrzyn G, Hernandez VJ. Multiple mechanisms of transcription inhibition by ppGpp at the lambdap(R) promoter. J. Biol.Chem. 2002;277:43785–43791. doi: 10.1074/jbc.M208768200. [DOI] [PubMed] [Google Scholar]
- 28.Haugen SP, Berkmen MB, Ross W, Gaal T, Ward C, Gourse RL. rRNA promoter regulation by nonoptimal binding of sigma region 1.2: an additional recognition element for RNA polymerase. Cell. 2006;125:1069–1082. doi: 10.1016/j.cell.2006.04.034. [DOI] [PubMed] [Google Scholar]
- 29.Sze CC, Bernardo LM, Shingler V. Integration of global regulation of two aromatic-responsive sigma(54)-dependent systems: a common phenotype by different mechanisms. J. Bacteriol. 2002;184:760–770. doi: 10.1128/JB.184.3.760-770.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang PJ, Craig EA. Identification and characterization of a new Escherichia coli gene that is a dosage-dependent suppressor of a dnaK deletion mutation. J. Bacteriol. 1990;172:2055–2064. doi: 10.1128/jb.172.4.2055-2064.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd edn. NY: Cold Spring Harbor Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 32.Brondsted L, Atlung T. Anaerobic regulation of the hydrogenase 1 (hya) operon of Escherichia coli. J. Bacteriol. 1994;176:5423–5428. doi: 10.1128/jb.176.17.5423-5428.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Łyżeń R, Wegrzyn G, Wegrzyn A, Szalewska-Pałasz A. Stimulation of the lambda pR promoter by Escherichia coli SeqA protein requires downstream GATC sequences and involves late stages of transcription initiation. Microbiology. 2006;152:2985–2992. doi: 10.1099/mic.0.29110-0. [DOI] [PubMed] [Google Scholar]
- 34.Elliott T, Geiduschek EP. Defining a bacteriophage T4 late promoter: absence of a “-35” region. Cell. 1984;36:211–219. doi: 10.1016/0092-8674(84)90091-6. [DOI] [PubMed] [Google Scholar]
- 35.Cashel M. Preparation of guanosine tetraphosphate (ppGpp) and guanosine pentaphosphate (pppGpp) from Escherichia coli ribosomes. Anal. Biochem. 1974;57:100–107. doi: 10.1016/0003-2697(74)90056-6. [DOI] [PubMed] [Google Scholar]
- 36.Burgess RR, Jendrisak JJ. A procedure for the rapid, large-scall purification of Escherichia coli DNA-dependent RNA polymerase involving Polymin P precipitation and DNA-cellulose chromatography. Biochemistry. 1975;14:4634–4638. doi: 10.1021/bi00692a011. [DOI] [PubMed] [Google Scholar]
- 37.Hager DA, Jin DJ, Burgess RR. Use of Mono Q high-resolution ion-exchange chromatography to obtain highly pure and active Escherichia coli RNA polymerase. Biochemistry. 1990;29:7890–7894. doi: 10.1021/bi00486a016. [DOI] [PubMed] [Google Scholar]
- 38.Miller JH. Cold Spring Harbor New York, Cold Spring Harbor Laboratory Press; 1972. Experiments in Molecular Genetics. [Google Scholar]
- 39.Węgrzyn G, Węgrzyn A, Pankiewicz A, Taylor K. Allele specificity of the Escherichia coli dnaA gene function in the replication of plasmids derived from phage lambda. Mol. Gen. Genet. 1996;252:580–586. doi: 10.1007/BF02172404. [DOI] [PubMed] [Google Scholar]
- 40.Potrykus K, Vinella D, Murphy H, Szalewska-Pałasz A, D'A;ri R, Cashel M. Antagonistic regulation of Escherichia coli ribosomal RNA rrnB P1 promoter activity by GreA and DksA. J. Biol. Chem. 2006;281:15238–15248. doi: 10.1074/jbc.M601531200. [DOI] [PubMed] [Google Scholar]
- 41.Perron K, Comte R, van Delden C. DksA represses ribosomal gene transcription in Pseudomonas aeruginosa by interacting with RNA polymerase on ribosomal promoters. Mol. Microbiol. 2005;56:1087–1102. doi: 10.1111/j.1365-2958.2005.04597.x. [DOI] [PubMed] [Google Scholar]
- 42.Kontur WS, Saecker RM, Davis CA, Capp MW, Record MT. Solute probes of conformational changes in open complex formation by E. coli RNA polymerase at the λPR promoter: evidence for unmasking of the active site in the isomerization step and for large-scale coupled folding in the subsequent conversion to RPo. Biochemistry. 2006;45:2161–2177. doi: 10.1021/bi051835v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Teich A, Meyer S, Lin HY, Andersson L, Enfors S, Neubauer P. Growth rate related concentration changes of the starvation response regulators sigmaS and ppGpp in glucose-limited fed-batch and continuous cultures of Escherichia coli. Biotechnol. Prog. 1999;15:123–129. doi: 10.1021/bp980102h. [DOI] [PubMed] [Google Scholar]
- 44.Chesbro W. The domains of slow bacterial growth. Can. J. Microbiol. 1988;34:427–435. doi: 10.1139/m88-075. [DOI] [PubMed] [Google Scholar]
- 45.Łoś M, Golec P, Łoś JM, Weglewska-Jurkiewicz A, Czyz A, Wegrzyn A, Wegrzyn G, Neubauer P. Effective inhibition of lytic development of bacteriophages lambda, P1 and T4 by starvation of their host, Escherichia coli. BMC Biotechnol. 2007;26:7–13. doi: 10.1186/1472-6750-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]