Abstract
Here it is demonstrated that the yeast Saccharomyces cerevisiae can take up and assemble at least 38 overlapping single-stranded oligonucleotides and a linear double-stranded vector in one transformation event. These oligonucleotides can overlap by as few as 20 bp, and can be as long as 200 nucleotides in length. This straightforward scheme for assembling chemically-synthesized oligonucleotides could be a useful tool for building synthetic DNA molecules.
INTRODUCTION
Chemically synthesized oligonucleotides are often joined into larger DNA fragments containing full-length genes. This was first demonstrated in 1970 when Khorana and colleagues synthesized the 77 nucleotide gene encoding a yeast alanine transfer RNA from 17 overlapping oligonucleotides (1). Since then, chemical oligonucleotide synthesis has improved tremendously (2) and a number of in vitro enzymatic strategies are available for the assembly of oligonucleotides into larger constructs (3–5).
The capacity of the yeast Saccharomyces cerevisiae to take up and recombine DNA fragments has made it a model eukaryote for studying numerous cellular processes and human diseases (6–8), and a platform for biofuel production (9) and drug discovery (10,11). But, this capacity has also made it a tractable organism for developing new technologies (12). DNA sequences can be genetically altered at will by transforming yeast with either double-stranded (ds) DNA fragments (13) or single-stranded (ss) oligonucleotides (14). In addition, homologous recombination in yeast can be used to build DNA fragments from overlapping constituent parts. This was first demonstrated when a plasmid was constructed from two dsDNA fragments containing homologous ends (15). Two non-homologous dsDNA fragments can also be bridged by ss oligonucleotides that join the ends of the two fragments (16). Previously, we showed that six overlapping dsDNA fragments could be assembled by yeast into an entire Mycoplasma genitalium genome (17). Subsequently, this process was improved and 25 overlapping fragments, between 17 and 35 kb in length, were assembled at once into this genome (18). Later work showed that six smaller fragments were also acceptable (19).
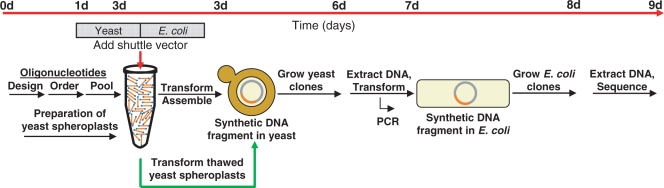
To turn yeast into a ‘factory’ able to produce whole genomes and large constructs of any reasonable sequence, what remains is to demonstrate the assembly of chemically synthesized oligonucleotides into appropriate dsDNA molecules in yeast (Figure 1). For this to succeed, a single yeast cell would need to take up all of the necessary oligonucleotides as well as a vector that overlaps the terminal oligonucleotides. These pieces would then need to be assembled within the nucleus. This is achievable and is demonstrated below. Thus, a methodology that could lead from oligonucleotides to whole chromosomes while requiring little more expertise than yeast transformation is introduced.
Figure 1.
Schematic overview and timeline for the assembly of overlapping ssDNA oligonucleotides (orange lines with blue circles) into a linear dsDNA yeast/E. coli shuttle vector (pRS313; grey) within the nucleus of a yeast cell. Following a single transformation event, a synthetic dsDNA fragment (orange) is produced. These fragments are recovered from yeast and then transferred to E. coli for more efficient amplification.
MATERIALS AND METHODS
Assembly vector preparation
The yeast/Escherichia coli shuttle vector, pRS313 (20), was linearized by restriction digestion with Bam HI then extracted from an agarose gel following electrophoresis. This linearized vector was then PCR-amplified using the Phusion Hot Start High-Fidelity DNA Polymerase with HF Buffer [New England Biolabs (NEB)] with primers pRS313 Universal Assembly For (5′-gatcctctagagtcgacctgcaggaattcgatatcaagcttatcg-3′) and pRS313 Universal Assembly Rev (5′-cgggtaccgagctcgaattcggagctccaattcgccctat-3′). One hundred microliter reactions were setup using 0.04 ng/µl linearized pRS313, 500 nM each primer, 200 µM each dNTP, 1× HF Buffer with 0.5 mM additional MgCl2, and 20 U/ml enzyme. Cycling parameters were 98°C for 30 s, then 30 cycles of 98°C for 10 s, 60°C for 30 s and 72°C for 4 min, followed by a single 72°C incubation for 5 min. PCR products were extracted from agarose gels following electrophoresis then purified using the QIAquick Gel Extraction Kit (Qiagen) according to the instructions provided. The PCR-amplified vector was quantified and diluted to 100 ng/µl.
Preparation of the oligonucleotides that were assembled in yeast
The oligonucleotides used for in vivo assembly were purchased from Integrated DNA Technologies (IDT) unmodified with standard desalting, at the 10 or 4 nmol scale (Ultramers), and suspended to 50 µM with Tris/EDTA (TE) buffer pH 8.0. Equal volumes of each oligonucleotide were pooled then diluted in TE pH 8.0 buffer to a per oligonucleotide concentration of 60 or 240 nM (Ultramers).
Yeast spheroplast transformation
The yeast spheroplast transformation procedure was carried out using a published protocol with the highly transformable VL6-48N yeast strain (21). A similar strain can be purchased from the American Type Culture Collection (ATCC no. MYA-3666). Frozen competent spheroplasts were stored at −80°C in 1 M Sorbitol and 15% DMSO, as previously described (22). Cells were grown to an OD600 of 0.5–1.0 prior to preparation of yeast spheroplasts. Twenty microliters of the oligonucleotide pool and 2 µl PCR-amplified pRS313 were mixed with ∼108 yeast spheroplasts. Following transformation, yeast spheroplasts were regenerated and selected on complete supplemental medium without histidine (CSM-His) and 1 M sorbitol agar plates for 3 days at 30°C.
PCR analysis of yeast clones
Individual yeast colonies were picked into 1.0 ml CSM-His liquid medium and incubated overnight at 30°C. DNA was extracted from these cells using the ChargeSwitch Plasmid Yeast Mini Kit (Invitrogen) according to the provided manual. PCR was carried out with Phusion polymerase with 1 µl DNA template and primers M13F (5′-tgtaaaacgacggccagt-3′) and M13R (5′-caggaaacagctatgacc-3′) as above, except primers were annealed at 55°C and product extension was for 30 or 60 s. Two microliters of each reaction were loaded onto a 0.8% E-gel (Invitrogen) and 72 V was applied for 30 min. DNA bands were visualized with a Typhoon 9410 Fluorescence Imager.
Sequence analysis of E. coli containing assembled products transferred from yeast
From each oligonucleotide transformation experiment, 96 individual yeast colonies were picked into 1 ml CSM-His liquid medium and incubated overnight at 30°C. Approximately 10 µl of each culture was pooled. DNA was extracted from this pool using the ChargeSwitch Plasmid Yeast Mini Kit (Invitrogen), as above. Three microliters were electroporated into 30 µl TransforMax EPI300 (Epicentre) electrocompetent E. coli cells in a 1 mm cuvette (BioRad) at 1200 V, 25 µF and 200 Ω using a Gene Pulser Xcell Electroporation System (BioRad). Cells were allowed to recover at 37°C for 1 h in 1 ml SOC medium then plated onto LB medium + 100 µg/ml carbenicillin. Following incubation at 37°C for 18 h, individual colonies were selected and grown in 1 ml LB medium + 100 µg/ml carbenicillin. Plasmid DNA was then extracted from these cells using a Qiagen miniprep kit. Both strands of the insert DNA were sequenced with the M13F and M13R primers, as indicated in the text. Standard sequencing reactions were carried out on a 3100 sequencer (Applied Biosystems). Trace files were aligned with the reference sequences (see below) by ClustalW Multiple alignment (23), contained within the BioEdit Sequence Alignment Editor software. The synthesis error rate was determined by dividing the number of errors in the complete insert clones by the total number of bases synthesized, and then multiplying by 100 to produce a percentage.
DNA sequences of the synthesized fragments
Each synthetic DNA fragment begins with 5′-GAATTCGAGCTCGGTACCCGgcggccgc-3′ and ends with 5′-gcggccgcGATCCTCTAGAGTCGACCTG-3′. Regions containing homology to PCR-amplified pRS313 are capitalized. NotI sites are italicized and are included to release the synthetic DNA fragment from the vector. Oligonucleotides 1–38 u (Supplementary Table S1) produce the DNA sequence found at 227–1340 bp of pRS316 (20). Oligonucleotides A–H and Arc–Hrc (Supplementary Table S2) produce the DNA sequence found at 6115–6398 bp of the mouse mitochondrial genome (24). Oligonucleotides 1–28 g (Supplementary Table S3) produce the DNA sequence found at 227–1310 bp of pRS316. Oligonucleotides HF1–HF6 (Supplementary Table S4) produce the DNA sequence found at 227–1270 bp of pRS316.
RESULTS
As many as 38 overlapping oligonucleotides can be assembled in yeast at once
To determine the capacity for oligonucleotide assembly in yeast, a 1170 bp DNA sequence was split into 38 60-mers (named 1–38 u) that overlap each other by 30 bp (Figure 2a). The terminal oligonucleotides 1 and 38 u also contain a 20 bp overlap to PCR-amplified pRS313, a shuttle vector that allows for selection of the assembled oligonucleotides in yeast and E. coli. Approximately 108 yeast cells were made competent for transformation as described earlier (21). They were then added to a DNA pool containing each of the 38 oligonucleotides and 200 ng PCR-amplified pRS313 vector (60 fmol or ∼360 molecules per yeast spheroplast). Each oligonucleotide was used at 20-fold molar excess relative to the vector. Following transformation, yeast cells were plated on selective medium. Incubation at 30°C for 72 h produced ∼1000 colonies. The number of colonies obtained could be increased by raising the oligonucleotide concentration (Supplementary Table S5). DNA was extracted from 12 individual colonies then analyzed by PCR with primers M13F and M13R to identify full-length assemblies (Figure 2b). Five out of 12 colonies screened produced the PCR amplicon predicted for proper in vivo assembly of all 38 pieces. This provided strong evidence that yeast has the capacity to take up and correctly assemble at least 38 overlapping oligonucleotides into a vector.
Figure 2.
Assembly of 38 overlapping 60-mer oligonucleotides in yeast. (a) The 38 oligonucleotides, named 1–38 u, have 30 bp overlaps and produce a 1170 bp synthetic DNA fragment following assembly. The terminal oligonucleotides overlap the vector (grey) by 20 bp (red x). Ten nucleotide gaps (green) are repaired inside the yeast cell. (b) PCR analysis of 12 randomly selected yeast clones following transformation and assembly of the oligonucleotides and vector depicted in (a). The primers used for this PCR analysis and for DNA sequencing are M13F and M13R and are shown in (a). The predicted amplicon size for a complete assembly is 1393 bp and is indicated by an asterisk. The presence (+) or absence (−) of the expected product is noted for each clone screened. L indicates the 1 kb DNA ladder (NEB).
To determine the accuracy of synthetic DNA construction by this method, 96 yeast colonies were randomly selected and grown to stationary phase in selective medium then pooled. Plasmid DNA was extracted from this pool and then electroporated into E. coli, a more suitable host for producing copies of the plasmid DNA. The assembled inserts from 79 individual E. coli colonies were sequenced then analyzed (Table 1). Fifty-two out of 79 (66%) of these clones had the oligonucleotides properly assembled into the full-length product. This confirmed our results above (Figure 2b). Furthermore, of these full-length products, six (11.5%) perfectly matched the designed sequence (see Supplementary Table S6 for a complete list of errors). Thus, this method (outlined in Figure 1) can be used to produce error-free synthetic DNA fragments.
Table 1.
Sequence analysis of clones following in vivo yeast assembly
| Experiment | Percent of clones with complete, partial, or no inserts |
Percent of clones with 0 to ≥4 errors within synthetic DNA fragment |
Error rate (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Complete | Partial | ≥4 None | 0 | 1 | 2 | 3 | ≥ 4 | ||
| 38 ungapped oligos | 65.8 | 15.2 | 19.0 | 11.5 | 21.2 | 36.5 | 21.2 | 9.6 | 0.181 |
| 28 gapped oligos | 64.3 | 10.7 | 25.0 | 16.7 | 0 | 27.8 | 22.2 | 33.3 | 0.249 |
| 200 base HF oligos | 25.8 | 38.7 | 35.5 | 59.1 | 22.7 | 18.2 | 0 | 0 | 0.054 |
Oligonucleotides containing 20 bp overlaps can be assembled in yeast
Reducing the size of the overlap between oligonucleotides allows for larger stretches of DNA to be synthesized from fewer constituent parts. However, any resulting gaps would need to be filled in vivo. Figure 2 demonstrates the successful assembly of the terminal oligonucleotides and the vector with only 20 bp overlaps. And, these results show that 10 bp gaps can be filled in vivo. To determine whether smaller overlaps could be used to assemble oligonucleotides in yeast, eight 60-mers (named A–H) were designed with 20 bp overlaps (Figure 3a). The reverse complements of these same oligonucleotides (named Arc‐Hrc) were also designed (Figure 3b). In this case, the 3′-ends of the terminal oligonucleotides, and not the 5′-ends, are to be recombined with the vector. Both sets of these oligonucleotides would produce the exact 340 bp synthetic DNA fragment. Yeast transformation of each set of oligonucleotides with the overlapping vector resulted in ∼1000 colonies. From each transformation, DNA was extracted from four yeast clones and then analyzed by PCR with primers M13F and M13R. Three out of four colonies screened for the assembly of oligonucleotides A‐H (Figure 3c) and four out of four colonies screened for the assembly of oligonucleotides Arc to Hrc (Figure 3d) produced a PCR amplicon predicted for a full-length assembly. This data shows that yeast can be used to assemble oligonucleotides that overlap by as few as 20 bp. This analysis also revealed that the 3′- or 5′-ends of the oligonucleotides can be recombined with the ends of the vector.
Figure 3.
Twenty base-pair overlaps are sufficient for oligonucleotide assembly in yeast. (a and b) Schematic demonstrating the assembly of eight 60-mers, named A–H, or their reverse complements, named Arc–Hrc. The oligonucleotides each contain 20 bp overlaps and were assembled into a vector to produce a 340 bp synthetic DNA fragment. The terminal oligonucleotides overlap the vector (grey) by 20 bp (red x). Twenty nucleotide gaps (green) were repaired inside the yeast cell. (c and d) PCR analysis of four randomly selected yeast clones following transformation and assembly of oligonucleotides A–H (c) as depicted in (a) or oligonucleotides A–rc–H–rc (d) as depicted in (b). The predicted amplicon size for a complete assembly is 563 bp and is indicated by an asterisk. M indicates the 100 bp DNA ladder (NEB). (e) Assembly of 28 60-mers, named 1–28 g, containing 20 bp overlaps, to produce a 1140 bp synthetic DNA fragment. (f) PCR analysis of 12 randomly selected yeast clones following transformation and assembly of the oligonucleotides and vector shown in (e). The predicted amplicon size for a complete assembly is 1363 bp and is indicated by an asterisk.
To determine whether a large number of oligonucleotides could be assembled with shorter overlaps, 28 60-mers (named 1–28g) that overlap by 20 bp were designed to produce a 1140 bp synthetic DNA fragment (Figure 3e). With the exception of 30 bp, this sequence is exactly the same as the one produced by oligonucleotides 1–38 u (Figure 2a). Yeast transformation of oligonucleotides 1–28 g with the vector resulted in 300 colonies. As above, the number of colonies obtained could be increased by adding more oligonucleotides (Supplementary Table S5). DNA was extracted from 12 of these, then analyzed by PCR with primers M13F and M13R to identify full-length assemblies (Figure 3f). Seven out of 12 colonies screened produced the PCR amplicon predicted for proper in vivo assembly of all pieces. This shows that yeast can assemble at least 28 oligonucleotides that share only 20 bp overlapping sequence. To better demonstrate this, and to determine the accuracy of synthetic DNA construction by this method, DNA was extracted from a pool of 96 yeast colonies and then electroporated into E. coli, as described above. The assembled inserts from 28 individual E. coli colonies were sequenced then analyzed (Table 1). Eighteen out of 28 clones (64.3%) had the oligonucleotides properly assembled into the full-length product. This confirmed our results above (Figure 3f). Of the 18 full-length clones, three of them (16.7%) perfectly matched the designed sequence (see Supplementary Table 7 for a complete list of errors). These results show that yeast has the capacity to accurately produce DNA fragments from at least 28 chemically synthesized oligonucleotides that overlap by as few as 20 bp.
Assembly with 200-mer oligonucleotides that have 20 bp overlaps
The results above show that yeast can fill 20 nucleotide gaps that are produced following oligonucleotide assembly (Figure 3). To determine whether larger gaps can be filled when longer oligonucleotides are used, six 200-mer high-fidelity Ultramer oligonucleotides (named HF1-HF6) having 20 bp overlaps were designed to produce a 1.1 kb DNA fragment (Figure 4a). This design requires yeast to fill 160 nucleotide gaps. With the exception of 70 bp, this sequence is exactly the same as the one produced by oligonucleotides 1–38 u (Figure 2a). Following yeast transformation of these oligonucleotides with the vector, ∼200 colonies were obtained. As above, DNA was extracted from a pool of 96 colonies and then electroporated into E. coli. Plasmid DNA was extracted from 10 E. coli colonies, and then NotI-digested to release the assembled insert from the vector (Figure 4b). Two out of 10 clones screened had the correct 1052 bp insert. DNA sequencing of 31 clones (including those analyzed in Figure 4b) revealed eight clones (26%) having the full-length product (Table 1). Furthermore, seven of these exactly matched the designed sequence. DNA sequencing of 14 additional clones with a full-length insert revealed six with the designed sequence. Therefore, a total of 13 out of 22 clones (59%) had the intended sequence when all 6 oligonucleotides were assembled (Table 1). These results demonstrate that yeast has the capacity to take up and assemble oligonucleotides that are as long as 200 nucleotides in length and can fill gaps that are as long as 160 bases. These results also demonstrate that the success of obtaining clones with the intended DNA fragments can be increased when higher fidelity DNA oligonucleotides are transformed.
Figure 4.
High-fidelity oligonucleotide assembly in yeast. (a) Schematic demonstrating the assembly of six 200-mers, named HF1–HF6. The oligonucleotides each contain 20 bp overlaps and were assembled into a vector to produce a 1100 bp synthetic DNA fragment. The terminal oligonucleotides overlap the vector (grey) by 20 bp (red x). One hundred sixty nucleotide gaps (green) were repaired inside the yeast cell. (b) NotI restriction analysis of 10 E. coli clones containing synthetic DNA fragments that were assembled in yeast. Products were separated by gel-electrophoresis on a 2% E-gel. The predicted size for a complete assembly (C) is 1052 bp and is indicated by an asterisk. This analysis also revealed no assembly (N), multiple assembly (M) and partial assembly (P) of oligonucleotides.
DISCUSSION
Oligonucleotide assembly in yeast, as described here, could be very useful in the synthesis of large DNA fragments, including genes and genomes. This study, together with our previous studies (17,18), demonstrates that entire genomes can be synthesized within a yeast cell beginning from a series of oligonucleotides that overlap by as few as 20 nucleotides. And, the genomes can be built following just three rounds of yeast transformation. The limits of DNA uptake and recombination have not been fully explored. It may already be possible that yeast has the capacity to assemble tens of kilobases of DNA sequence in one transformation event from 100 or more ssDNA oligonucleotides. Using longer oligonucleotides (Figure 4) or annealing complementary adjacent oligonucleotides prior to in vivo assembly may be viable approaches for producing longer DNA fragments in a single step, especially if the number of pieces taken up by a yeast cell becomes a limiting factor. Indeed, it is very likely that many of the overlapping oligonucleotides become annealed outside the yeast cell during the transformation procedure. This is especially likely once the molecular crowding agent, polyethylene glycol, is added.
Although oligonucleotide synthesis is drastically better than it was 40-years-ago, this process continues to produce a fraction of unintended DNA sequence (Table 1), which will be more prevalent in longer synthetic DNA fragments. The synthesis error-rate for the assembly of oligonucleotides 1–38 u is 0.181% and 0.249% for the assembly of 1–28g (Table 1; based on data from Supplementary Tables S6 and S7). This is comparable to previously reported data obtained with in vitro protocols (25,26). The use of high-fidelity oligonucleotides (Ultramers, IDT) reduced the error rate to 0.054% (Table 1; based on data from Supplementary Table S8), which is only slightly higher than what can be achieved with error-correction methods (26). Since the error-rate of oligonucleotide synthesis increases with size, the use of smaller high-fidelity oligonucleotides, such as 60-mers, would even further increase the percentage of correct clones obtained. The accuracy of DNA assembled by in vitro approaches would also be improved by the use of high-fidelity oligonucleotides.
Three oligononucleotide assembly schemes were designed and described here. When choosing a scheme, oligonucleotide expenses should be considered along with DNA sequencing rates. The use of high fidelity Ultramer oligonucleotides will decrease sequencing costs. However, these oligonucleotides are significantly more expensive than their lower fidelity counterparts. DNA sequencing costs can be reduced even further by sequencing only the clones that contain a full-length synthetic fragment (as determined by PCR or restriction digestion). Oligonucleotide costs can be further reduced by making use of shorter overlaps. However, longer overlaps may be preferred in instances where unique overlaps cannot be obtained by 20 bp or less. It is expected that oligonucleotide chemistry will continue to improve and soon reliably produce error-free stretches of ssDNA at a more reasonable cost. Once these advances are in place, the one-step synthesis method described here could be used to rapidly construct long DNA sequences that perfectly match the ones that are designed.
Several approaches were considered to improve the oligonucleotide assembly method described here. Yeast spheroplasts can take 2–3 days to prepare. However, it has previously been demonstrated that competent yeast spheroplasts can be prepared and stored frozen (22). The use of frozen yeast spheroplasts would facilitate this method by having the DNA assembly host organism ready-to-go. In the transformation of oligonucleotides 1–38 u, 1–28 g and HF1‐HF6 into both fresh and frozen spheroplasts, colonies were obtained in all cases (Supplementary Table S9). Although the transformation efficiency is reduced, enough colonies are produced to allow the identification of the assembled DNA fragments in yeast. One drawback of the method described here is the 3–4-day wait-time for cloning and growing yeast clones that contain the assembled DNA constructs. In general, however, E. coli is the preferred host organism for propagating these recombined DNA molecules. Thus, it would be interesting to determine whether assembled fragments could be immediately cloned in E. coli following yeast transformation, without selection and propagation in yeast. This should be possible if the recombination efficiency is high and the assembled DNA molecules can be recovered from yeast for immediate transfer to E. coli.
Here the issue of process complexity in gene synthesis is addressed. Overlapping oligonucleotides are assembled and cloned in a single step. But, a method such as this one may also be useful when traditional in vitro assembly methods fail (likewise, in vitro approaches could be useful if this in vivo approach fails). The oligonucleotides used in gene synthesis are often optimized so that the overlapping sequences have similar melting temperatures (27–30). In the gene-synthesis experiments shown here, no effort was made to optimize the oligonucleotide sequences. For example, in the assembly of oligonucleotides 1–28 g, the melting temperatures at the overlaps differed by as much as 16.8°C (Supplementary Table S10). Furthermore, many of the oligonucleotides used in this study had complete homology to yeast genomic sequences. Assembly of synthetic DNA sequences with oligonucleotides having strong homology to the yeast genome can be explained by the preference for homologous recombination at free ends of DNA (31). Living biological cells carry out an extraordinary amount of processes that cannot be duplicated elsewhere. Chemical synthesis in yeast from overlapping oligonucleotides is likely no exception. Interestingly, in the assembly of the six 200-mer oligonucleotides (Figure 4), 44% of the DNA fragments are synthesized by yeast (960 of 2200 nucleotides are added in vivo).
The work described here extends beyond gene synthesis and may be applicable to other schemes in yeast that utilize oligonucleotide transformation for gene knockouts or manipulations (14,32) and linker-mediated recombination (16). It could also be useful for creating combinatorial libraries and for selecting a particular phenotype in yeast.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Synthetic Genomics, Inc. Funding for open access charge: Synthetic Genomics, Inc.
Conflict of interest statement. The research reported in this article is in the field in which Synthetic Genomics, Inc (SGI) is developing products. SGI has funded the research reported in the article and has filed a patent application.
Supplementary Material
ACKNOWLEDGEMENTS
The author thanks Ham Smith, Clyde Hutchison and Gwyn Benders for helpful discussions, Chuck Merryman for his critical review of the manuscript, and Monzia Moodie for technical assistance.
REFERENCES
- 1.Agarwal KL, Buchi H, Caruthers MH, Gupta N, Khorana HG, Kleppe K, Kumar A, Ohtsuka E, Rajbhandary UL, Van de Sande JH, et al. Total synthesis of the gene for an alanine transfer ribonucleic acid from yeast. Nature. 1970;227:27–34. doi: 10.1038/227027a0. [DOI] [PubMed] [Google Scholar]
- 2.Reese CB. Oligo- and poly-nucleotides: 50 years of chemical synthesis. Org. Biomol. Chem. 2005;3:3851–3868. doi: 10.1039/b510458k. [DOI] [PubMed] [Google Scholar]
- 3.Xiong AS, Peng RH, Zhuang J, Gao F, Li Y, Cheng ZM, Yao QH. Chemical gene synthesis: strategies, softwares, error corrections, and applications. FEMS Microbiol. Rev. 2008;32:522–540. doi: 10.1111/j.1574-6976.2008.00109.x. [DOI] [PubMed] [Google Scholar]
- 4.Xiong AS, Peng RH, Zhuang J, Liu JG, Gao F, Chen JM, Cheng ZM, Yao QH. Non-polymerase-cycling-assembly-based chemical gene synthesis: strategies, methods, and progress. Biotechnol. Adv. 2008;26:121–134. doi: 10.1016/j.biotechadv.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Czar MJ, Anderson JC, Bader JS, Peccoud J. Gene synthesis demystified. Trends Biotechnol. 2008;27:63–72. doi: 10.1016/j.tibtech.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 6.Kow YW, Bao G, Reeves JW, Jinks-Robertson S, Crouse GF. Oligonucleotide transformation of yeast reveals mismatch repair complexes to be differentially active on DNA replication strands. Proc. Natl Acad. Sci. USA. 2007;104:11352–11357. doi: 10.1073/pnas.0704695104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petranovic D, Nielsen J. Can yeast systems biology contribute to the understanding of human disease? Trends Biotechnol. 2008;26:584–590. doi: 10.1016/j.tibtech.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 8.Mustacchi R, Hohmann S, Nielsen J. Yeast systems biology to unravel the network of life. Yeast. 2006;23:227–238. doi: 10.1002/yea.1357. [DOI] [PubMed] [Google Scholar]
- 9.Alper H, Moxley J, Nevoigt E, Fink GR, Stephanopoulos G. Engineering yeast transcription machinery for improved ethanol tolerance and production. Science. 2006;314:1565–1568. doi: 10.1126/science.1131969. [DOI] [PubMed] [Google Scholar]
- 10.Ro DK, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho KA, Eachus RA, Ham TS, Kirby J, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 11.Simon JA, Bedalov A. Yeast as a model system for anticancer drug discovery. Nat. Rev. Cancer. 2004;4:481–492. doi: 10.1038/nrc1372. [DOI] [PubMed] [Google Scholar]
- 12.Kumar A, Snyder M. Emerging technologies in yeast genomics. Nat. Rev. Genet. 2001;2:302–312. doi: 10.1038/35066084. [DOI] [PubMed] [Google Scholar]
- 13.Orr-Weaver TL, Szostak JW, Rothstein RJ. Yeast transformation: a model system for the study of recombination. Proc. Natl Acad. Sci. USA. 1981;78:6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moerschell RP, Tsunasawa S, Sherman F. Transformation of yeast with synthetic oligonucleotides. Proc. Natl Acad. Sci. USA. 1988;85:524–528. doi: 10.1073/pnas.85.2.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma H, Kunes S, Schatz PJ, Botstein D. Plasmid construction by homologous recombination in yeast. Gene. 1987;58:201–216. doi: 10.1016/0378-1119(87)90376-3. [DOI] [PubMed] [Google Scholar]
- 16.Raymond CK, Sims EH, Olson MV. Linker-mediated recombinational subcloning of large DNA fragments using yeast. Genome Res. 2002;12:190–197. doi: 10.1101/gr.205201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibson DG, Benders GA, Andrews-Pfannkoch C, Denisova EA, Baden-Tillson H, Zaveri J, Stockwell TB, Brownley A, Thomas DW, Algire MA, et al. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008;319:1215–1220. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- 18.Gibson DG, Benders GA, Axelrod KC, Zaveri J, Algire MA, Moodie M, Montague MG, Venter JC, Smith HO, Hutchison CA., 3rd One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome. Proc. Natl Acad. Sci. USA. 2008;105:20404–20409. doi: 10.1073/pnas.0811011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shao Z, Zhao H, Zhao H. DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Res. 2009;37:e16. doi: 10.1093/nar/gkn991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kouprina N, Larionov V. Selective isolation of genomic loci from complex genomes by transformation-associated recombination cloning in the yeast Saccharomyces cerevisiae. Nat. Protoc. 2008;3:371–377. doi: 10.1038/nprot.2008.5. [DOI] [PubMed] [Google Scholar]
- 22.Orr-Weaver TL, Szostak JW, Rothstein RJ. Genetic applications of yeast transformation with linear and gapped plasmids. Methods Enzymol. 1983;101:228–245. doi: 10.1016/0076-6879(83)01017-4. [DOI] [PubMed] [Google Scholar]
- 23.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bayona-Bafaluy MP, Acin-Perez R, Mullikin JC, Park JS, Moreno-Loshuertos R, Hu P, Perez-Martos A, Fernandez-Silva P, Bai Y, Enriquez JA. Revisiting the mouse mitochondrial DNA sequence. Nucleic Acids Res. 2003;31:5349–5355. doi: 10.1093/nar/gkg739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith HO, Hutchison CA, 3rd, Pfannkoch C, Venter JC. Generating a synthetic genome by whole genome assembly: phiX174 bacteriophage from synthetic oligonucleotides. Proc. Natl Acad. Sci. USA. 2003;100:15440–15445. doi: 10.1073/pnas.2237126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bang D, Church GM. Gene synthesis by circular assembly amplification. Nat. Methods. 2008;5:37–39. doi: 10.1038/nmeth1136. [DOI] [PubMed] [Google Scholar]
- 27.Hoover DM, Lubkowski J. DNAWorks: an automated method for designing oligonucleotides for PCR-based gene synthesis. Nucleic Acids Res. 2002;30:e43. doi: 10.1093/nar/30.10.e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rouillard JM, Lee W, Truan G, Gao X, Zhou X, Gulari E. Gene2Oligo: oligonucleotide design for in vitro gene synthesis. Nucleic Acids Res. 2004;32:W176–W180. doi: 10.1093/nar/gkh401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bode M, Khor S, Ye H, Li MH, Ying JY. TmPrime: fast, flexible oligonucleotide design software for gene synthesis. Nucleic Acids Res. 2009;37:W214–W221. doi: 10.1093/nar/gkp461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Villalobos A, Ness JE, Gustafsson C, Minshull J, Govindarajan S. Gene designer: a synthetic biology tool for constructing artificial DNA segments. BMC Bioinformatics. 2006;7:285. doi: 10.1186/1471-2105-7-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leem SH, Noskov VN, Park JE, Kim SI, Larionov V, Kouprina N. Optimum conditions for selective isolation of genes from complex genomes by transformation-associated recombination cloning. Nucleic Acids Res. 2003;31:e29. doi: 10.1093/nar/gng029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Storici F, Lewis LK, Resnick MA. In vivo site-directed mutagenesis using oligonucleotides. Nat. Biotechnol. 2001;19:773–776. doi: 10.1038/90837. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.