Abstract
Experimental analysis and manipulation of protein–DNA interactions pose unique biophysical challenges arising from the structural and chemical homogeneity of DNA polymers. We report the use of yeast surface display for analytical and selection-based applications for the interaction between a LAGLIDADG homing endonuclease and its DNA target. Quantitative flow cytometry using oligonucleotide substrates facilitated a complete profiling of specificity, both for DNA-binding and catalysis, with single base pair resolution. These analyses revealed a comprehensive segregation of binding specificity and affinity to one half of the pseudo-dimeric interaction, while the entire interface contributed specificity at the level of catalysis. A single round of targeted mutagenesis with tandem affinity and catalytic selection steps provided mechanistic insights to the origins of binding and catalytic specificity. These methods represent a dynamic new approach for interrogating specificity in protein–DNA interactions.
INTRODUCTION
Specific interactions between proteins and DNA embody a rigorous molecular recognition challenge (1). DNA molecules present homogeneous and locally static structural and electrochemical surfaces which support a restricted set of mechanisms for the extraction of binding information. This contrasts with protein–protein interactions, where the diverse structures and chemistries of polypeptide surfaces facilitate a high level of binding specificity (2). As such, the energetic differences which sustain specificity in protein–DNA interactions are subtle relative to those in other interacting biomolecules and difficult to resolve experimentally (3). Additionally, relationships between binding and catalytic specificity in the many classes of DNA-binding proteins which deliver enzymatic modifications to specific targets have remained elusive.
Technical limitations have prevented routine quantitative assessments of specificity in protein–DNA interactions. While high throughput techniques to analyze protein–DNA binding have been described (4–10), these methods have not achieved widespread use because of their inherent complexity, costly reagent or apparatus set-up, and a failure to provide selection-based applications which support dynamic testing of protein variants. Moreover, no current technique provides the capacity to assess relationships between binding interactions and subsequent catalytic events. An analytical platform that enables quantitative interrogation of binding and catalytic specificity and which also facilitates the rapid analysis of protein variants would represent a significant advance for studies of DNA-interacting proteins. Translating such a platform to high throughput library screening would supplement a flexible analytical method with selection capabilities useful for mechanistic studies of protein–DNA interactions as well as protein engineering applications.
Yeast surface display (YSD) is a widely used platform for the manipulation of intermolecular binding interactions and has been successfully applied to engineer desired properties into multiple types of proteins (11,12). Its utility has been further extended by the development of rapid flow cytometric methods that allow quantification of binding affinity with results paralleling that of standard biophysical methods, but without a requirement for generation of highly purified protein (13). Efficient transformation protocols and yeast’s intrinsic capacity for homologous recombination of transfected DNA allow facile production of large and complex molecular libraries (14). The relatively robust, neutral and non-reactive nature of the yeast cell wall is ideal for flow cytometry applications, and is compatible with a wide variety of conditions necessary for efficient catalysis by surface displayed enzymes. Additionally, the eukaryotic translational machinery and its associated chaperones and quality control mechanisms provide a stringent folding checkpoint, ensuring that molecular variants emerging from YSD-based selection are stable and well folded (15,16). This is particularly important in efforts to engineer molecules whose downstream applications require efficient expression in mammalian cells.
The LAGLIDADG family is a group of proteins with DNA recognition properties adapted for specific target recognition and, in many cases, DNA cleavage (17,18). DNA targets recognized by LAGLIDADG proteins range in length from 16–22 base pairs, making them some of the most specific DNA recognition molecules known in nature and a unique model for the study of protein–DNA interactions. Additionally, LAGLIDADG proteins have emerged as an important class of molecules for delivering endonuclease activity—or potentially other modalities—to select loci in biotechnology and therapeutic applications (19–22). Widespread use of LAGLIDADG proteins in genome targeting applications hinges on achieving a comprehensive understanding of the mechanistic contributions to specificity. While interactions between some members and their native DNA targets have been well characterized structurally and thermodynamically (23), biochemical analyses and engineering strategies have depended heavily on readouts of endonuclease activity (22,24–32). As such, the longitudinal contributions of intermolecular interactions that govern specificity remain unappreciated, as no study has systematically correlated binding specificity with endonuclease activity. Conversely, current experimental strategies to correlate structural, binding, and catalytic properties are limited in their adaptability to high throughput analytical or selection platforms (10,33,34).
Here we report the adaptation of the YSD platform to enable the study of protein–DNA interactions both at the level of substrate binding and catalysis. Flow cytometric methods to quantify interactions with DNA substrates bearing single mutations were developed and used to profile the binding and cleavage properties of a prototypical homing endonuclease. We demonstrate how such analyses can be applied to uncover local determinants of specificity as well as regional contributions to substrate affinity. Our results were then used to inform a selection strategy that incorporated affinity and cleavage steps in tandem. In a single round of selection, this process generated variants whose biophysical properties were rapidly assessed on the yeast surface, the results of which provided new insights into the mechanistic correlations of binding and catalytic specificity for LAGLIDADG protein function.
MATERIALS AND METHODS
DNA constructs and substrates for binding and cleavage assays
The restriction site embedded ORF (REOAni) was designed using the EMBOSS ‘Silent’ web application (http://emboss.sourceforge.net/) and codon optimized for yeast expression (Blue Heron Biotechnology). Biotinylated and/or fluorophore-conjugated double-stranded oligonucleotides (ds-oligos) and their complements were mixed at equimolar concentrations and annealed or generated using PCR and purified from single-stranded contaminants by Exo1 digestion (New England Biolabs) and size exclusion through a G-50 or G-100 column (GE Healthcare), then analyzed for purity by gel electrophoresis (determined to be >98%). See Supplementary Data online for oligonucleotide sequences used in all applications.
Yeast growth, transformation, library construction and plasmid recovery
Saccharomyces cerevisiae strain EBY100 was transformed using the lithium-acetate (LiAc) method (35). For library construction, error-prone PCR was performed over the STS3/4 region of the I-AniI ORF using the GeneMorph-II Random Mutagenesis kit (Stratagene) according to the manufacturer’s protocol. Library size for the STS3/4 library was determined by serial dilution to be 0.5 × 106 unique transformants. Mutation distribution and frequencies were verified by sequencing an unselected library and determined to be in the range of 0.5–1.0 mutations per kilobase with no major biasing of the type or positions of mutations. Yeast propagation was performed in the presence of 2% raffinose +0.1% glucose at 30°C for at least 12 h prior to induction. Cells were induced in 2% galactose for 2–3 h at 30°C followed by 18–26 h at 20°C. Plasmids were isolated from yeast populations using the Zymoprep-II kit (Zymo Research) and electroporated into Escherichia coli DH10B (Invitrogen) for amplification and/or sequencing. Sequencing was performed on 40–60 clones for a given selection output.
Flow cytometry
Flow cytometric binding analyses were performed using a buffer containing 10 mM HEPES, 10 mM NaCl, 180 mM KCl, 5 mM CaCl2, 0.1% galactose, 0.2% BSA, pH 7.5. For Kd determination, roughly 2–5 × 105 cells/well were stained in a final volume of 100 µl, corresponding to an approximate concentration of 100 pM (assuming 104–105 molecules per yeast surface). Serial dilutions of substrate ranging from ∼5 µM to 0.1 nM were used for staining. Samples were incubated at 4°C for 2–4 h to achieve equilibrium then washed twice in excess staining buffer and counterstained with streptavidin-phycoerythrin (PE) (BD biosciences), fluorescein isothiocyanate (FITC)-conjugated anti-Myc (ICL Labs), and an amine-reactive viability dye, Alexa fluor 350 succinimidyl ester (Molecular Probes). All analytical flow cytometry samples were acquired on a BD LSRIITM cytometer (BD biosciences) and analyzed using FloJo software (Tree Star).
Binding affinity calculation
Epitope-normalized values for substrate binding were determined for each sample. This value was established as the median PE fluorescence in a 10% cell gate of FITC fluorescence. This gate position, which generated normally distributed PE fluorescence data from ∼2000 cells per sample, was held constant across the entire experimental analysis. Median PE values were plotted versus ds-oligo concentration and the resulting distribution was fit using iterative least-squares modeling to the equation for equilibrium binding:
In the VisualEnzymics (SoftZymics) module for IGOR Pro 6 (WaveMetrics), where [L] is the concentration of substrate and Y is the measured median PE value for a given Myc-normalized sample. Non-linear regression analysis was applied using the Levenberg-Marquart (LM) Robust algorithm to isolate the maximum bound ligand, Bmax and the dissociation constant, Kd. Samples were weighted towards the lower ds-oligo concentrations which showed the least variation across samples. The constant term, A, was included to adjust for the median fluorescence value of unstained cells for a given cytometer’s photomultiplier tube voltage setting.
Substrate binding competition assay
His-tagged recombinant I-AniI was immobilized in nickel-NTA coated HisSorb plates by incubation of a 100 nM solution in TBS (50 mM Tris–HCl pH 7.5, 150 mM NaCl) with 0.2%BSA for 2 h at room temperature, followed by four washes with TBS containing 0.05% Tween-20. Each well was incubated for 2 h with a mixture of 100 nM labeled wild-type target and 3 μM unlabeled competition substrate in 200 μl TBS with 0.02 mg/ml poly(dI–dC) and 10 mM CaCl2. The plates were washed four times with TBS and the fluorescent signal retained in each well was quantified using a SpectraMax M5/M5e micro-plate reader (Molecular Devices). Additional negative control experiments performed in the absence of the enzyme indicated that no significant detectable background fluorescence. Relative binding affinities were calculated using the following equation: {[(F(n) – F(x)] × F(t)}/{[(F(n) − F(t)] × F(x)}, where F(x), F(t) and F(n) indicate fluorescent intensities obtained from wells in which the immobilized protein was incubated with the unlabeled single base-pair substitutions, target sites and negative control sequence, respectively (36).
In vitro cleavage assay
Assuming a maximum of 105 molecules per yeast cell surface (11), the final concentration of enzyme was estimated at 3 nM in a 50 µl reaction containing 1 × 106 displaying yeast. Samples were incubated with 50 nM Alexa-647-conjugated ds-oligo in a buffer containing 150 mM KCl, 10 mM NaCl, 10 mM HEPES, 5 mM MgCl2 (or CaCl2 where indicated), 5 mM DTT, 0.5 mg/ml BSA, pH 8.25 and placed at 37°C (or indicated temperatures for thermal titration) for 1 h. Supernatants were extracted once with phenol and run on a 12–15% non-denaturing polyacrylamide gel. Quantification was performed with an Odyssey infrared imaging system (Li-Cor Biosciences).
On-cell ds-oligo cleavage
Approximately 2 × 105 induced cells were stained first with 1 : 300 dilution biotinylated anti-HA (Covance), then with pre-conjugated streptavidin-PE:biotin-DNA-A647 in the same buffer used for binding assays, absent of divalent cations. Samples were washed twice in the buffer described for the in vitro cleavage assay (lacking DTT) and then transferred to cleavage buffer containing 10 mM of either CaCl2 or MgCl2, and placed at 37°C for the indicated time points. For kinetic analyses, cells were prepared as in the first two staining steps and transferred to ice-cold cleavage buffer lacking divalent ions. Immediately prior to acquisition, cells were spiked with an excess of cold buffer containing divalent ions and placed in the acquisition chamber pre-warmed to 37°C. Cells were acquired continuously for 8–10 min.
Cell sorting
For cell sorting on the basis of substrate binding, 10–50 × 106 cells per sample were stained with 30–50 nM ds-oligo in a final volume of 1 ml, then washed twice and counterstained with SA-PE, anti-Myc-FITC and Alexa fluor 350 succinimidyl ester and hierarchically gated as described above but processed on a BD FACSAriaTM II cell sorter. Maximal phase and purity masking were employed. Ten-fold sampling of the estimated library size was processed to ensure coverage of the total variability and ∼5000–50 000 cells were sorted at each round. For cleavage based sorting, samples were prepared as described for the on-cell assay yet scaled up 10–50-fold depending on the size of the input population.
RESULTS
Flow cytometric interrogation of DNA-binding specificity
A potential hurdle in applying YSD to intracellular proteins is the risk that their expression on the cell surface may compromise native functions. Exposure to non-native cellular compartments could abolish folding or incur post-translational modifications that affect function. To address whether a monomeric LAGLIDADG protein could be expressed on the yeast cell surface, we fused a full-length I-AniI homing endonuclease to the secreted protein Aga2p (11) (Figure 1a) and developed assays to confirm that native binding and catalytic functions were maintained.
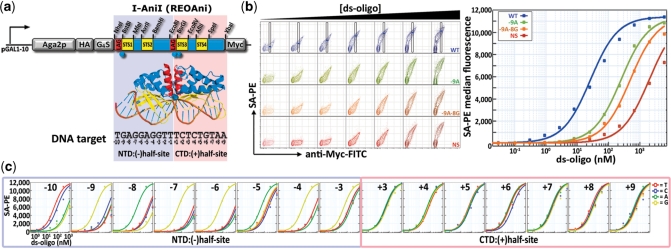
Figure 1.
Profiling protein–DNA binding specificity. (a) Schematic representation of the Aga2p-REOAni expression construct. Silent restriction sites unique within the vector and embedded adjacent to domain boundaries are indicated. A corresponding structural diagram (PDB file: 2QOJ) with emphasis on the motifs that comprise the N-terminal (STS1, STS2) and C-terminal (STS3, STS4) DNA-binding domains and annotated target sequence are shown below. NTD, N-terminal domain; CTD, C-terminal domain; LAG, LAGLIDADG helix; STS, strand-turn-strand; G4S, 3 × (Gly)4-Ser linker motif. (b) Flow cytometry contour plots for equilibrium titration staining with increasing concentrations of four ds-oligo substrates. For the wild-type target, Myc epitope normalization gates are shown with the ds-oligo:SA-PE median fluorescence intensity levels marked. Non-linear regression curves (see ‘Materials and Methods’ section) for binding affinity measurements using epitope-normalized fluorescence values from flow cytometry stains are shown in the adjacent panel. (c) Equilibrium binding curves from a representative experiment with target site substrate analogs bearing each single mutation across I-AniI’s native target sequence. The curve representing I-AniI’s interaction with its native target is reproduced on each plot for comparison.
Substrate interactions were quantified using equilibrium ligand binding approximations analogous to those used in the exploration of protein–protein interactions in this surface display system (13). Biotinylated ds-oligo substrates were used as staining reagents in flow cytometry assays on yeast cells displaying I-AniI. For a given concentration of DNA substrate, samples were incubated to equilibrium and the amount of bound substrate was detected by a secondary staining step with streptavidin-PE (Figure 1b). An antibody to the C-terminal Myc epitope was added coincident with this secondary detection step to evaluate substrate-binding capacity as a function of surface expression. Staining with ds-oligos encompassing I-AniI’s wild-type target site generated a signal that fit a standard one-site binding equation with an approximate dissociation constant (Kd) of 30 nM. This value for I-AniI is consistent with those obtained by traditional biophysical methods for Kd determination (23), as has been generally observed during the characterization of protein–protein interaction affinities on the yeast surface (13). Significant signal reductions were observed when yeast expressing I-AniI were stained with targets bearing a single base pair substitution predicted from structural data to disrupt direct hydrogen bond contacts (37).
Equilibrium binding interactions were next evaluated for a panel of ds-oligo substrate analogs representing the three alternative bases at all target site positions (Figure 1c). Our analysis revealed that I-AniI’s N-terminal domain discriminates with exclusive specificity at multiple positions of the corresponding (−) half-site. A comparison of the binding specificity profile of I-AniI’s N-terminal domain with contacts identified in the crystal structure indicates that hydrogen bond interactions are highly exclusive determinants of target site specificity in this region of the interface. In contrast with the strict direct binding readout observed for the N-terminal domain, there is a striking lack of binding specificity within the C-terminal domain that can be attributed to any one position along the (+) half-site. Cross-validation of the flow cytometric assay for equilibrium binding profiling using a substrate competition assay confirmed that the surface displayed enzyme exhibits similar specificity characteristics as recombinant I-AniI (Supplementary Figure S1). These results imply that the majority of high-resolution binding discrimination is accomplished by I-AniI’s N-terminal domain, and highlight the value of a direct quantitative assay for describing local binding specificity contributions in protein–DNA interactions.
Profiling catalytic specificity
Monitoring DNA hydrolysis on the cell surface via flow cytometry would enable both high-throughput analyses of catalytic specificity as well as clonal isolation of active enzymes from libraries of variants. To this end, an on-cell fluorophore release assay was devised for flow cytometric interrogation of cleavage events (Figure 2a). Substrates containing 5′-biotin and (on the complementary strand) 5′-Alexa fluor-647 were generated by PCR and conjugated to streptavidin-PE at ratio that preserves biotin-binding sites. These complexes were then tethered to the yeast surface through biotinylated antibodies to the HA or Myc epitope tags of the Aga2p-I-AniI fusion. Tethering in this manner immobilizes bifluorescent DNA substrates proximal to I-AniI, poised for cleavage, yet does so independently of interactions with the DNA-binding interface (Supplementary Figure S2). When a productive complex is placed in conditions which support catalysis, the specific release of the non-tethered fluorophore, A647, generates a measurable deviation in the linear ratio of the two signals (Figure 2b), while a catalytically inactive interaction or any spurious non-cleaved substrate release events maintain a scaled linearity between the two fluorophores. Limiting titrations of yeast expressing active I-AniI into a population expressing an inactive variant were performed to confirm that cleavage of the tethered substrate was catalyzed by endonucleases displayed on the same cell—a necessary pre-condition for clonal isolation (Supplementary Figure S3).
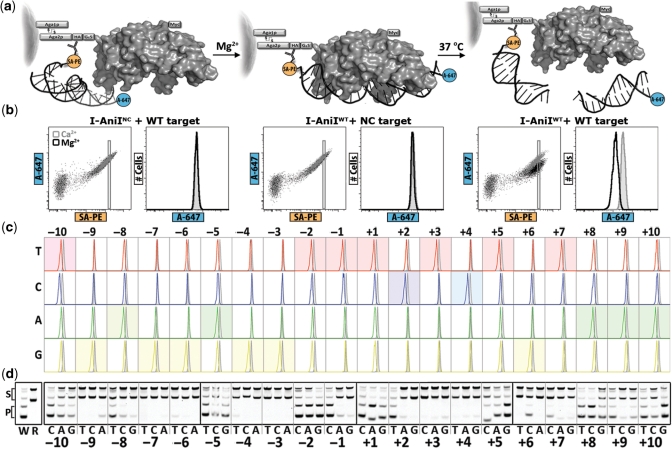
Figure 2.
Profiling endonuclease catalytic specificity. (a) Schematic representation of the on-cell cleavage assay before (left), during (middle) and after (right) transition to conditions which support substrate cleavage. PE, phycoerythrin; A647, Alexa fluor 647. For clarity, tethering and cleavage in the schematic is shown to occur in cis, however, it is most likely that cleavage events occur in trans, as only minimal orientation effects have been observed when using HA versus Myc epitopes to tether substrate. (b) Flow cytometry dot plots and corresponding histograms monitoring the fluorescence profile of wild-type, WT, versus a non-cleaving, NC, mutant in the on-cell cleavage assay in the presence of divalent cations which either inhibit (Ca2+) or support (Mg2+) catalysis. (c) Flow cytometry histograms from a representative profiling experiment where each PCR-generated substrate was conjugated to SA-PE and tested for cleavage by overlaying the A647 signal following incubation in cleavage buffer with Ca2+ or Mg2+ ions. The shaded wild-type profile is reproduced at each position for comparison with the unshaded one-off substrates. (d) In vitro cleavage assay performed on PCR-generated targets confirming that the fluorescence shift in A647 during on-cell specificity profiling correlates with the formation of cleaved product in a classical endonuclease reaction in solution. W, ds-oligo with I-AniI’s wild-type target sequence; R, substrate with a random DNA sequence.
As in the substrate-binding experiments, specificity profiling at the level of target cleavage was performed using a panel of singly-mutated substrate analogs. Cleavage activity closely paralleled binding specificity in the N-terminal domain; positions with high binding specificity (positions −9, −7, −6, −4) also displayed stringent catalytic specificity, while those with diminished binding specificity (positions −10, −8, −5) also showed the most promiscuous readout of catalysis (Figure 2c). Position +3 was unique in that binding discrimination was not as complete as at positions more distal to the active site, yet catalytic specificity does not appear to be compromised. These results were confirmed using a traditional in vitro assay for endonuclease activity, verifying the on-cell assay as a high-throughput technique for investigating endonuclease specificity (Figure 2d). Binding and cleavage correlations broke down significantly in the C-terminal domain. Substrates with mutations in the (+) half-site which bind to I-AniI with imperceptible differences in affinity showed dramatic discrepancies in cleavability. Base changes at positions +3, +4, +6 and +7 all appear to be efficiently discriminated at the level of catalysis without imparting resolvable binding penalties. Thus, mechanistic control of cleavage specificity by the C-terminal domain is not easily accounted for by a simple direct readout mechanism, suggesting a global asymmetry in the nature of I-AniI’s interaction with its substrate.
Probing regional binding affinities
The lack of binding specificity in the C-terminal domain’s interaction with the (+) half-site may result from qualitative and/or quantitative mechanistic contributions. This distinction is especially relevant when considering specificity redesign in this region of the interaction. Since flow cytometric measurements of A647 release as described earlier encompass two sequential events in the endonuclease-target interaction—catalysis and product release—we applied real-time monitoring of fluorophore loss to test whether global quantitative differences in binding affinity to the cleaved half-site products exist on either side of the complex. Bifluorescent substrates with A647 conjugated to either end of the target sequence were tethered to surface displayed I-AniI (Figure 3a). The rate of fluorescence loss was found to be significantly faster when A647 was conjugated to the (+) half-site, indicating that this side of the complex is released more rapidly than the (−) half-site following catalysis (Figure 3b). This observation indicates that the C-terminal domain (+) half-site interaction is of lower basal affinity relative to the other side of the complex, as cleavage activity was determined to be independent of either the substrate orientation or tethering epitope (Figure 3c).
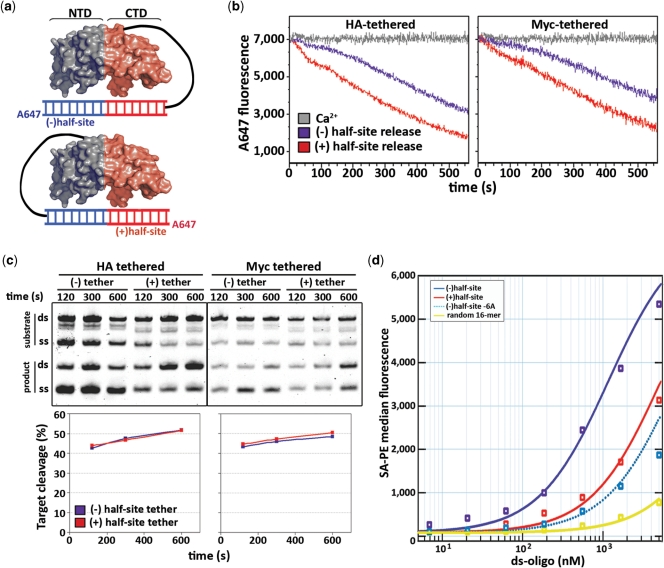
Figure 3.
Half-site product release and truncated substrate binding to isolate N- and C-terminal domain contributions to affinity. (a) Tethering scheme for evaluating product release events in real time by placing the biotin moiety and A647 fluorophore on either side of the DNA substrate. The black line connecting the substrate and the epitope tag (either HA or Myc, independent of which end of the substrate is tethered to Alexa 647) represents the tethering apparatus as described in Figure 2a. (b) Kinetic traces of (−) versus (+) half-site cleavage/release rates, defined by the position of the Alexa 647 fluorophore, when tethered either to HA or Myc epitopes demonstrating a much faster fluorophore loss signal when conjugated to the (+) half-site. (c) Despite a faster fluorophore release signal when monitoring cleavage and release of (+) half-site, the relative rates of substrate cleavage were comparable in either substrate orientation and only minor differences were observed using different epitopes for tethering. (d) Equilibrium binding curves for a representative half-site substrate binding experiment in which truncated substrates were used—in combination with unlabelled blocking substrates representing the opposite half-site—to isolate the relative binding affinities of the two halves of the pseudo-dimeric complex; a 5-fold higher affinity of the N-terminal domain: (−) half-site interaction [Kd of ∼800 nM versus 4000 nM for the C-terminal domain: (+) half-site interaction] was observed, while a single mutation at position −6 in the (−) half-site drops its affinity below that of the C-terminal domain.
To confirm these findings, we isolated regional contributions to substrate binding by performing equilibrium binding analysis with truncated substrates consisting of only the (−) and (+) half-sites. A substantially higher affinity between I-AniI’s N-terminal domain and the (−) half-site was observed, as the Kd value for this side of the interaction was 5-fold lower than that between the C-terminal domain and the (+) half-site (Figure 3d). This large discrepancy in binding affinity, where >80% of the binding energy is contained within the interaction between the N-terminal domain and the (−) half-site, suggests that the low affinity of the interaction between I-AniI’s C-terminal domain and the (+) half-site results in an apparent lack of binding specificity in this region of the complex.
Selection for high-affinity endonucleases
The results above demonstrate that I-AniI’s C-terminal domain supplies catalytic specificity in spite of minimal binding affinity and specificity towards its native target. We therefore sought to identify high-affinity C-terminal domain variants with the expectation that the identity and position of recovered mutations in this region, and their effects on catalytic performance, might enable a more detailed understanding of the observations described above. To achieve this, we carried out a targeted mutagenesis and tandem affinity/activity selection procedure, applying the analytical tools described earlier in conjunction with flow sorting. Silent restriction sites embedded at motif and domain boundaries (Figure 1a) enabled targeted variation of a region comprising a 250 base pair stretch from the LAGLIDADG motif to STS4 of the C-terminal domain (38).
A library of I-AniI variants was first sorted by flow cytometry for the top performing binders to the wild-type target site, generating a population of cells which stained brighter with DNA substrates following a single round of mutation/selection (Figure 4a). Following sequencing, clones were derived bearing individual mutations and analyzed by equilibrium binding titration to confirm their impact on binding affinity, with all clones analyzed demonstrating increases in binding affinity to the native target sequence (Supplementary Figure S4). In relating the positions of the mutated residues to the crystal structure of the complex, we found that they were skewed towards positions contacting or immediately proximal to the phosphoribosyl backbone of the DNA strand which culminates in the active site of the C-terminal domain (Figure 4b). Mutations at R172 and Q171 would likely produce side chains contacting the DNA backbone at the +1 and +2 positions, respectively, immediately 5′ of the hydrolyzed phosphodiester bond. E148 is a critical active site residue which coordinates a Mg2+ ion at the scissile phosphate belonging to position +3. Indeed, the effect of conservatively mutating residue E148 to an aspartate residue (E148D), thereby retracting a Mg2+ ion-coordinating carboxyl group by the length of a single C–C bond, is a 5-fold improvement in binding affinity. C150, S152 and K155 are situated along the DNA backbone 3′ of the active site at positions +4 +5 and +6. This pattern suggests that the orientation of the native enzyme–substrate complex results in binding penalties along the phosphoribosyl backbone that limit affinity in the C-terminal domain.
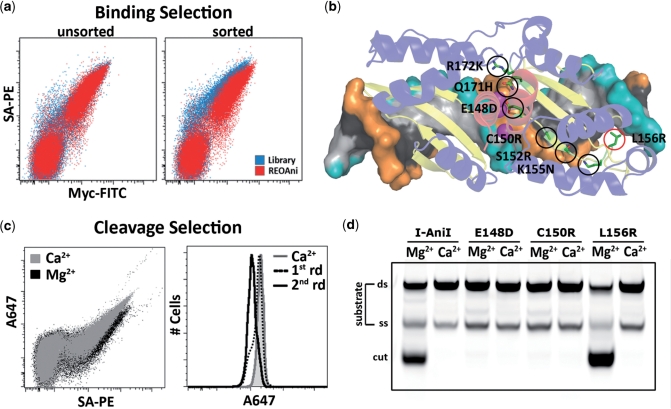
Figure 4.
Selection of high-affinity I-AniI variants reveals a positional correlation of binding affinity versus catalytic contributions to specificity. (a) Flow cytometry plots representing the pre- and post-sort populations of a strand-turn-strand 3 (STS3)/STS4 mutagenized library relative to the wild-type enzyme when selected for binding to the native target sequence. (b) Structural representation of positions where mutations (side chains shown in green, highlighted with black circles if contacting the phosphoribosyl backbone, red circles if contacting the DNA bases) were enriched following selection. Active site Mg2+ ions depicted as magenta spheres, and the phosphoribosyl backbones of each strand are shown in cyan and orange. (c) Flow cytometry plot and histograms of control (Ca2+) and experimental (Mg2+) conditions during sorting for catalytic activity performed on the output population selected for high binding affinity. (d) In vitro endonuclease assay confirming that variants enriched or depleted during activity-based selections were catalytically active and inactive, respectively.
Flow sorting for catalysis was next used to isolate variants from the high-affinity pool that maintained endonuclease activity, generating a population of cells which released A647 in a Mg2+-dependent manner (Figure 4c). Cells isolated from this process were enriched with the non-mutated wild-type I-AniI ORF and the variant bearing the L156R mutation, yet did not contain any of the high-affinity variants with mutations at positions contacting the C-terminal phosphoribosyl backbone. An in vitro cleavage assay confirmed the effect of the recovered mutations on catalysis relative to wild-type I-AniI (Figure 4d). The discovery that all the mutations which enhance affinity through modulating contacts with the phosphoribosyl backbone are uniformly inactive, while the one recovered high-affinity active variant is positioned distal to the active site and contacts the DNA bases, supports the concept that specificity in the C-terminal domain is coupled to catalysis and independent of binding discrimination. The observation that the L156R mutation enhances both affinity and catalytic activity yet does not alter regional target binding or cleavage specificity (Supplementary Figure S5) suggests that, in contrast with the N-terminal domain, the C-terminal domain uses a mechanism for target discrimination that does not correlate with contributions to binding affinity.
DISCUSSION
The YSD-based analytical tools described herein offer a flexible quantitative method for describing DNA-binding and cleavage specificity without the need for protein purification or complex substrate arrays. Contributions to specificity from individual protein–DNA contacts are easily discriminated at single nucleotide resolution by flow cytometric interrogation. While previous attempts using mammalian cell surface display were able to resolve single base pair differences in binding affinity (10), conversion of such a system to selection on a catalytic parameter was unsuccessful due to the toxicity of the conditions necessary for efficient endonuclease activity. The techniques described here represent the first application, to our knowledge, where high-resolution analysis of protein–DNA binding and cleavage specificity can be seamlessly translated to a high throughput selection platform.
Our study demonstrates the application of YSD techniques in the study and manipulation of the binding and cleavage properties of a prototypical LAGLIDADG homing endonuclease, I-AniI. By profiling the specificity contributions of each position in the native DNA substrate of I-AniI, we uncovered a surprising functional asymmetry between I-AniI’s two structurally symmetric domains. Local binding specificity and regional binding affinity was found to be concentrated to the interface between the N-terminal domain and the (−) half-site of the substrate, yet analysis of cleavage specificity indicated that mechanisms which supply catalytic discrimination are present throughout the complex, including areas where no obvious alterations in binding affinity are present.
The observed discordance in binding and catalytic specificity at the C-terminal domain could be accounted for by affinity-based discrimination that is below the resolution limit of the YSD assay or the existence of an affinity-independent mechanism that directly influences the enzyme active site. To differentiate between these possibilities, we randomly mutagenized the C-terminal domain of I-AniI, and sorted the resulting library for variants with increased affinity, but without regard for catalysis. If a solely affinity-based discrimination mechanism were operative, an even distribution of affinity increasing mutations over the protein/DNA interface surface would be expected in the output, as beneficial affinity improving mutations could occur at any protein/DNA contact. Remarkably, all but one of the affinity-increasing mutations were localized to the DNA backbone proximal to the active site, and all of these were incompatible with catalysis. This result suggests that the interaction between the C-terminal domain’s active site and the nearby phosphoribosyl backbone is a critical conformational checkpoint in catalytic discrimination.
The conformation checkpoint described above appears to limit the overall affinity of the interaction between I-AniI and its native DNA target. Consistent with this, the single distal mutation, L156R, which enhanced the affinity of the C-terminal domain did not degrade or alter catalytic specificity. Thus, in contrast with the affinity-dependent mechanism of catalytic specificity that our data implies in the N-terminal domain: (−) half site interaction, these data suggest that a stringent conformational checkpoint endows catalytic specificity to the C-terminal domain. A related study recently identified a hyperactive I-AniI variant, called ‘Y2’, which contains two mutations, F13Y and S111Y, in the N-terminal domain which enhance the overall affinity of the complex (39). The crystal structure of this variant demonstrated an alteration in the geometry of the N-terminal domain’s active site. We tested the specificity of the Y2 variant displayed on the yeast surface (Supplementary Figure S6). Our data demonstrates that the enhanced affinity of the N-terminal domain results in a modest reduction in the specificity within the complex, yet that the general specificity patterns follow that of the wild-type enzyme. More detailed kinetic analyses of these variants should provide a more thorough understanding of the asymmetric nature of specificity in the complex between I-AniI and its substrate.
In light of the growing field of LAGLIDADG engineering and re-specification, further investigation into the mechanistic origins of specificity and their impact on in vivo activity is warranted. Evaluating regional and local effects of increased affinity may enable the development of more appropriate position specific scoring matrices (PSSMs) to identify optimal genomic HE targets, and thereby assist in efforts to redirect HE specificity. Computational all-atom modeling and redesign efforts will be aided tremendously by profiling the local and global binding effects of direct protein–DNA contacts (40). YSD also provides a highly flexible platform for testing large numbers of variants generated through computational or directed evolution approaches to redirect target specificity.
Detailed quantitative descriptions of DNA-binding specificity will have a direct impact on design, engineering, and directed evolution of protein–DNA interactions. Experimental systems which facilitate comprehensive characterization of the specificity of such interactions will become increasingly relevant as therapeutic and biotechnological approaches utilizing DNA-binding and/or cleaving molecules become more widespread. The methods described here facilitated a rapid and detailed analysis of the substrate binding and cleavage relationships in a representative enzyme–substrate interface that is the focus of a concerted engineering effort aimed at redefining target specificity. Their application has demonstrated both parsimony and discordance in the interrelationships between DNA binding and catalysis within a single enzyme.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [RL1CA133832 and UL1DE019582 to A.M.S., PL1HL092557 to David J. Rawlings and RL1CA133833 to B.L.S]; and by followships from the Cancer Research Institute and Natural Sciences and Engineering Research Council of Canada to J.J. Funding for open access charge: National Institutes of Health grant #RL1CA133832.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank David Baker, Ray Monnat Jr, Jim Havranek, Summer Thyme, Umut Ulge, and David Rawlings for their input and critical reading of the manuscript, and all members of the Northwest Genome Engineering Consortium (NGEC) (http://research.seattlechildrens.org/centers/immunity_vaccines/ngec/) for their many insightful discussions.
REFERENCES
- 1.von Hippel PH. From “simple” DNA-protein interactions to the macromolecular machines of gene expression. Annu. Rev. Biophys. Biomol. Struct. 2007;36:79–105. doi: 10.1146/annurev.biophys.34.040204.144521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grigoryan G, Reinke AW, Keating AE. Design of protein-interaction specificity gives selective bZIP-binding peptides. Nature. 2009;458:859–864. doi: 10.1038/nature07885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jen-Jacobson L, Engler LE, Jacobson LA. Structural and thermodynamic strategies for site-specific DNA binding proteins. Structure. 2000;8:1015–1023. doi: 10.1016/s0969-2126(00)00501-3. [DOI] [PubMed] [Google Scholar]
- 4.Mukherjee S, Berger MF, Jona G, Wang XS, Muzzey D, Snyder M, Young RA, Bulyk ML. Rapid analysis of the DNA-binding specificities of transcription factors with DNA microarrays. Nat. Genet. 2004;36:1331–1339. doi: 10.1038/ng1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hallikas O, Taipale J. High-throughput assay for determining specificity and affinity of protein-DNA binding interactions. Nat. Protoc. 2006;1:215–222. doi: 10.1038/nprot.2006.33. [DOI] [PubMed] [Google Scholar]
- 6.Warren CL, Kratochvil NC, Hauschild KE, Foister S, Brezinski ML, Dervan PB, Phillips GN, Jr, Ansari AZ. Defining the sequence-recognition profile of DNA-binding molecules. Proc. Natl Acad. Sci. USA. 2006;103:867–872. doi: 10.1073/pnas.0509843102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 8.Meng X, Thibodeau-Beganny S, Jiang T, Joung JK, Wolfe SA. Profiling the DNA-binding specificities of engineered Cys2His2 zinc finger domains using a rapid cell-based method. Nucleic Acids Res. 2007;35:e81. doi: 10.1093/nar/gkm385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Puckett JW, Muzikar KA, Tietjen J, Warren CL, Ansari AZ, Dervan PB. Quantitative microarray profiling of DNA-binding molecules. J. Am. Chem. Soc. 2007;129:12310–12319. doi: 10.1021/ja0744899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Volna P, Jarjour J, Baxter S, Roffler SR, Monnat RJ, Jr, Stoddard BL, Scharenberg AM. Flow cytometric analysis of DNA binding and cleavage by cell surface-displayed homing endonucleases. Nucleic Acids Res. 2007;35:2748–2758. doi: 10.1093/nar/gkm182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 1997;15:553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- 12.Shusta EV, Holler PD, Kieke MC, Kranz DM, Wittrup KD. Directed evolution of a stable scaffold for T-cell receptor engineering. Nat. Biotechnol. 2000;18:754–759. doi: 10.1038/77325. [DOI] [PubMed] [Google Scholar]
- 13.Gai SA, Wittrup KD. Yeast surface display for protein engineering and characterization. Curr. Opin. Struct. Biol. 2007;17:467–473. doi: 10.1016/j.sbi.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swers JS, Kellogg BA, Wittrup KD. Shuffled antibody libraries created by in vivo homologous recombination and yeast surface display. Nucleic Acids Res. 2004;32:e36. doi: 10.1093/nar/gnh030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirsch C, Gauss R, Horn SC, Neuber O, Sommer T. The ubiquitylation machinery of the endoplasmic reticulum. Nature. 2009;458:453–460. doi: 10.1038/nature07962. [DOI] [PubMed] [Google Scholar]
- 17.Stoddard BL. Homing endonuclease structure and function. Q Rev. Biophys. 2005;38:49–95. doi: 10.1017/S0033583505004063. [DOI] [PubMed] [Google Scholar]
- 18.Knizewski L, Ginalski K. Bacterial DUF199/COG1481 proteins including sporulation regulator WhiA are distant homologs of LAGLIDADG homing endonucleases that retained only DNA binding. Cell Cycle. 2007;6:1666–1670. doi: 10.4161/cc.6.13.4471. [DOI] [PubMed] [Google Scholar]
- 19.Cohen-Tannoudji M, Robine S, Choulika A, Pinto D, El Marjou F, Babinet C, Louvard D, Jaisser F. I-SceI-induced gene replacement at a natural locus in embryonic stem cells. Mol. Cell Biol. 1998;18:1444–1448. doi: 10.1128/mcb.18.3.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gouble A, Smith J, Bruneau S, Perez C, Guyot V, Cabaniols JP, Leduc S, Fiette L, Ave P, Micheau B, et al. Efficient in toto targeted recombination in mouse liver by meganuclease-induced double-strand break. J. Gene Med. 2006;8:616–622. doi: 10.1002/jgm.879. [DOI] [PubMed] [Google Scholar]
- 21.Windbichler N, Papathanos PA, Catteruccia F, Ranson H, Burt A, Crisanti A. Homing endonuclease mediated gene targeting in Anopheles gambiae cells and embryos. Nucleic Acids Res. 2007;35:5922–5933. doi: 10.1093/nar/gkm632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Redondo P, Prieto J, Munoz IG, Alibes A, Stricher F, Serrano L, Cabaniols JP, Daboussi F, Arnould S, Perez C, et al. Molecular basis of xeroderma pigmentosum group C DNA recognition by engineered meganucleases. Nature. 2008;456:107–111. doi: 10.1038/nature07343. [DOI] [PubMed] [Google Scholar]
- 23.Eastberg JH, McConnell Smith A, Zhao L, Ashworth J, Shen BW, Stoddard BL. Thermodynamics of DNA target site recognition by homing endonucleases. Nucleic Acids Res. 2007;35:7209–7221. doi: 10.1093/nar/gkm867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Epinat JC, Arnould S, Chames P, Rochaix P, Desfontaines D, Puzin C, Patin A, Zanghellini A, Paques F, Lacroix E. A novel engineered meganuclease induces homologous recombination in yeast and mammalian cells. Nucleic Acids Res. 2003;31:2952–2962. doi: 10.1093/nar/gkg375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chames P, Epinat JC, Guillier S, Patin A, Lacroix E, Paques F. In vivo selection of engineered homing endonucleases using double-strand break induced homologous recombination. Nucleic Acids Res. 2005;33:e178. doi: 10.1093/nar/gni175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arnould S, Chames P, Perez C, Lacroix E, Duclert A, Epinat JC, Stricher F, Petit AS, Patin A, Guillier S, et al. Engineering of large numbers of highly specific homing endonucleases that induce recombination on novel DNA targets. J. Mol. Biol. 2006;355:443–458. doi: 10.1016/j.jmb.2005.10.065. [DOI] [PubMed] [Google Scholar]
- 27.Ashworth J, Havranek JJ, Duarte CM, Sussman D, Monnat RJ, Jr, Stoddard BL, Baker D. Computational redesign of endonuclease DNA binding and cleavage specificity. Nature. 2006;441:656–659. doi: 10.1038/nature04818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doyon JB, Pattanayak V, Meyer CB, Liu DR. Directed evolution and substrate specificity profile of homing endonuclease I-SceI. J. Am. Chem. Soc. 2006;128:2477–2484. doi: 10.1021/ja057519l. [DOI] [PubMed] [Google Scholar]
- 29.Rosen LE, Morrison HA, Masri S, Brown MJ, Springstubb B, Sussman D, Stoddard BL, Seligman LM. Homing endonuclease I-CreI derivatives with novel DNA target specificities. Nucleic Acids Res. 2006;34:4791–4800. doi: 10.1093/nar/gkl645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith J, Grizot S, Arnould S, Duclert A, Epinat JC, Chames P, Prieto J, Redondo P, Blanco FJ, Bravo J, et al. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 2006;34:e149. doi: 10.1093/nar/gkl720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arnould S, Perez C, Cabaniols JP, Smith J, Gouble A, Grizot S, Epinat JC, Duclert A, Duchateau P, Paques F. Engineered I-CreI derivatives cleaving sequences from the human XPC gene can induce highly efficient gene correction in mammalian cells. J. Mol. Biol. 2007;371:49–65. doi: 10.1016/j.jmb.2007.04.079. [DOI] [PubMed] [Google Scholar]
- 32.Scalley-Kim M, McConnell-Smith A, Stoddard BL. Coevolution of a homing endonuclease and its host target sequence. J. Mol. Biol. 2007;372:1305–1319. doi: 10.1016/j.jmb.2007.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aagaard C, Awayez MJ, Garrett RA. Profile of the DNA recognition site of the archaeal homing endonuclease I-DmoI. Nucleic Acids Res. 1997;25:1523–1530. doi: 10.1093/nar/25.8.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gimble FS, Moure CM, Posey KL. Assessing the plasticity of DNA target site recognition of the PI-SceI homing endonuclease using a bacterial two-hybrid selection system. J. Mol. Biol. 2003;334:993–1008. doi: 10.1016/j.jmb.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 35.Gietz RD, Schiestl RH. Large-scale high-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007;2:38–41. doi: 10.1038/nprot.2007.15. [DOI] [PubMed] [Google Scholar]
- 36.Zhao L, Pellenz S, Stoddard BL. Activity and specificity of the bacterial PD-(D/E)XK homing endonuclease I-Ssp6803I. J. Mol. Biol. 2009;385:1498–1510. doi: 10.1016/j.jmb.2008.10.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bolduc JM, Spiegel PC, Chatterjee P, Brady KL, Downing ME, Caprara MG, Waring RB, Stoddard BL. Structural and biochemical analyses of DNA and RNA binding by a bifunctional homing endonuclease and group I intron splicing factor. Genes Dev. 2003;17:2875–2888. doi: 10.1101/gad.1109003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wells JA, Vasser M, Powers DB. Cassette mutagenesis: an efficient method for generation of multiple mutations at defined sites. Gene. 1985;34:315–323. doi: 10.1016/0378-1119(85)90140-4. [DOI] [PubMed] [Google Scholar]
- 39.Takeuchi R, Certo M, Caprara MG, Scharenberg AM, Stoddard BL. Optimization of in vivo activity of a bifunctional homing endonuclease and maturase reverses evolutionary degradation. Nucleic Acids Res. 2009;37:877–890. doi: 10.1093/nar/gkn1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morozov AV, Havranek JJ, Baker D, Siggia ED. Protein-DNA binding specificity predictions with structural models. Nucleic Acids Res. 2005;33:5781–5798. doi: 10.1093/nar/gki875. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.