Abstract
Four-stranded DNA and RNA quadruplexes or G4 motifs are non-B DNA conformations that are presumed to form in vivo, although only few explicit evidence has been reported. Using bioinformatics the presence of putative DNA G-quadruplexes within critical promoter regions has been demonstrated and a regulatory role in transcription has been suspected. However, in genomic DNA the presence of the complementary strand interferes with the potential to form a quadruplex motif. Contrarily RNA G4 motifs have no such limitation and consequently strong interference with gene expression is suspected. Nevertheless, experimental evidence is scarce. Here we show a well-defined structure–function relationship of synthetic quadruplex sequences in 5′-UTRs in multiple mammalian cell-lines. We establish a universal ‘translational suppressor’ effect of these motifs on gene expression at the translational level and show for the first time that specific features such as loop-length and the number of ‘GGG’-repeats further determine the suppressive impact. Moreover, a consistent and predictable repression of gene expression is observed for naturally occurring RNA G4 motifs, augmenting the functional relevance of these unusual nucleic acid structures.
INTRODUCTION
DNA and RNA sequences with a minimum four interspersed ‘GGG’ repeats can potentially fold back to form stable G-quadruplex (G4) motifs, a four-stranded conformation composed of stacked guanine tetrads interacting via Hoogsteen hydrogen bonds which is further stabilized by chelating monovalent metal ions such as sodium or potassium (1,2). Initial computational searches identified putative sequences which can form G4 motifs throughout the genome in various mammalian (3–5) and bacterial (6) species. More rigorous analyses of the human genome suggested a statistically significant enrichment in gene promoters (7,8) and untranslated regions (UTRs) (9). Based on these findings, regulatory functions of DNA quadruplexes have been suggested in gene expression (10,11) and genomic maintenance (12–14). Although in genetic mechanisms such as replication, recombination (15), and transcription transient single-stranded DNA is observed, the presence of the complementary strand should disfavor quadruplex formation (16,17). Interestingly the single-stranded guanine-rich telomeric overhang, which has no duplex competition against structure formation, has been shown to form a quadruplex motif under in vitro conditions, while the in vivo existence is still controversial (18). The telomeric repeat has been studied in great detail with respect to its potential to form quadruplex structures (1,2). The mammalian telomeric overhang flips over and invades the telomeric duplex sequence generating an unique conformation called ‘t-loop’ (19). An increasing number of proteins have been identified that specifically interact with DNA quadruplexes, resulting in enhanced or disrupted formation of quadruplex motifs (20–28). Since the induction of quadruplexes seems to interfere with telomere length maintenance and hence with the replicative life-span, various classes of small molecules targeting genomic quadruplexes have been developed, displaying anti-proliferative activities due to inhibition of proto-oncogene expression (29) and telomerase activity (30). In contrast, very little is known about the properties and influence of RNA quadruplex formation in vivo. The formation of four-stranded RNA structures is well established in vitro, demonstrating that G-rich RNAs form quadruplexes with stabilities equal to their DNA counterparts (31–33). Computational searches for potential RNA quadruplexes in human mRNAs have identified a prevalence in 5′-UTRs (9), but only little experimental evidence concerning its effect on gene expression is available. Two recent studies demonstrate a suppressive influence of individual RNA quadruplexes found in the NRAS (34) and ZIC-1 (35) 5′-UTRs. We recently demonstrated a pronounced effect of artificially introduced quadruplexes on gene expression in bacterial mRNAs (31), however, an extensive study of the impact of RNA quadruplexes in mammalian mRNAs is lacking. In addition to characterizing naturally occurring sequence motifs (34,35), the use of artificial sequences allows for studying how distinct quadruplex features such as loop compositions and the number of G-rich repeats contribute to the suppressive effects of these unusual nucleic acid structures on gene expression (31). Here we systematically characterize the influence of synthetic and natural quadruplex-forming sequences on gene expression in several mammalian cell lines.
MATERIALS AND METHODS
Circular dichroism spectroscopy
All samples were prepared at 5 µM strand concentration (metabion Gmbh, HPLC purified) in RNase free 10 mM Tris–Cl, pH 7.4, 10 mM KCl, except Figure S3 (1 mM KCl). Samples were heated to 95°C for 5 mins and slowly cooled overnight before scanning in a 1 ml capped-cuvette at 15°C using Jasco 715 spectropolarimeter (Jasco Hachioji, Tokyo, Japan) fitted with a Peltier temperature controller. Circular dichroism (CD) scans were recorded in pentaplets at 50 nm min–1 with a 2 s response time, 1 nm pitch and 1 nm bandwidth, averaged, buffer subtracted and zero-corrected at 320 nm. For Tm calculation, samples were annealed from 95 to 15°C and melted back, at a controlled rate of 1°C min−1.
Construction of plasmids
The G-quadruplex sequences (Tables 1 and 2) were introduced into the psiCHECK-2 (Promega, Supplementary Figure 1) by whole plasmid PCR with primers having G-rich 5′-overhangs (Supplementary Figure 5) using the sequences shown in Supplementary Table 1. A detailed protocol for generating the quadruplex insertions can be found in ref. (36).
Table 1.
Characterized artificial quadruplex sequences
| Construct name | Sequence (5′–3′) |
|---|---|
| wt | ACUAUA·GGCUAGCCACCAUGGCU |
| 3G3U | (GGGU)2GGG |
| 4G3U | (GGGU)3GGG |
| 4G3U2 | (GGGUU)3GGG |
| 4G3U3 | (GGGUUU)3GGG |
| 5G3U | GGGU(GGGU)3GGG |
| 5G3U2 | GGGUU(GGGUU)3GGG |
| 5G3U3 | GGGUUU(GGGUUU)3GGG |
| 6G3U | GGGUGGGU(GGGU)3GGG |
| 6G3U2 | GGGUUGGGUU(GGGUU)3GGG |
| 6G3U3 | GGGUUUGGGUUU(GGGUUU)3GGG |
RNA quadruplex motif sequences inserted 11 bp upstream (dot) of the start codon of the hRluc gene (wt). The start codon is shown in bold. The prefixes 3, 4, 5 or 6 in the construct names represent the number of total ‘GGG’-repeat runs in the sequence and were further prefixed with ‘con’ for the respective control sequences (U instead of underlined G).
Table 2.
Characterized natural quadruplex sequences
| Construct name | Sequence (5′–3′) |
|---|---|
| 4NRAS | GGGAGGGGCGGGUCUGGG |
| 4MAPK2 | GGGAGGGAGGGAGGG |
| 5CHST2 | GGGCGGGUUGGGUCGGGUCGGG |
| 6PCGF2 | GGGAUGGGAGGGAGGGAGGGAAGGG |
Sequences of naturally occurring 5′-UTR G-quadruplex motifs were named according to the gene they belong to and prefixed with total number of ‘GGG’-repeat runs (4, 5 or 6) in the sequence. Additional prefix of ‘con’ is used to describe the respective motif control wherein U is inserted instead of G (underlined).
Natural 5′-UTR quadruplex sequences
Human genome 5′-UTR sequences (25 193; one FASTA record per refgene) were downloaded from UCSC table browser (http://genome.ucsc.edu/cgi-bin/hgTables) as on May 2008. We could retrieve 2013 putative RNA G-quadruplex forming sequences using customized algorithm (6), which is in good accordance with 2334 quadruplex reported using different algorithm and 5′-UTR sequence retrieval option recently (9). We then manually searched for distinct multiple ‘GGG’-repeat sequences and the 4MAPK2, 5CHST2 and 6PCGF2 quadruplex sequences were arbitrarily chosen.
Cell culture and dual luciferase assay
Cells were grown to ∼50% confluency in flat-bottomed 96-well plate under standard conditions of 37°C, 5% CO2 in Dulbecco’s modified Eagles medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum (FCS), 2 mM glutamine, non-essential amino acids and the antibiotics penicillin and streptomycin, before transient transfection with 50 ng of plasmid constructs (Tables 1 and 2) using Lipofectamine 2000 (Invitrogen) or Turbofect (Fermentas), according to the manufacturer’s protocol. Twenty-four hours post-transfection, cells were retrieved and processed using Dual-luciferase Reporter Assay kit (E1910, Promega) according to the manufacturer’s protocol. The luciferase activity was determined using a TECAN M200 reader.
Quantitative real-time PCR assay
HEK293 cells were grown to ∼50% confluency in flat-bottomed 6-well plate, under conditions stated above and transfected with Turbofect (Fermentas), according to the manufacturer’s protocol. Twenty-four hours later, the total cellular RNA was isolated using RNeasy Plus Mini Kit (74134, Qiagen). For quantification of mRNA levels, 1 µg of total RNA was used in a 25 µl reaction using Titan One Tube RT-PCR System (11888382001, Roche) and SYBR-green (kindly provided by the Marx group, University of Konstanz) on a Chromo4 Real-Time Detector (Biorad) using primer pairs 5′-CCAACCCTGTTCAGCTTCTTC-3′ and 5′-ACCTTGGCCTCGAAGAATGG-3′ for hluc mRNA and 5′-TGATCGAGTCCTGGGACGA-3′ and 5′-ACAATCTGGACGACGTCGGG-3′ for hRluc mRNA. All reactions were done in triplicate.
RESULTS AND DISCUSSION
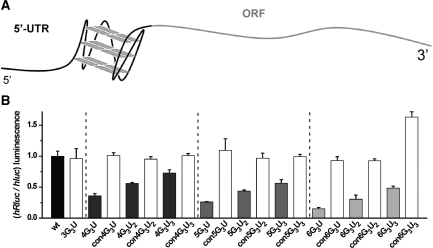
In order to investigate the impact of quadruplex formation on gene expression, we chose to introduce artificially designed G4 motif sequences into a reporter gene setup (Figures 1A and Supplementary Figure 1). The use of such non-natural sequences allows for studying the effect of stability-determining features, circumventing the sequence restrictions that apply when using natural motifs. Since the thermodynamic stability of G4 motifs is known to depend on the loop lengths, we first tested the influence of quadruplexes containing one- (4G3U), two- (4G3U2), or three- (4G3U3) nucleotide loops (Table 1), inserted eleven nucleotides upstream of the start codon (corresponding to +124 bp with respect to the 5′-end of the mRNA, for a detailed description of the construct see also Supplementary Table 1) of a luciferase reporter. For DNA quadruplexes, the thermodynamic stability increases with decreasing loop lengths, with mononucleotide loops exhibiting the highest stabilities (37,38). We found that the expression of the Renilla reporter luciferase in HEK293 cells decreased to residual 36, 56 and 73%, respectively, compared to the parental reporter construct (Figure 1B). To confirm that perturbation of gene expression is solely based on quadruplex structure formation and not due to the mere insertion of the additional G-rich sequence, we investigated a control sequence for each quadruplex studied. For this purpose, we constructed similar G/U-rich control sequences (Table 1) which are unable to form a quadruplex by disrupting the middle tetrads by G to U mutations. Importantly, the expression was restored in all three control sequences (Figure 1B). In addition, we inserted a control sequence containing only three ‘GGG’-repeats (3G3U, Table 1) which by itself is not able to form an intramolecular quadruplex, but could potentially participate in an intermolecular RNA quadruplex. We found no influence on expression for the construct 3G3U compared to wt-hRluc or the other G/U-rich control sequences (Figure 1B) and hence we conclude that the changes in expression are imminently due to intramolecular RNA quadruplex formation.
Figure 1.
Effect of 5′-UTR RNA quadruplexes on gene expression. (A) Scheme of an RNA quadruplex located in the 5′-UTR (black) of the reporter gene. A typical RNA quadruplex structure comprising all-parallel strand orientation is shown. (B) The expression of various quadruplex-containing constructs and the respective controls (Table 1) in HEK293 cells, measured using a dual luciferase assay and transient transfection, is shown. Error bars represent standard deviation of three independent experiments.
In order to comprehend the expression differences observed for the different loop sizes, we decided to structurally and thermodynamically characterize the investigated RNA sequences using CD spectroscopy. For DNA quadruplexes it has been established that the thermodynamic stability is inversely proportional to loop sizes (37,38). For RNA quadruplexes there are only few data available (31), hence we investigated the stability of the quadruplexes 4G3U3, 4G3U2 and 4G3U. We found that the inserted RNA sequences form parallel-stranded quadruplexes (Figure 2A–C) displaying gradually increasing stability with decreasing loop length from 4G3U3 to 4G3U (Figure 2D). The latter finding confirms previous data demonstrating a higher stability of RNA quadruplexes with decreasing loop sizes, behaving similarly to DNA quadruplexes (31). We have also carried out CD spectroscopy with the control RNA sequences that should be unable to adopt quadruplexes. We found no structure formation (Figure 2A–C) which agrees with the absence of effects on gene expression as discussed above. In conclusion, a direct correlation of the quadruplex stability with its influence on gene expression has been demonstrated in mammalian 5′-UTRs.
Figure 2.
In vitro stability and in vivo abundancy of the quadruplex motifs. The CD scans (black) show a positive band around 264 nm and negative maxima around 240 nm for 4G3U (A), 4G3U2 (B) and 4G3U3 (C), characteristic for a parallel stranded quadruplex motif, while the respective control sequences; con4G3U (A), con4G3U2 (B) and con4G3U3 (C) does not show any quadruplex characteristic peaks (grey). In CD melting experiments (D), denaturing (hollow) and annealing (solid) curves are almost identical and show a Tm of >95, 73 and 61°C for 4G3U (dark grey), 4G3U2 (grey) and 4G3U3 (light grey) RNA-quadruplex motifs, respectively. (E) Relative hRluc mRNA levels for different constructs determined by real-time PCR assays [hRluc(CT)] is normalized to hluc mRNA levels [hluc(CT)], as reported earlier (35). Error bars represent standard deviation of three independent experiments.
Next, we investigated at which level of gene expression the quadruplex formation takes effect. For this purpose, we quantified mRNA-levels by real time PCR in order to clarify whether the insertion of the G-rich sequences interfered with mRNA abundance by reducing transcriptional efficiency or mRNA stability. Identical mRNA levels were detected for all tested expression constructs (Figure 2E) in accordance to the results reported recently for the ZIC-1 quadruplex, pointing at a mechanism acting at the level of translational initiation (35). This result is also supported by a report of quadruplex formation in the FMR1 mRNA resulting in stalled 40S ribosomal subunits (39,40). Additional experiments are necessary to decipher the exact mechanism of quadruplex interference with translation.
Although four interspersed ‘GGG’-repeats are the minimal requirement to form a G4 motif, naturally occurring G4 motifs often contain five, six or even more ‘GGG’-repeats (1). In order to investigate the influence of the number of ‘GGG’-repeats we examined additional series of 5′-UTR luciferase constructs (Table 1, 5G3UX and 6G3UX). Interestingly, we found that gene expression further decreased consecutively from four to six ‘GGG’-repeat runs consistently in each loop-length series (Figure 1B and Supplementary Figure 2), with 6G3UX being more repressive than the five- and four-repeat series, respectively. The most potent suppressor sequence 6G3U reduced expression by 85%. Importantly, the control sequences containing G to U mutations as well restored gene expression in the 5- and 6-repeat series, with the clone con6G3U3 showing an unanticipated high expression. In order to investigate whether this unexpected effect of increasing the ‘GGG’ repeat number resulting in even more pronounced suppression, we carried out additional CD melting studies. Surprisingly, we found 5G3U and 6G3U RNA-quadruplex motifs are thermodynamically more stable than its shorter companion, the 4G3U motif (Supplementary Figure 3). This observation is surprising as only one G4 motif with a similar conformation is possible even with five or six ‘GGG’ repeats present and hence is expected to have a similar thermodynamic stability. Although the CD spectra are very similar to the 4G3U sequence, pointing at an all-parallel RNA quadruplex (Supplementary Figure 3), additional structural stabilization of the four-stranded structure by the additional ‘GGG’ repeats cannot be ruled out. In light of this finding, it is tempting to speculate that this could be a possible reason why many naturally occurring biologically relevant DNA [c-myc, bcl-2, VEGF, HIF-1α) (2) and RNA (Zic-1 (35), 5CHST2, and 6PCGF2, see below] quadruplex sequences contain more than four ‘GGG’-repeat runs.
Since Balasubramanian and co-workers have recently shown that pronounced quadruplex-based repression was evidenced only when the quadruplex motif was located within the first 50 nt of the mRNA 5′-end (41) compared to the position +124 nt as described earlier, we investigated whether moving the quadruplex upstream towards the 5′-end of the mRNA would have any effect on the suppression of gene expression. For this purpose, we inserted 4G3U as well as 6G3U along with their control sequences at two additional positions (+12 and +95, Supplementary Table 1) in our hRluc reporter construct to evaluate any positional effects on expression. We found a gradual but minor decrease in suppression for both 4G3U and 6G3U, but not in control constructs, when we moved the motif towards the 5′-end (Figure 3A). In conclusion, we observed pronounced suppression of gene expression in all characterized positions in contrast to the study carried out by Balasubramanian and co-workers. This discrepancy may be due to a different experimental setup with respect to the plasmid backbone, 5′-UTR sequence or read-out [a combined in-vitro transcription / translation assay was used before to quantify interference with gene expression (41)].
Figure 3.
Positional effect and naturally occurring RNA quadruplexes. (A) Normalized luminescence of hRluc luciferase of 4G3U, 6G3U, con4G3U or con6G3U construct at different positions in the 5′-UTR. Numbers in parentheses indicate the insertion position from the 5′-end. (B) Normalized luminescence of hRluc luciferase of naturally occurring RNA G4 motifs and respective control sequences (Supplementary Table 2). Error bars are standard deviations of three independent experiments.
Next, in order to clarify whether the results obtained with our artificially engineered sequences are transferable to naturally occurring quadruplex motifs in mRNAs, we reinvestigated the NRAS G4 motif sequence characterized by Balasubramanian and co-workers together with three additional natural 5′-UTR quadruplex sequences (termed 4NRAS, 5CHST2, 4MAPK2 and 6PCGF2, Supplementary Table 2). For this purpose, we introduced the natural quadruplex motifs into the synthetic luciferase 5′-UTR (+124 from transcription start site / −11 from start codon, Supplementary Table 1). We were especially interested whether the general tendencies observed with our synthetic quadruplex sequences with respect to loop lengths and number of GGG repeats would allow for predicting the influence of the natural motifs on gene expression. Reliably, for the 4NRAS RNA G4 motif (with respective loop-lengths of 1, 2 and 3 nucleotides) the repression (50%, Figure 3B) closely matched the synthetic analogue 4G3U2 (44%, Figure 3B). In case of the other natural motifs 4MAPK2, 5CHST2 and 6PCGF2, the repression (70, 59 and 78%, Figure 3B) also compared well to the synthetic motifs closely resembling loop lengths and ‘GGG’-repeat numbers (4G3U, 5G3U2 and 6G3U2 with reductions of 64, 57 and 70%, respectively, Figure 1B). Given the overall consistency of repression of naturally occurring 5′-UTR G4 motifs to corresponding synthetic RNA G4 motifs under similar conditions, our study presents a rationale to estimate the effect of naturally occurring RNA G4 motifs on gene expression. Small variations are expected to arise from the different nucleotide composition in loops, which are known to affect the stability of the G4 motif and consequently gene expression (37). Finally, to determine whether the observed quadruplex-mediated repression was specific to the adenovirus-transformed HEK293 cell line, we repeated the experiments using the poorly differentiated human glioblastoma cell line LN18, the human cervical cancer line HeLa, and the mouse fibroblast line B18. Importantly, consistent but in some cases less pronounced results were obtained (Supplementary Figure 4). In general, the suppressive effects seem to be slightly less prominent using these fundamentally different cell lines with the effect of different loop-lengths being less pronounced as well. Overall, the effects of the different quadruplexes are observed in these alternate cell lines as well. This finding further strengthens the evidence that quadruplex formation in 5′-UTRs affect expressions by a rather unspecific mechanism correlating with thermodynamic stability.
Taken together, the presented work represents the most systematic in vivo study of synthetic and naturally occurring quadruplex motifs located in 5′-UTRs in mammalian cell lines. The results demonstrate a coherent impact of RNA quadruplex formation on gene expression and suggest a universal ‘translational-suppressor’ effect for these non-canonical RNA conformations. We found a predictable correlation of the suppressive effects of RNA quadruplexes with its authoritative features such as loop length and repeat numbers, representing an outstanding example of a facile structure—function relationship of unusual nucleic acid conformations in cellular mechanisms. The present study should allow for predicting the suppressive effects of similar RNA quadruplexes in 5′-UTRs discovered in natural sequence contexts. Moreover, the present system should prove useful for a setup to characterize small molecules selectively targeting RNA quadruplexes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Lichtenberg-Professorship by VolkswagenStiftung (to J.S.H.); Alexander von Humboldt-Foundation fellowship (to K.H.). Funding for open access charge: University of Konstanz.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the group of Prof. M. Leist for the LN18 cell line, the group of Prof. M. Groettrup for HeLa and B18 cell lines and Astrid Joachimi for excellent technical assistance.
REFERENCES
- 1.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patel DJ, Phan AT, Kuryavyi V. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verma A, Halder K, Halder R, Yadav V, Rawal P, Thakur R, Mohd F, Sharma A, Chowdhury S. Genome-wide computational and expression analyses reveal G-quadruplex DNA motifs as conserved cis-regulatory elements in human and related species. J. Med. Chem. 2008;51:5641–5649. doi: 10.1021/jm800448a. [DOI] [PubMed] [Google Scholar]
- 6.Rawal P, Kummarasetti VBR, Ravindran J, Kumar N, Halder K, Sharma R, Mukerji M, Das SK, Chowdhury S. Genome-wide prediction of G4 DNA as regulatory motifs: role in Escherichia coli global regulation. Genome Res. 2006;16:644–655. doi: 10.1101/gr.4508806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao Y, Du Z, Li N. Extensive selection for the enrichment of G4 DNA motifs in transcriptional regulatory regions of warm blooded animals. FEBS Lett. 2007;581:1951–1956. doi: 10.1016/j.febslet.2007.04.017. [DOI] [PubMed] [Google Scholar]
- 9.Huppert JL, Bugaut A, Kumari S, Balasubramanian S. G-quadruplexes: the beginning and end of UTRs. Nucleic Acids Res. 2008;36:6260–6268. doi: 10.1093/nar/gkn511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du Z, Zhao Y, Li N. Genome-wide analysis reveals regulatory role of G4 DNA in gene transcription. Genome Res. 2008;18:233–241. doi: 10.1101/gr.6905408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qin Y, Hurley L. Structures, folding patterns, and functions of intramolecular DNA G-quadruplexes found in eukaryotic promoter regions. Biochimie. 2008;90:1149–1171. doi: 10.1016/j.biochi.2008.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halder K, Halder R, Chowdhury S. Genome-wide analysis predicts DNA structural motifs as nucleosome exclusion signals. Mol. Biosyst. 2009 doi: 10.1039/b905132e. (in press) [DOI] [PubMed] [Google Scholar]
- 13.Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat. Struct. Mol. Biol. 2006;13:1055–1059. doi: 10.1038/nsmb1171. [DOI] [PubMed] [Google Scholar]
- 14.Wong HM, Huppert JL. Stable G-quadruplexes are found outside nucleosome-bound regions. Mol. Biosyst. 2009 doi: 10.1039/b905848f. (in press) [DOI] [PubMed] [Google Scholar]
- 15.Mani P, Yadav V, Das S, Chowdhury S. Genome-wide analyses of recombination prone regions predict role of DNA structural motif in recombination. PLoS ONE. 2009;4:e4399. doi: 10.1371/journal.pone.0004399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar N, Maiti S. A thermodynamic overview of naturally occurring intramolecular DNA quadruplexes. Nucleic Acids Res. 2008;36:5610–5622. doi: 10.1093/nar/gkn543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Risitano A, Fox KR. Stability of intramolecular DNA quadruplexes: comparison with DNA duplexes. Biochemistry. 2003;42:6507–6513. doi: 10.1021/bi026997v. [DOI] [PubMed] [Google Scholar]
- 18.Parkinson G, Lee M, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 19.Griffith J, Comeau L, Rosenfield S, Stansel R, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 20.Fry M. Tetraplex DNA and its interacting proteins. Front Biosci. 2007;12:4336–4351. doi: 10.2741/2391. [DOI] [PubMed] [Google Scholar]
- 21.Giraldo R, Rhodes D. The yeast telomere-binding protein RAP1 binds to and promotes the formation of DNA quadruplexes in telomeric DNA. EMBO J. 1994;13:2411–2420. doi: 10.1002/j.1460-2075.1994.tb06526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Froelich-Ammon SJ, Dickinson BA, Bevilacqua JM, Schultz SC, Cech TR. Modulation of telomerase activity by telomere DNA-binding proteins in Oxytricha. Genes Dev. 1998;12:1504–1514. doi: 10.1101/gad.12.10.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun H, Karow JK, Hickson ID, Maizels N. The Bloom’s Syndrome Helicase Unwinds G4 DNA. J. Biol. Chem. 1998;273:27587–27592. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]
- 24.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 25.De Boeck G, Forsyth RG, Praet M, Hogendoorn PC. Telomere-associated proteins: cross-talk between telomere maintenance and telomere-lengthening mechanisms. J. Pathol. 2009;217:327–344. doi: 10.1002/path.2500. [DOI] [PubMed] [Google Scholar]
- 26.Cech TR. Life at the end of the chromosome: telomeres and telomerase. Angew. Chem. Int. Ed. 2000;39:34–43. doi: 10.1002/(sici)1521-3773(20000103)39:1<34::aid-anie34>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 27.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 28.Zaug AJ, Podell ER, Cech TR. Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc. Natl Acad. Sci. USA. 2005;102:10864–10869. doi: 10.1073/pnas.0504744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doherty K, Sharma S, Gupta R, Brosh R. Tetraplex binding molecules as anti-cancer agents. Recent Pat. Anticancer Drug Discov. 2006;1:185–200. doi: 10.2174/157489206777442232. [DOI] [PubMed] [Google Scholar]
- 30.Mergny J-L, Riou J-F, Mailliet P, Teulade-Fichou M-P, Gilson E. Natural and pharmacological regulation of telomerase. Nucleic Acids Res. 2002;30:839–865. doi: 10.1093/nar/30.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wieland M, Hartig JS. RNA quadruplex-based modulation of gene expression. Chem. Biol. 2007;14:757–763. doi: 10.1016/j.chembiol.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 32.Shafer RH, Smirnov I. Biological aspects of DNA/RNA quadruplexes. Biopolymers. 2000;56:209–227. doi: 10.1002/1097-0282(2000/2001)56:3<209::AID-BIP10018>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 33.Pan B, Shi K, Sundaralingam M. Base-tetrad swapping results in dimerization of RNA quadruplexes: implications for formation of the i-motif RNA octaplex. Proc. Natl Acad. Sci. USA. 2006;103:3130–3134. doi: 10.1073/pnas.0507730103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumari S, Bugaut A, Huppert JL, Balasubramanian S. An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 2007;25:25. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arora A, Dutkiewicz M, Scaria V, Hariharan M, Maiti S, Kurreck J. Inhibition of translation in living eukaryotic cells by an RNA G-quadruplex motif. RNA. 2008;14:1290–1296. doi: 10.1261/rna.1001708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wieland M, Hartig JS. Investigation of mRNA quadruplex formation in Escherichia coli. Nature Protocols. 2009 doi: 10.1038/nprot.2009.111. in press. [DOI] [PubMed] [Google Scholar]
- 37.Bugaut A, Balasubramanian S. A sequence-independent study of the influence of short loop lengths on the stability and topology of intramolecular DNA G-quadruplexes. Biochemistry. 2008;47:689–697. doi: 10.1021/bi701873c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hazel P, Huppert J, Balasubramanian S, Neidle S. Loop-length-dependent folding of G-quadruplexes. J. Am. Chem. Soc. 2004;126:16405–16415. doi: 10.1021/ja045154j. [DOI] [PubMed] [Google Scholar]
- 39.Heitz D, Rousseau F, Devys D, Saccone S, Abderrahim H, Le Paslier D, Cohen D, Vincent A, Toniolo D, la Valle G, et al. Isolation of sequences that span the fragile X and identification of a fragile X-related CpG island. Science. 1991;251:1236–1239. doi: 10.1126/science.2006411. [DOI] [PubMed] [Google Scholar]
- 40.Ofer N, Weisman-Shomer P, Shklover J, Fry M. The quadruplex r(CGG)n destabilizing cationic porphyrin TMPyP4 cooperates with hnRNPs to increase the translation efficiency of fragile X premutation mRNA. Nucleic Acids Res. 2009;37:2712–2722. doi: 10.1093/nar/gkp130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumari S, Bugaut A, Balasubramanian S. Position and stability are determining factors for translation repression by an RNA G-quadruplex-forming sequence within the 5′ UTR of the NRAS proto-oncogene. Biochemistry. 2008;47:12664–12669. doi: 10.1021/bi8010797. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.